Abstract
At any time, each cell of the protozoan parasite Trypanosoma brucei expresses a single species of its major antigenic protein, the variant surface glycoprotein (VSG), from a repertoire of >2,000 VSG genes and pseudogenes. The potential to express different VSGs by transcription and recombination allows the parasite to escape the antibody-mediated host immune response, a mechanism known as antigenic variation. The active VSG is transcribed from a sub-telomeric polycistronic unit called the expression site (ES), whose promoter is 40–60 kb upstream of the VSG. While the mechanisms that initiate recombination remain unclear, the resolution phase of these reactions results in the recombinational replacement of the expressed VSG with a donor from one of three distinct chromosomal locations; sub-telomeric loci on the 11 essential chromosomes, on minichromosomes, or at telomere-distal loci. Depending on the type of recombinational replacement (single or double crossover, duplicative gene conversion, etc), several DNA-repair pathways have been thought to play a role. Here we show that VSG recombination relies on at least two distinct DNA-repair pathways, one of which requires RMI1-TOPO3α to suppress recombination and one that is dependent on RAD51 and RMI1. These genetic interactions suggest that both RAD51-dependent and RAD51-independent recombination pathways operate in antigenic switching and that trypanosomes differentially utilize recombination factors for VSG switching, depending on currently unknown parameters within the ES.
Introduction
Monoallelic expression of multigene families occurs in a variety of cellular processes, including mating-type switching in yeasts, immunoglobulin gene diversification in B-cell development, odorant receptors, and surface antigen variation in several pathogens [1]-[7]. Trypanosoma brucei is a protozoan pathogen that causes African sleeping sickness. Only one allele of the variant surface glycoprotein (VSG) is expressed at a time in each trypanosome, yet the genome may contain more than 2,000 VSG genes and pseudogenes. About 15 sub-telomeric polycistronic expression sites (ES) contain a VSG ∼ 50 kb downstream of their promoters [8]-[10] and about 1 kb upstream of the telomeric repeat array. Only one ES can transcribe a VSG at any time and the rest are transcriptionally silent. Most VSGs are not associated with an ES (minichromosomal and ‘telomere-distal’, also referred to as ‘chromosome-internal’), and they lack promoters. Antigenic switching is caused mainly by switching the expressed VSG through DNA recombination. Infrequently, a new VSG can be activated by switching the transcriptional status among the active and silent ES (reviewed in [4], [11], [12]). Recombination-mediated VSG switching occurs preferentially by gene conversion (GC) rather than crossover [13]-[16], despite the fact that there are no clear advantages in switching through GC in vivo.
One of the major factors that control mitotic crossover is the RTR complex (also known as the BTB complex). This complex consists of a RecQ-family helicase (BLM in mammals and SGS1 in budding yeast), a Topoisomerase III α, and RMI1/2 (BLAP75/18 in mammals and RMI1 in budding yeast). RMI is the third component that has been recently identified in several organisms [17]-[23]. The functions of the RTR complex have been documented extensively in yeasts and mammalian systems [21], [22], [24]-[31]. The complex safeguards genome integrity by influencing various aspects, including DNA replication, mitotic and meiotic recombination, and telomere dynamics. One of major functions of the RTR complex is to remove recombination intermediates that may accumulate during these processes. Genetic studies from budding yeast showed that the growth defect of top3 is caused by the accumulation of recombination intermediates and the defect could be relieved by mutations in SGS1 or in the RAD51-pathway [28], [32], [33]. Recombination intermediates accumulated when TOP3 enzyme activity was compromised [34]. A Holliday Junction (HJ) is structurally similar to a recombination intermediate, and the RTR complex is required for the removal of both structures. In vitro studies showed that human RMI1 (hRMI1) stimulates the double Holliday Junction (dHJ) dissolution activity of TOPO3α and the dHJ unwinding activity of BLM-TOPO3α [35], [36]. In yeast, RMI1 stimulates the ssDNA binding and relaxation activity of TOP3 in vitro [37]. hRMI1 interaction with TOPO3α appears to control TOPO3α enzyme activity, as hRMI1 mutants defective for interaction with TOPO3α, not for DNA, lose the ability to stimulate the dHJ dissolution activity of TOPO3α in vitro [38].
