Abstract
Graft-versus-host disease (GVHD) accounts for increased morbidity and mortality after HLA-identical unrelated hematopoietic cell transplantation (HCT). To test the hypothesis that the MHC encodes functional variation other than the classical HLA genes, we measured risks associated with donor-recipient MHC microsatellite (Msat) marker mismatching in 819 HCT recipients and their HLA-A, -B, -C, -DRB1, -DQB1 allele-matched unrelated donors. Suggestive trends of association with transplant outcome were observed for five Msats. Donor-recipient mismatching for the extended class I D6S105, class III D6S2787 and class II D6S2749 markers was each associated with an increased risk of death (hazard ratio, 1.32; 95% confidence interval, 1.02–1.71; P = .03; hazard ratio, 1.26; 95% confidence interval, 1.03–1.53; P = .02; hazard ratio, 1.37; 95% confidence interval, 1.08–1.72; P = .007, respectively) whereas mismatching for the class I D6S2811 marker was associated with a decreased risk of death (hazard ratio, 0.80; 95% confidence interval, 0.66–0.98; P = .03). Mismatching for the class I D6S265 marker was associated with a decreased risk of grades III-IV acute GVHD (odds ratio, 0.67; 95% confidence interval, 0.45–0.98; P = .04). These results suggest that Msats may be informative for mapping MHC-resident genetic variation of clinical importance in HCT.
Keywords: MHC, microsatellite, hematopoietic cell transplantation
INTRODUCTION
Despite matching of donor and recipient HLA-A, -B, -C, -DRB1 and -DQB1 alleles, HLA-identical transplant recipients suffer from life-threatening complications after unrelated hematopoietic cell transplantation (HCT) [1–3]. HLA genes are encoded within the gene-dense MHC region, residence for over 400 genes, many of which have immune-related function [4–6]. The MHC region has been the subject of intense study due to its pivotal role as the “transplantation barrier” and its association of HLA genes with disease susceptibility [4]. The identification of non-HLA MHC loci among the hundreds of candidate genes presents a methodological challenge, because complete sequence characterization of the 7.6 Mb extended MHC region is prohibitive.
Recent studies of MHC microsatellite (Msat) markers have furthered the understanding of the genetic organization and the extent and patterns of linkage disequilibrium (LD) within the HLA region [7–11]. MHC Msats are not themselves functional, however their inherent polymorphism and LD with HLA loci make them a robust disease-mapping tool in autoimmune and infectious disease models and in HCT [8, 12–14]. Broad application of Msats has also been proposed as an efficient means for screening potential unrelated donors for HCT [15–17, http://www.euromado.org/].
We hypothesized that unidentified donor-recipient variation could be associated with reduced or increased risks of graft-versus-host disease (GVHD), relapse or death after HLAmatched unrelated donor HCT, and we used Msats to query the MHC for new transplantation determinants. We report the findings from a large dataset of the International Histocompatibility Working Group (IHWG) in HCT, a consortium of international transplant centers and histocompatibility laboratories (http://www.ihwg.org).
MATERIALS AND METHODS
Study Population
A total of 819 transplants met the following study criteria: 1) first transplantation from an HLA-A,-B,-C,-DRB1,-DQB1 allele-matched unrelated donor; 2) myeloablative conditioning regimen; 3) marrow (T cell-replete or T cell-depleted) or peripheral blood stem cell (PBSC) HCT for the treatment of acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myeloid leukemia (CML), or myelodysplastic syndrome (MDS); 4) availability of complete clinical outcome and retrospective high resolution HLA-A, -B, -C, -DRB1, -DQB1 typing data (Table 1). The following contributors provided clinical samples and data to this study: National Marrow Donor Program (NMDP) and Center for International Blood and Marrow Transplant Research (n = 384), Australian Bone Marrow Donor Registry (n = 27), the Société Française de Greffe de Moelle (SFGM) and the Registre France Greffe de Moelle (RFGM) (n = 78), Netherlands Stem Cell Transplantation Registry (Typhon) (n = 16) and Fred Hutchinson Cancer Research Center (FHCRC) (n = 314).
Table 1.
Characteristics of Study Population, n = 819 Donor-Recipient Pairs
| Median recipient age, y (range) | 36.2 (1–62.8) |
| Year of transplantation - n (%) | |
| 1985–1990 | 76 (9.3%) |
| 1991–1996 | 484 (59.1%) |
| 1997–2003 | 259 (31.6%) |
| Recipient/donor gender, n (%) | |
| Male/male | 331 (40.4%) |
| Male/female | 153 (18.7%) |
| Female/male | 173 (21.1%) |
| Female/female | 156 (19.0%) |
| Unknown | 6 (0.7%) |
| Diagnosis/disease severity*, n (%)/ n | |
| ALL/ high-risk, intermediate-risk | 62 (7.6%)/12, 50 |
| AML/ high-risk, intermediate-risk | 183 (22.3%)/77, 106 |
| CML/ high-risk, intermediate-risk, low-risk | 544 (66.4%)/23, 122, 399 |
| MDS/ high-risk, intermediate-risk | 30 (3.7%)/15, 15 |
| Recipient/donor CMV serology, n (%) | |
| Positive/positive | 129 (15.8%) |
| Positive/negative | 207 (25.3%) |
| Negative/positive | 102 (12.5%) |
| Negative/negative | 281 (34.3%) |
| Unknown | 100 (12.2%) |
| Conditioning regimen, n (%) | |
| Total body irradiation-containing | 726 (88.6%) |
| Other | 93 (11.4%) |
| Source of cells - n (%) | |
| Bone marrow | 738 (90.1%) |
| Peripheral blood stem cells | 13 (1.6%) |
| Unknown | 68 (8.3%) |
| T cell depletion, n (%) | |
| Yes | 93 (11.4%) |
| No | 726 (88.6%) |
ALL indicates acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; CMV, cytomegalovirus.
Disease severity categories were defined as low-risk (CML chronic phase), intermediate-risk (CML accelerated phase or blast crisis/remission; acute leukemia transplanted in remission; MDS -refractory anemia); or high-risk (blast crisis CML; acute leukemia transplanted in relapse; MDS other than refractory anemia).
All research samples and data were collected according to institutional review board (IRB)-approved guidelines and protocols of each of the participating institutions. For samples from NMDP, all surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program. Informed consent was waived by the NMDP IRB for all deceased recipients. To address bias introduced by inclusion of only a proportion of surviving patients (those who consented) but all deceased recipients, a sample of deceased patients was selected using a weighted randomized scheme that adjusts for over-representation of deceased patients in the consented cohort.
The self-described patient and donor racial backgrounds were as follows: North American and European Caucasian 89% and 71%; Hispanic 1.7% and 1.1%; Black 1.0% and 0.7%; Asian 0.6% and 0.2%; Native American 0.2% and 0.6%; other or unknown 7.4% and 26%.
Genotyping of HLA and Microsatellites
Genotyping of donor-recipient HLA-A, -B, -C, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, - DQB1, -DPA1 and -DPB1 alleles was performed by direct sequencing and PCR-sequence specific oligonucleotide probe (SSOP) methods using 13th IHWG workshop or locally designed protocols [18–20, http://ihwg.org/tmanual/TMcontents, http://bioinformatics.nmdp.org/HLA/HR/hr_methods_idx.html].
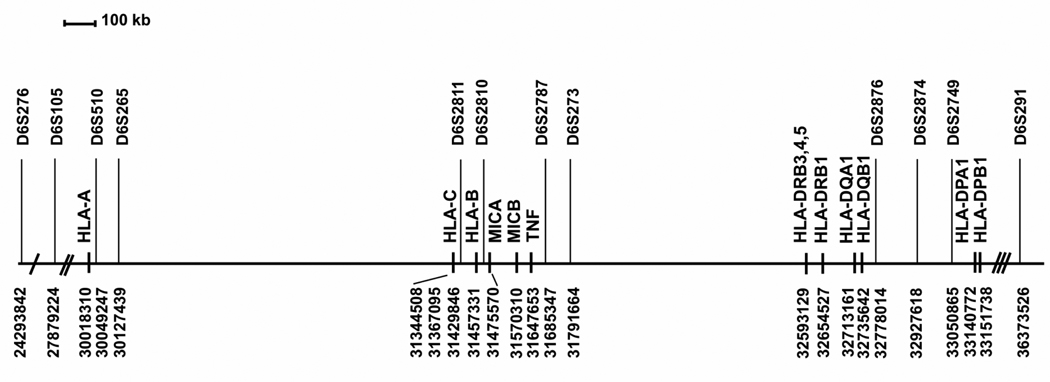
The following Msats were studied: D6S276, D6S105, D6S510, D6S265, D6S2811, D6S2810, D6S2787, D6S273, D6S2876, D6S2874, D6S2749 and D6S291 (Figure 1, Table 2). Msat genotyping of the samples from the Australian Bone Marrow Donor Registry, NMDP, Typhon and FHCRC was performed by the IHWG HCT Coordinating Laboratory (FHCRC, Seattle), and samples from the SFGM and RFGM was genotyped by four laboratories (FRABET, FRABOI, FRABIG and FRAMYE) as previously described (http://www.ihwg.org/components/hctr.htm). A DNA reference panel containing 50 DNAs (13th IHWG Core Cell and Gene Bank, www.ihwg.org/cellbank/) was typed as a quality control. This reference panel data was used to create an IHWG Msat dictionary, which normalizes Msat alleles to a common nomenclature thus allowing comparison of data from laboratories using different typing platforms [21]. Both the recipient and their corresponding donor sample were included in the same Msat genotyping experiment to avoid possible experiment-to-experiment variation that could affect the scoring of the Msat match status between recipient and donor. Each genotyping experiment also included four to six previously typed IHWG reference panel DNAs as quality control for Msat allele assignments. Msat alleles were defined according to the base pair (bp) sizes of the amplified Msat fragment.
Figure 1.
Physical map of the MHC region and the location of Msat markers used in this study. Position of the Msats and HLA-A, -B, -C, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1, -DPB1, MICA, MICB and TNF are listed in map order from telomere to centromere on chromosome 6p21.3. Coordinates refer to National Center for Bioinformatics (NCBI) chromosome 6 build 36.2 (www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&chr=6, www.ncbi.nlm.nih.gov/sites/entrez?db=unists). The 3.1 Mb region between HLA-A to HLA-DPB1 is drawn to scale. The genetic distance is 3.6 Mb between D6S276 and D6S105 (single hatch), 2.1 Mb between D6S105 and HLA-A (double hatch) and 3.2 Mb between HLA-DPB1 and D6S291 (triple hatch). The kilobase (kb) pair distance between markers is provided in Table 1.
Table 2.
Microsatellite Markers Evaluated in the Study Population
| Marker* | Alias | Distance to Next Marker (kb) |
Number of Transplant Pairs Genotyped |
|---|---|---|---|
| D6S276 | 3600 | 814 | |
| D6S105 | 2100 | 813 | |
| HLA-A | 31 | 819 | |
| D6S510 | 78 | 818 | |
| D6S265 | 1200 | 813 | |
| HLA-C | 23 | 819 | |
| D6S2811 | HLABCCA2 | 63 | 817 |
| HLA-B | 27 | 819 | |
| D6S2810 | MIB | 228 | 734 |
| D6S2787 | BAT2CA | 106 | 817 |
| D6S273 | 863 | 810 | |
| HLA-DRB1 | 81 | 819 | |
| HLA-DQB1 | 42 | 819 | |
| D6S2876 | G51152 | 149 | 818 |
| D6S2874 | TAP1CA | 123 | 818 |
| D6S2749 | RING3CA | 101 | 818 |
| HLA-DPB1 | 3200 | 809 | |
| D6S291 | 810 |
Markers are listed in the order of their mapping on chromosome 6p21.3. HLA loci are included for the purpose of orientation with respect to the Msats studied.
Of the 819 pairs in this dataset, the genotyping for HLA-DRB3, DRB4 or DRB5 loci was performed for 612 pairs, and the genotyping for DQA1 and DPA1 loci for 576 pairs. Out of the 612 pairs, DRB3 gene was present in 401 pairs, DRB4 in 327 pairs and DRB5 in 197 pairs.
Assessment of Hardy-Weinberg Equilibrium (HWE)
The principle of Hardy-Weinberg Equilibrium (HWE) is the relationship between the frequencies of alleles and the genotypes of a population [22]. Inherent in HWE is the assumption that a sufficiently large population has random mating and no migration, mutation, or selection. Under these conditions, genotypes are constant and their frequencies are in genetic equilibrium. Violation of Hardy Weinberg assumptions cause deviation from expectation, and can arise from non-random mating, genetic drift, mutation, or natural selection. In studies of genetic association, technical failure of allele genotyping leading to excess homozygosity or heterozygosity may also lead to violation of HWE [23]. In the current study, observed HLA-A, -B, -C, -DRB1, -DQB1 and Msat genotype frequencies in Caucasians were tested for HWE as previously described [11].