We showed previously that trypanosomes lacking TOPO3α significantly increased VSG switching and this increase was largely due to the elevated levels of VSG GC and VSG crossover [39]. The data suggested that the RTR complex is probably conserved in T. brucei and may play roles in VSG switching by controlling undesirable recombination intermediates arising between the active VSG and silent VSG donors. Here we identify the T. brucei RMI1 homologue and demonstrate that TbRMI1 interacts with TbTOPO3α and promotes productive VSG switching. We show that, similarly to the previously described TOPO3α defect, RMI1 deficiency increases VSG switching rate by promoting VSG gene conversion and crossover. Genetic interactions between TbRMI1 and TbRAD51 further reveal that antigenic variation is under control of multiple recombination pathways, depending on where the recombination occurs, and that VSG GC and ES GC are likely to be initiated by different triggers.
Results
Deletion of Tb927.3.1830, a T. brucei DUF1767 domain-containing protein, causes a growth defect
We demonstrated previously that T. brucei TOPO3α plays critical roles in recombination-mediated VSG switching [39]. RMI1 homologues that work together with TOPO3α-BLM (TOP3-SGS1 in budding yeast) have recently been identified in several model organisms [18]-[20], [23], [30]. RMI1 homologues share a signature domain at the N-terminus, called DUF1767, whose function is unknown. By BLAST searches, we found two T. brucei proteins that contain a DUF1767 domain at the N-terminus, Tb927.3.1830 and Tb927.8.2040.
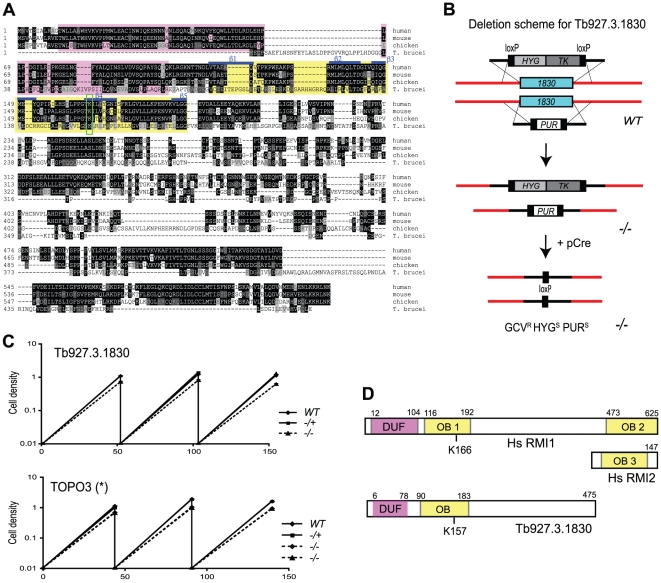
Figure 1A shows sequence alignment of Tb927.3.1830 with human, mouse, and chicken RMI1 homologues. RMI1 proteins possess an ‘oligonucleotide and oligosaccharide binding’ (OB)-fold (yellow box) next to the DUF1767 domain (pink box). In higher eukaryotes, RMI1 contains another OB-fold domain (OB2) in the C-terminus and associates with RMI2 (BLAP18) via the interaction between OB2 of RMI1 and OB3 of RMI2 [21],[22]. RMI1 deletion mimics TOP3 or TOPO3α deficiency in other organisms. Yeast rmi1 mutant showed growth defects, similar to top3 mutants [19], [23]. Tbtopo3α exhibited a minor growth defect [39]. To determine whether trypanosomes lacking any of these potential RMI1 homologues phenocopy Tbtopo3α's growth defect, we constructed mutants lacking Tb927.3.1830 (Figure 1B) or Tb927.8.2040. Wild type, heterozygous and homozygous mutants were examined for cell growth. Deletion of Tb927.3.1830 mimicked Tbtopo3α, exhibiting a minor growth defect (Figure 1C), while Tb927.8.2040 deletion grew normally (data not shown), indicating that Tb927.3.1830 could be a RMI1 homologue but Tb927.8.2040 is probably not. Similar to Tbtopo3α mutant [39], Tb927.3.1830 deletion shows sensitivity to hydroxyurea, a drug that blocks replication (data not shown).
Figure 1. Deletion of Tb927.3.1830 causes a growth defect.