Statistical Analysis
Relapse was defined by morphologic or cytogenetic evidence of disease in the peripheral blood or bone marrow as previously described [2,3]. Acute GVHD was defined as previously described [2,3,24]. For the GVHD analysis, Msat mismatching was defined as the presence of recipient Msat alleles not shared by the donor. For relapse and survival analyses, a pair was mismatched if the recipient encoded a Msat allele not shared by the donor, and/or the donor an allele not shared by the recipient.
Cox regression models were used to assess the impact of Msat mismatching on outcome and were fit for the time-to-event endpoints of relapse and overall survival. Logistic regression models were used to examine the association of mismatching with grades III-IV acute GVHD. All models were adjusted for patient age, disease severity, use of total body irradiation (TBI), use of T-cell depletion, number of mismatches at Msat loci other than the Msat of interest, and number of HLA-DPB1 disparities.
To test the hypothesis that the total number of mismatched Msats between HLA-A and HLA-DPB1 was associated with outcome, the number of mismatched Msats was modeled as a continuous linear variable (a value from 0 to 11). Treating the number of mismatched Msats in this way assumes that all Msats contribute equally to outcome and that any increase in a specified number of mismatches is associated with the same change in hazard (or odds) of failure. By grouping the Msats according to their location within class I (D6S510, D6S265, D6S2810), class III (D6S2787, D6S273), or class II (D6S2876, D6S2874, D6S2749), the assumption that mismatching at each Msat contributes equally to outcome was relaxed to assume that each Msat within a region contributes equally, but not necessarily the same from region to region. The number of mismatched Msats within a region was modeled as a continuous linear variable (a value of 0–3 for class I, 0–2 for class III and 0–3 for class II). The D6S2810 was excluded from these analyses because 10% of the study population was not genotyped for this marker.
Two-sided P-values from the regression models were obtained from the Wald test. Due to the unknown level of correlation between mismatching at the various Msats and the high degree of association between clinical outcomes, no adjustments were made for multiple comparisons. Therefore, P-values between .01 and .05 should be considered as suggestive, rather than conclusive, evidence of a true difference.
RESULTS
Hardy Weinberg Equilibrium (HWE) of Msats
The most polymorphic Msat marker was D6S2811 (23 alleles) and the least polymorphic were D6S291 and D6S2749 (11 alleles each) (data not shown). The frequency of Msat alleles did not differ significantly between recipient and donor samples.
Deviation from HWE was tested for each HLA locus and Msat marker among patients and among donors since the pairs were matched for HLA loci and therefore, not independent. No HWE deviation was observed for HLA loci. Deviation from HWE was detected for D6S2810 and D6S276 in both donor and patient samples and for D6S2811 in patients and D6S2749 in donors. Accordingly, we investigated the contributions of homozygosity and heterozygosity at D6S276, D6S2810, D6S2811 and D62749. Only D6S276 demonstrated excess homozygosity in patient and donor samples (D6S276*141,141 was observed 14 times and expected 6.94 times in donor samples; it was observed 15 times and expected 6.84 times in patient samples). The majority of the Msats that deviated from HWE included only a small subset of combination of alleles that resulted in increased (donor D6S2810*277,281 [observed 14/expected 7.63]; patient D6S276*141,143 [observed 15/expected 6.84]) or decreased heterozygosity (donor D6S2749*231,233 [observed 22/expected 35.83]; donor D6S276*141,143 [observed 26/expected 41.64]; donor D6S2810*279,281 [observed 3/expected 12.30]; patient D6S2810*281:285 [observed 3/expected 9.78]; patient D6S2811*108,110 [observed 1/expected 7.60]). Among the heterozygous allele combinations that deviated from HWE, several allele pairs differed only by one dinucleotide repeat (2 bp). To investigate the possibility that deviation was caused by technical error, quality DNA controls were retyped and the raw genotyping data was re-analyzed for 90% of the samples; no obvious technical problems such as failure to amplify a second allele were identified. For this reason none of these Msats were removed from data analysis.
Donor-Recipient Msat Matching
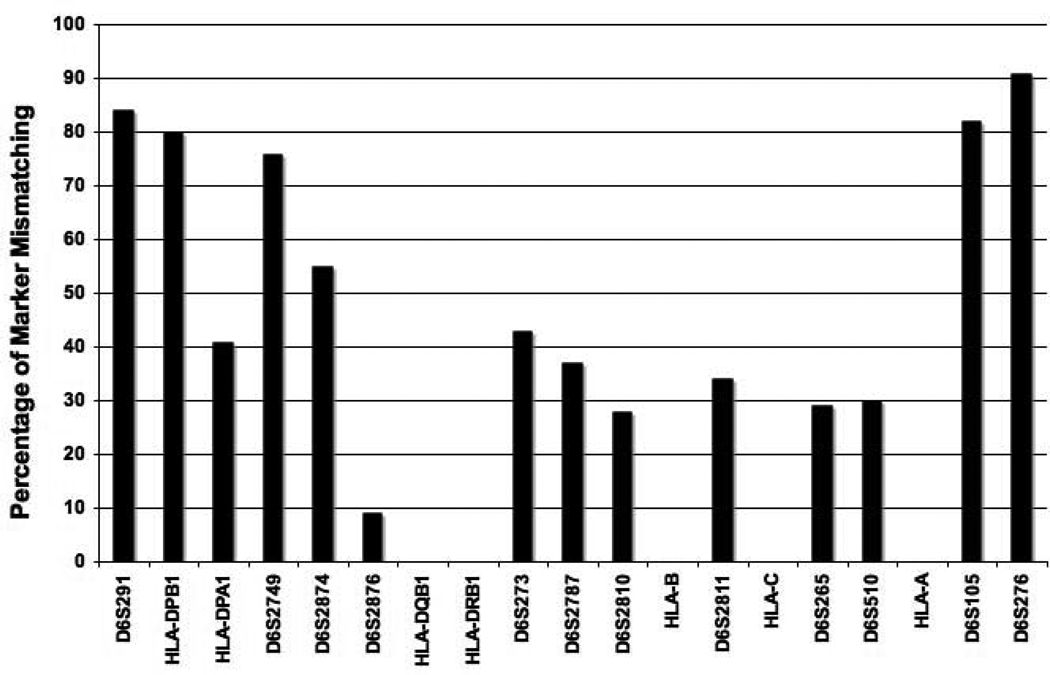
The frequency of donor-recipient Msat mismatching among the 819 HLA-A, -B, -C, -DRB1, -DQB1 matched pairs ranged from 9% (D6S2876) to 91% (D6S276) (Figure 2).
Figure 2.
Rate of mismatching among HLA-A, -B, -C, -DRB1, -DQB1 matched donor-recipient study pairs for MHC region Msats. HLA loci are included for the purpose of orientation with respect to the Msats studied. Of the 819 study pairs, 704 (86 %) were each genotyped for all 12 Msats, 109 (13 %) for all Msats except D6S2810 and 6 pairs (3 %) for 10 or fewer Msats because of PCR failure. Of the 704 pairs, 11 (1.6%) were matched for all 12 Msat loci; the remaining 693 pairs were mismatched for 1 (1.2 %), 2 (2.1 %), 3 (2.1 %), 4 (6.2 %), or 5 or more (86.8%) Msats.
LD patterns with HLA loci and the Msats included in this study have been previously described [11]. A higher degree of LD was observed with Msat markers that map in close proximity to the classical HLA loci. When the six Msats that reside between classical HLA-A to - DRB1 loci were evaluated for mismatching (D6S510, D6S265, D6S2811, D6S2810, D6S2787 and D6S273), 157 pairs (22.3%) were matched for all 6 Msat loci; the remaining 537 pairs were mismatched for 1 (17.8 %), 2 (21.3 %), 3 (15.1 %), 4 (9.8 %), or 5 or more (13.8%) Msats.
Grades III-IV Acute GVHD
A trend for lower probability of grades III-IV acute GVHD was observed with mismatching for the D6S265 class I marker (odds ratio [OR], 0.67; 95% confidence interval [CI], 0.45–0.98; P =. 04) (Table 3). No statistically significant association between the total number of mismatched Msats (regardless of the region in which the Msat resides) and the probability of grades III-IV acute GVHD was observed (P = .60). When Msats were grouped according to their location within class I, class III or class II, there was a suggestion of an association between the number of class I mismatched Msats and the probability of acute GVHD, with less acute GVHD associated with increasing number of mismatches (P = .06). The number of mismatched class III or class II Msats were not associated with the probability of acute GVHD (P = .65 and P = .38, respectively). Among the 11 patients (7 low-risk, 2 high-risk and 2 intermediate-risk recipients) who were completely matched with their respective donors for all 12 Msats, three developed grade III acute GVHD (but were alive at last contact), four died (day 110, 126, 154, 245) after HCT and none relapsed.
Table 3.
Adjusted Effect of Microsatellite (Msat) Mismatching on Probability of Grades III-IV Acute GVHD and Overall Mortality After Hematopoietic Cell Transplantation from HLA-identical Unrelated Donors*
| Proportion (%) of Events | |||||
|---|---|---|---|---|---|
| Clinical | Msat | Msat | |||
| Endpoint | Msat | Matched | Mismatched | OR or HR (95% CI)† | P |
| Grades III-IV | |||||
| acute GVHD | D6S276 | 48/143 (34) | 204/657 (31) | 0.83 (0.56–1.24) | .37 |
| D6S105 | 64/217 (30) | 188/583 (32) | 1.07 (0.76–1.52) | .69 | |
| D6S510 | 192/600 (32) | 61/204 (30) | 0.92 (0.64–1.33) | .67 | |
| D6S265 | 200/601 (33) | 52/199 (26) | 0.67 (0.45–0.98) | .04 | |
| D6S2811 | 180/544 (33) | 73/259 (28) | 0.76 (0.54–1.07) | .11 | |
| D6S2810 | 171/535 (32) | 59/187 (32) | 0.94 (0.65–1.35) | .74 | |
| D6S2787 | 177/545 (33) | 75/258 (29) | 0.83 (0.59–1.17) | .29 | |
| D6S273 | 153/512 (30) | 97/284 (34) | 1.23 (0.89–1.71) | .21 | |
| D6S2876 | 236/739 (32) | 17/65 (26) | 0.71 (0.39–1.31) | .28 | |
| D6S2874 | 155/496 (31) | 98/308 (32) | 1.00 (0.73–1.38) | .99 | |
| D6S2749 | 72/266 (27) | 181/538 (34) | 1.36 (0.97–1.91) | .08 | |
| D6S291 | 66/203 (33) | 186/593 (31) | 0.99 (0.70–1.40) | .94 | |
| Overall | |||||
| mortality | D6S276 | 40/76 (53) | 458/738 (62) | 1.38 (0.98–1.94) | .07 |
| D6S105 | 79/150 (53) | 417/663 (63) | 1.32 (1.02–1.71) | .03 | |
| D6S510 | 351/576 (61) | 151/242 (62) | 1.00 (0.82–1.47) | .92 | |
| D6S265 | 358/581 (62) | 139/232 (60) | 0.86 (0.70–1.07) | .19 | |
| D6S2811 | 339/537 (63) | 162/280 (58) | 0.80 (0.66–0.98) | .03 | |
| D6S2810 | 336/530 (63) | 113/204 (55) | 0.81 (0.65–1.00) | .06 | |
| D6S2787 | 294/511 (58) | 206/306 (67) | 1.26 (1.03–1.53) | .02 | |
| D6S273 | 272/460 (59) | 225/350 (64) | 1.18 (0.98–1.43) | .08 | |
| D6S2876 | 450/741 (61) | 51/77 (66) | 0.99 (0.72–1.35) | .95 | |
| D6S2874 | 219/366 (60) | 282/452 (62) | 1.17 (0.97–1.41) | .10 | |
| D6S2749 | 106/194 (55) | 395/624 (63) | 1.37 (1.08–1.72) | .007 | |
| D6S291 | 77/127 (61) | 419/683 (61) | 1.13 (0.87–1.46) | .39 | |
Adjusted for patient age, disease severity, use of total body irradiation, use of T-cell depletion, number of mismatches at Msat other than marker of interest and number of HLA-DPB1 disparities.
Odds ratio (OR; acute GVHD) and hazard ratio (HR; mortality) measure risks associated with Msat mismatching relative to Msat matching in interest; CI, confidence interval.
Relapse
No statistically significant or suggestive associations between Msat mismatching and relapse were observed. There was no statistically significant association between the number of mismatched Msats (regardless of the location of Msat) and the hazard of relapse (P = .18) or between the number of class I, class II or class III Msat mismatches and the hazard of relapse (P = .43, P = .29, P = .68 for class I, class III, and class II, respectively).
Overall Mortality
There was a trend for a lower risk of death with mismatching for D6S2811 (hazard ratio [HR], 0.80; 95% CI, 0.66–0.98; P = .03) and a higher risk of death with mismatching for D6S105 (HR, 1.32; 95% CI, 1.02–1.71; P = .03), D6S2787 (HR, 1.26; 95% CI, 1.04–1.53; P = .02) and D6S2749 (HR, 1.37; 95% CI, 1.09–1.72; P = .007) (Table 3).
The hazard of mortality increased as the total number of mismatched Msats increased regardless of the location of the Msat within the MHC (P = .01). Categorizing the number of Msat mismatches (arbitrarily) as 0–1, 2–3, 4–8, and 9–11, the hazard of mortality increased for each of these categories compared to the group with 0–1 mismatched Msats [HRs 1.25 (P = .53), 1.71 (P = .10), and 2.10 (P = .03), respectively].
When Msats were grouped according to their location within class I, class III, or class II, the hazard of mortality increased with increasing number of mismatched class III and class II Msats (P = .008 and P = .01, respectively), but not within class I Msats (P = .10).
DISCUSSION
Acute GVHD may arise when donor T cells recognize differences in recipient HLA. Alternatively, decreased GVHD risk has been observed when there is release of inhibition mediated by donor natural killer immunoglobulin-like receptors (KIR), either through donorrecipient ligand mismatching [25] or through absence of recipient ligand [26]. In the current study, we tested the hypothesis that undetected MHC variation among HLA matched transplants may influence the risk of GVHD, leading to either increased or decreased risk. To this end, we used Msat markers as a gene-mapping tool. Although Msats themselves are not functional, they may be in LD with the putative at-risk gene(s). Our results demonstrate that differences between donor and recipient Msat alleles influence GVHD risk and survival. Mapping of candidate genes will be facilitated in the future with the availability of a fine MHC map [5,6,27,28]. Dissection of the underlying cellular mechanisms that account for susceptibility to or protection against GVHD remains an important research question in the future.
In genetic association studies, it is customary to test the genotyping data for departures from HWE as deviations could help to identify genotyping errors [22, 23]. Deviation from HWE for a subset of markers could also arise from chance alone [22, 23]. In the current study, four Msats were found to deviate from HWE. Of these, D6S276 had an excess of homozygotes for one of the alleles, and D6S2810, D6S2749, and D6S2811 showed either increased or decreased heterozygosity of certain genotype combinations compared to their expectations under HWE. Re-evaluation of the raw electrophoresis data indicated no obvious technical explanation for any of the deviating Msats. We interpret these findings as potentially indicative of a highly selected population of HLA matched transplant recipients and donors.
The associations observed in this study will require validation in independent populations because the number of polymorphic Msat loci increases the potential that some of the statistically significant associations were due to chance alone. Donor-recipient HLA-A, -B, -C, -DRB1 and -DQB1 mismatching did not contribute to the risks in this study population, however, immune response gene variation, chromosome 22 Msat and killer immunoglobulin-like receptor/HLA interactions are well-recognized [25, 26, 29–40]. Since these genes are not encoded on chromosome 6 (with the exception of tumor necrosis factor [TNF] gene), their allele frequencies are expected to be similar among the Msat mismatched groups within the study population. Other MHC region genes such as HLA-DPB1 have been shown to be associated with GVHD, rejection and mortality after unrelated HCT [19, 41–44]. In the current study, mismatching at HLA-DPB1 was included in the regression models. Moreover, HLA-DPB1 mismatching did not have a demonstrable effect on the associations of Msat mismatching with outcome. Out of the 113 HLA-DPB1 matched pairs that had corresponding data for HLA-DPA1 locus, there were only 10 (9%) HLA-DPA1 mismatched pairs. Mismatching in HLA-DRB3, -DRB4, -DRB5 and -DQA1 was minimal (6.5%, 3.7%, 1% and 0.5%, respectively) (data not shown). Although genotyping data for HLA-DRB3, -DRB4, -DRB5, -DQA1 and -DPA1 were not available for all transplant pairs (Table 2), the contribution of these loci to clinical outcome is likely very small due to the high LD and consequently low mismatch frequency [45–47].
HLA-A2 positive patients may be at increased risk of acute GVHD after related donor transplantation because of HA-1 and HA-2 minor histocompatibility antigens [48]. In our study population HLA-A2 was present at equal frequencies in both Msat-matched and Msat-mismatched pairs for all Msats with the exception of D6S265 and D6S510. For both D6S265 and D6S510, HLA-A2 was present at 74% frequency in Msat mismatched pairs and at 39% frequency in Msat matched pairs. Despite the skewing of the HLA-A2 proportions, the rate of GVHD among the D6S265 mismatched pairs was 25% in HLA-A2 positive pairs and 26% in HLA-A2 negative pairs. Hence, it is unlikely that HLA-A2 affected the observed association of D6S265 mismatching with GVHD risk.
Our observations suggest that HLA-matched unrelated donors and recipients may have Msat-linked genetic variation outside of the classical HLA loci, and that this variation may influence the risks of acute GVHD and mortality. Although a dense map of the MHC [5,6,27,28] is currently available, LD between these single nucleotide polymorphisms and Msats is unknown. In the current study Msat marker mismatching was defined as any detectable difference between the recipient and donor. Whether Msat alleles that differ by 2 bp or more are linked to the same functional gene is not known, and remains an important research question. It was not our intention to examine Msat-HLA haplotype associations to clinical outcome because of the reported inaccuracies of the population statistics for individual haplotype inference [49]. These analyses will be feasible in the future when new laboratory techniques are available to define Msat haplotypes across the MHC [50].
The current study found suggestive associations with transplant outcome in four regions: D6S105 in the extended class I; D6S265 to D6S2811 in class I; D6S2787 in class III, and D6S2749 in class II. Although no data is currently available on the LD between D6S2811 and MICA or MICB genes, a potential role for MIC is intriguing. MIC-A and MIC-B genes map 18 kb and 113 kb centromeric to D6S2811. MIC gene products serve as ligands for the NKG2D activating receptor on NK cells, CD8+ T cells and gamma/delta T cells [51]. Donor-recipient matching for the class I region inclusive of MIC has been associated with improved survival after unrelated HCT [52]. Another interesting gene cluster, TNF, is located 38 kb telomeric to D6S2787. The polymorphic TNF alpha and beta genes have been implicated in GVHD and transplant related mortality [30,31,36–39]. Finally, several genes surrounding D6S265, D6S2749 and D6S105 markers have been described [5,6,53,54], however any putative role in transplantation is speculative at this time.
GVHD is a complex disease that is the consequence of synergistic effects of multiple genes. To address whether additive effects of MHC region polymorphisms contributed to the risks of GVHD, relapse or mortality, we analyzed the Msats according to the total number of mismatched Msats, and to the region within the MHC. The analysis of the cumulative effect of Msat mismatching revealed a suggestive association of increasing numbers of class III (D6S273 and D6S2787) or class II (D6S2876, D6S2874 and D6S2749) mismatched Msats with increased hazard of mortality. The majority of the class III and class II mismatches included D6S2787 (72%) and D6S2749 (82%) (data not shown), each of which was associated with increased risk of death (Table 3). The cumulative effect of class III or II mismatching with outcome might be explained by a higher proportion of mismatching for D6S2787 or D6S2749, respectively, compared to the other Msats within class III (D6S273) and II (D6S2876 and D6S2874) regions. Alternatively, these other class III and class II Msats may be hitchhiking with D6S2787 or D6S2749.
Four of the Msat markers (D6S273, D6S2810, D6S265 and D6S510) described herein were studied by Li et al [14]. The frequency of donor-recipient matching for these loci was higher among Japanese transplant pairs than in our study population (64%, 89%, 81%, 97% versus 57%, 72%, 71%, 70%, respectively), possibly indicating more highly conserved haplotypes within Japanese population (www.hapmap.org). No statistically significant associations between mismatching for any of the 13 Msats and acute GVHD, chronic GVHD, survival or relapse were uncovered by Li et al, however, matching for the class III TNFd marker among recipients with grades III-IV acute GVHD was associated with decreased survival [14]. Whether differences in Japanese and Caucasian MHC haplotypes (www.hapmap.org) can explain the associations to transplantation outcome remain to be determined in future studies. The current study provides the impetus for fine mapping of MHC sequence variation in ethnically diverse transplant populations.
Acknowledgements
Authors thank Dr. Mary Carrington (National Cancer Institute) and Dr. Richard Single (University of Vermont) for helpful discussions; Dr. John Hansen and Eric Mickelson (Fred Hutchinson Cancer Research Center) for cell lines; Sharie Cheng and Mark Gatterman for outstanding technical support.
This work was supported by grants CA72978 (EWP); CA18029, AI49213 and AI33484 (MM, TAG and EWP), FRM-ARS2000 (LA, AD, VD, KG), Office of Naval Research (N00014-93-1-0658, N00014-95-1-0055, N00014-96-2-0016) and Health Resources and Services Administration (240-97-0036) (SS).
Appendix
The following members of the IHWG HCT Component contributed data to this manuscript:
Australia
AUSCHR/ Frank Christiansen and Campbell Witt, Department of Clinical Immunology and Biochemical Genetics, Path West and Royal Perth Hospital, Perth
Peter Bardy, Greg Bennett and Ian Humphries, Red Cross Blood Transfusion Service, Adelaide
Heather Dunckley, Tissue Typing, National Transplantation Services, Australian Red Cross Blood Service, Sydney
Rhonda Holdsworth, Ian Nicholson, Simon Knowles, Mike Varney, Carmel Kanaan, Mary Diviney, Brian Tait, Victorian Transplantation and Immunogenetics Service, Australian Red Cross Blood Service, Melbourne
France
Colette Raffoux, French Registry of Hematopoïetic cells Donors, Hospital Saint Louis, Paris
Eliane Gluckman, Hospital Saint Louis, Paris
FRAABS/ Lena Absi, Laboratoire HLA, EFS Auvergne Loire, St. Etienne
Denis Guyotat, Institut Cancereux de la Loire, Service d'hématologie, St Priest en Jarez
FRABET/ Valerie Dubois, Laboratoire d'Histocompatibilité, EFS Rhone Alpes, Lyon
FRABIG/ Jean-Denis Bignon and Katia Gagne, Laboratoire d'Histocompatibilité et d'Immunogénétique, EFS Pays de Loire, Nantes
FRABOI/ Monique Bois, HLA EFS Centre Atlantique, Poitiers
FRAMYE/ Anne Dormoy, Laboratoire d'Histocompatibilité, Etablissement Français du Sang-Alsace, Strasbourg
The Netherlands
NLDOUD/ Machteld Oudshoorn, Europdonor Foundation, Leiden
Jan J Cornelissen, Department of Hematology, Erasmus University Medical Center-Daniel Den Hoed, Rotterdam
United States
CIBMTR/ Mary Horowitz, Center for International Blood and Marrow Transplant Research, Medical College of Wisconsin, Milwaukee, WI
Michael Haagenson, Center for International Blood and Marrow Transplant Research, Minneapolis, MN
NMDP*/ Stephen Spellman, National Marrow Donor Program, Minneapolis, MN
USAPET/ Effie Petersdorf, Mari Malkki, Mark Gatterman, Gary Schoch and Ted Gooley, Fred Hutchinson Cancer Research Center, Seattle
Mary Carrington, NCI-Frederick Cancer Research and Development Center, Frederick, MD, USA
* Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Office of Naval Research or the National Marrow Donor Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: MM and EWP designed research. All authors contributed data. MM, TAG, and EWP analyzed data. MM, TAG, FTC, MDH, SS and EWP prepared the manuscript.
REFERENCES
- 1.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339:1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 2.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 3.Petersdorf EW, Anasetti C, Martin PJ, et al. Limits of HLA mismatching in unrelated hematopoietic cell transplantation. Blood. 2004;104:2976–2980. doi: 10.1182/blood-2004-04-1674. [DOI] [PubMed] [Google Scholar]
- 4.Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 5.The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 6.Horton R, Wilming L, Rand V, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 7.Gourraud PA, Mano S, Barnetche T, Carrington M, Inoko H, Cambon-Thomsen A. Integration of microsatellite characteristics in the MHC region: a literature and sequence based analysis. Tissue Antigens. 2004;64:543–555. doi: 10.1111/j.1399-0039.2004.00317.x. [DOI] [PubMed] [Google Scholar]
- 8.Carrington M, Marti D, Wade J, et al. Microsatellite markers in complex disease: mapping disease associated regions within the human Major Histocompatibility Complex. In: Goldstein DB, Schlötterer C, editors. Microsatellites: Evolution and Applications. Oxford, England: Oxford University Press; 1999. pp. 225–237. [Google Scholar]
- 9.Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffreys AJ, Murray J, Neumann R. High-resolution mapping of cross-overs in human sperm defines a minisatellite associated recombination hotspot. Mol Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 11.Malkki M, Single R, Carrington M, Thomson G, Petersdorf E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 2005;66:114–124. doi: 10.1111/j.1399-0039.2005.00453.x. [DOI] [PubMed] [Google Scholar]
- 12.Hanifi Moghaddam P, Zwinderman A, Kazemi M, et al. D6STNFa microsatellite locus correlates with CTLp frequency in unrelated bone marrow donor-recipient pairs. Hum Immunol. 1998;59:295–301. doi: 10.1016/s0198-8859(98)00022-6. [DOI] [PubMed] [Google Scholar]
- 13.Witt C, Moghaddam PH, van der Meer R, et al. Matching for TNF microsatellites is strongly associated with matching for other non-HLA MHC sequences in unrelated bone marrow donor-recipient pairs. Hum Immunol. 1999;60:862–866. doi: 10.1016/s0198-8859(99)00026-9. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Kawata H, Katsuyama Y, et al. Association of polymorphic MHC microsatellites with GVHD survival and leukemia relapse in unrelated hematopoietic stem cell transplant donor/recipient pairs matched at five HLA loci. Tissue Antigens. 2004;63:362–368. doi: 10.1111/j.0001-2815.2004.00200.x. [DOI] [PubMed] [Google Scholar]
- 15.Carrington M, Wade J. Selection of transplant donors based on MHC microsatellite data. Hum Immunol. 1996;50:151–154. doi: 10.1016/0198-8859(96)00118-8. [Erratum Hum Immunol. 1996;51:106–109] [DOI] [PubMed] [Google Scholar]
- 16.Foissac A, Fort ML, Giraldo P, Abbal M, Raffoux C, Cambon-Thomsen A. Microsatellites in the HLA region potential applications in bone marrow transplantation. Transplant Proc. 1997;29:2374–2375. doi: 10.1016/s0041-1345(97)00408-9. [DOI] [PubMed] [Google Scholar]
- 17.Foissac A, Fort M, Clayton J, et al. Microsatellites in the HLA region HLA prediction and strategies for bone marrow donor registries. Transplant Proc. 2001;33:491–492. doi: 10.1016/s0041-1345(00)02107-2. [DOI] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Gooley TA, Anasetti C, et al. Optimizing outcome after unrelated marrow transplantation by comprehensive matching of HLA class I and II alleles in the donor and recipient. Blood. 1998;92:3515–3520. [PubMed] [Google Scholar]
- 19.Petersdorf EW, Gooley T, Malkki M, et al. The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br J Haematol. 2001;112:988–994. doi: 10.1046/j.1365-2141.2001.02655.x. [DOI] [PubMed] [Google Scholar]
- 20.Hurley CK, Fernandez-Vina M, Hildebrand WH, et al. A high degree of HLA disparity arises from limited allelic diversity: analysis of 1775 unrelated bone marrow transplant donor-recipient pairs. Hum Immunol. 2007;68:30–40. doi: 10.1016/j.humimm.2006.09.004. Epub 2006 Oct 25. [DOI] [PubMed] [Google Scholar]
- 21.Mickelson E, Damodaran A, He P, et al. Standardization of microsatellite data: Comparative analysis of results from 13th IHWS participating laboratories and establishment of a microsatellite reference panel and standard nomenclature. In: Hansen JA, editor. Immunobiology of the Human MHC; Proceedings of the 13th International Histocompatibility Workshop and Conference; Seattle, WA: IHWG Press; 2007. pp. 1277–1314. [Google Scholar]
- 22.Hartl DL, Clark AG. Principles of Population Genetics. 3rd ed. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- 23.Leal SM. Detection of genotyping errors and pseudo-SNPs via deviations from Hardy-Weinberg Equilibrium. Genet. Epidemiol. 2005;29:204–214. doi: 10.1002/gepi.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor 2transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006;12:876–884. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Hsu KC, Gooley T, Malkki M, et al. KIR Ligands and Prediction of Relapse after Unrelated Donor Hematopoietic Cell Transplantation for Hematologic Malignancy. Biol Blood Marrow Transplant. 2006;12:828–836. doi: 10.1016/j.bbmt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 27.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miretti MM, Walsh EC, Ke X, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullighan CG, Bardy PG. Directions in the Genomics of Allogeneic Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2007;13:127–144. doi: 10.1016/j.bbmt.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Middleton PG, Taylor PR, Jackson G, Proctor SJ, Dickinson AM. Cytokine gene polymorphisms associating with severe acute graft-versus-host disease in HLA-identical sibling transplants. Blood. 1998;92:3943–3948. [PubMed] [Google Scholar]
- 31.Holler E, Rogler G, Brenmoehl J, et al. Prognostic significance of NOD2/CARD15 variants in HLA-identical sibling hematopoietic stem cell transplantation: effect on long-term outcome is confirmed in 2 independent cohorts and may be modulated by the type of gastrointestinal decontamination. Blood. 2006;107:4189–4193. doi: 10.1182/blood-2005-09-3741. [DOI] [PubMed] [Google Scholar]
- 32.Lin MT, Storer B, Martin PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349:2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 33.Mullighan C, Heatley S, Doherty K, et al. Non-HLA immunogenetic polymorphisms and the risk of complications after allogeneic hemopoietic stem-cell transplantation. Transplantation. 2004;77:587–596. doi: 10.1097/01.tp.0000111769.45088.a2. [DOI] [PubMed] [Google Scholar]
- 34.Rocha V, Franco RF, Porcher R, et al. Host defense and inflammatory gene polymorphisms are associated with outcomes after HLA-identical sibling bone marrow transplantation. Blood. 2002;100:3908–3918. doi: 10.1182/blood-2002-04-1033. [DOI] [PubMed] [Google Scholar]
- 35.Socie G, Loiseau P, Tamouza R, et al. Both genetic and clinical factors predict the development of graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Transplantation. 2001;72:699–706. doi: 10.1097/00007890-200108270-00024. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi H, Furukawa T, Hashimoto S, et al. Contribution of TNF-alpha and IL-10 gene polymorphisms to graft-versus-host disease following allo-hematopoietic stem cell transplantation. Bone Marrow Transplant. 2000;26:1317–1323. doi: 10.1038/sj.bmt.1702724. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa Y, Kashiwase K, Akaza T, et al. Polymorphisms in TNFA and TNFR2 affect outcome of unrelated bone marrow transplantation. Bone Marrow Transplant. 2002;29:569–575. doi: 10.1038/sj.bmt.1703409. [DOI] [PubMed] [Google Scholar]
- 38.Keen LJ, Defor TE, Bidwell JL, et al. Interleukin-10 and tumor necrosis factor alpha region haplotypes predict transplant-related mortality after unrelated donor stem cell transplantation. Blood. 2004;103:3599–3602. doi: 10.1182/blood-2002-11-3568. [DOI] [PubMed] [Google Scholar]
- 39.Bogunia-Kubik K, Polak M, Lange A. TNF polymorphisms are associated with toxic but not with a GVHD complications in the recipients of allogeneic sibling haematopoietic stem cell transplantation. Bone Marrow Transplant. 2003;32:617–622. doi: 10.1038/sj.bmt.1704200. [DOI] [PubMed] [Google Scholar]
- 40.Kikuchi K, Naruse TK, Onizuka M, et al. Mapping of susceptibility and protective loci for acute GVHD in unrelated HLA-matched bone marrow transplantation donors and recipients using 155 microsatellite markers in chromosome 22. Immunogenetics. 2007;59:99–108. doi: 10.1007/s00251-006-0186-2. [DOI] [PubMed] [Google Scholar]
- 41.Loiseau P, Esperou H, Busson M, et al. DPB1 disparities contribute to severe GVHD and reduced patient survival after unrelated donor bone marrow transplantation. Bone Marrow Transplant. 2002;30:497–502. doi: 10.1038/sj.bmt.1703658. [DOI] [PubMed] [Google Scholar]
- 42.Shaw BE, Marsh SG, Mayor NP, Russell NH, Madrigal JA. HLA-DPB1 matching status has significant implications for recipients of Unrelated Donor Stem Cell Transplants. Blood. 2005;7:1220–1226. doi: 10.1182/blood-2005-08-3121. [DOI] [PubMed] [Google Scholar]
- 43.Shaw BE, Potter MN, Mayor NP, et al. The degree of matching at HLA-DPB1 predicts for acute graft-versus-host disease and disease relapse following haematopoietic stem cell transplantation. Bone Marrow Transplant. 2003;31:1001–1008. doi: 10.1038/sj.bmt.1704029. [DOI] [PubMed] [Google Scholar]
- 44.Fleischhauer K, Locatelli F, Zecca M, et al. Graft rejection after unrelated donor hematopoietic stem cell transplantation for thalassemia is associated with non-permissive HLA-DPB1 disparity in host-versus-graft direction. Blood. 2006;107:2984–2992. doi: 10.1182/blood-2005-08-3374. [DOI] [PubMed] [Google Scholar]
- 45.Klitz W, Maiers M, Spellman S, et al. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens. 2003;62:296–307. doi: 10.1034/j.1399-0039.2003.00103.x. [DOI] [PubMed] [Google Scholar]
- 46.Knipper AJ, Hakenberg P, Enczmann J, et al. HLA-DRB1,3,4,5 and -DQB1 allele frequencies and HLA-DR/DQ linkage disequilibrium of 231 German caucasoid patients and their corresponding 821 potential unrelated stem cell transplants. Hum Immunol. 2000;61:605–614. doi: 10.1016/s0198-8859(00)00114-2. [DOI] [PubMed] [Google Scholar]
- 47.Begovich AB, Moonsamy PV, Mack SJ, et al. Genetic variability and linkage disequilibrium within the HLA-DP region: analysis of 15 different populations. Tissue Antigens. 2001;57:424–439. doi: 10.1034/j.1399-0039.2001.057005424.x. (Erratum in: Tissue Antigens 2001;58:431. [DOI] [PubMed] [Google Scholar]
- 48.Goulmy E, Schipper R, Pool J, et al. Mismatches of minor histocompatibility antigens between HLA-identical donors and recipients and the development of graft-versus-host disease after bone marrow transplantation. N Engl J Med. 1996;334:281–285. doi: 10.1056/NEJM199602013340501. [DOI] [PubMed] [Google Scholar]
- 49.Tishkoff SA, Pakstis AJ, Ruano G, Kidd KK. The accuracy of statistical methods for estimation of haplotype frequencies: an example from the CD4 locus. Am J Hum Genet. 2000;67:518–522. doi: 10.1086/303000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Z, Hood L, Malkki M, Petersdorf EW. Long-range multilocus haplotype phasing of the MHC. Proc Natl Acad Sci U S A. 2006;103:6964–6969. doi: 10.1073/pnas.0602286103. [Erratum in: Proc Natl Acad Sci U S A. 2006;103:9374] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins RW. Human MHC class I chain related (MIC) genes: their biological function and relevance to disease and transplantation. Eur J Immunogenet. 2004;31:105–114. doi: 10.1111/j.1365-2370.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- 52.Kitcharoen K, Witt CS, Romphruk AV, Christiansen FT, Leelayuwat C. MICA, MICB, and MHC beta block matching in bone marrow transplantation: relevance to transplantation outcome. Hum Immunol. 2006;67:238–246. doi: 10.1016/j.humimm.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 53.Thorpe KL, Abdulla S, Kaufman J, Trowsdale J, Beck S. Phylogeny and structure of the RING3 gene. Immunogenetics. 1996;44:391–396. doi: 10.1007/BF02602785. [DOI] [PubMed] [Google Scholar]
- 54.Dorak MT, Burnett AK, Worwood M. Hemochromatosis gene in leukemia and lymphoma. Leuk Lymphoma. 2002;43:467–477. doi: 10.1080/10428190290011930. [DOI] [PubMed] [Google Scholar]