(A) Alignment of Tb927.3.1830 with human, mouse and chicken RMI1. DUF1767 and OB-fold domains are indicated in pink and yellow boxes, respectively. Conserved lysine residues are in green box in the OB1 domain. Blue bars show locations of five β-strands of OB1. (B) Deletion scheme for Tb927.3.1830. The entire open reading frames (ORFs) of Tb927.3.1830 were sequentially deleted using cassettes containing HYG-TK or PUR flanked by loxP sites. The markers were removed by transient expression of Cre-recombinase [39], [64]. (C) Tbrmi1 exhibits a minor growth defect, similar to Tbtopo3α. Wild-type, rmi1-/+ and rmi1-/- cells were diluted to 10,000 cells/ml and cells were counted after two days of incubation. This was repeated twice. Growth phenotypes of Tbtopo3α mutants were taken from the previous study [39]. Error bars are shown, but are small. (D) Diagrams of human RMI1/2 [41] and Tb927.3.1830.
Human RMI core complex contains three OB domains, N-terminal OB1 and C-terminal OB2 in RMI1, and OB3 in RMI2 (Figure 1D). The OB1 of hRMI1 interacts with TOPO3α-BLM to form the BTB complex and the OB2 with OB3 of RMI2 to form the RMI subcomplex [22], [40]. X-ray crystal structure studies have shown that the RMI core complex interaction is essential for its in vivo function [40], [41]. The OB1 domain is essential for stimulation of the dHJ dissolution activity of the BLM-TOPO3α [40]. K166 (green box in Figure 1A) appears to be important for the stability of OB1 structure of hRMI1 [40]. rmi1-K166A mutant is unable to interact with TOPO3α and defective in stimulating the dissolution activity of BLM-TOPO3α [38]. This lysine residue seems to be conserved in T. brucei Tb927.3.1830 (K157 in green box in Figure 1A). But Tb927.3.1830 appears to lack OB2 domain and it is unknown whether there are multiple RMI proteins present in T. brucei.
Tb927.3.1830 interacts with TbTOPO3α
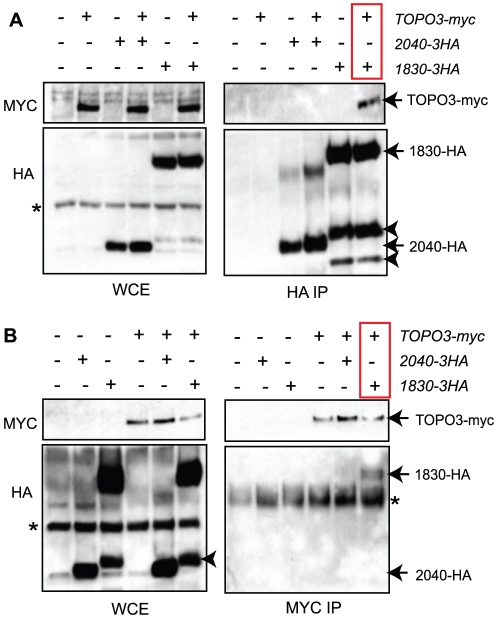
To confirm whether Tb927.3.1830 is a T. brucei RMI1 homologue, we performed co-immunoprecipitation experiments in cells expressing endogenous copies of Tb927.3.1830-3xHA or Tb927.8.2040-3xHA, and/or TOPO3α-3xMYC, each tagged at the C-terminus, using a PCR method [42]. The functionalities of TOPO3α-3xMYC and Tb927.3.1830-3xHA were confirmed by complementation of growth defects (data not shown). The functionality of Tb927.8.2040-3xHA could not be assessed as the mutant showed no phenotype but the tagging was made in a Tb927.8.2040 heterozygote and these grew normally. Lysates were prepared and immunoprecipitated with either anti-HA or anti-MYC antibodies. Precipitated proteins were analyzed by western blot. TOPO3α-3xMYC was pulled down only in Tb927.3.1830-3xHA immunoprecipitates, but not in Tb927.8.2040-3xHA (Figure 2A, red box). Consistent with this, TOPO3α-MYC coimmunoprecipitated with Tb927.3.1830, but not Tb927.8.2040 (Figure 2B, red box). These data confirm that Tb927.3.1830 is a RMI1 homologue, so Tb927.3.1830 will be referred to as TbRMI1 henceforth.
Figure 2. Tb927.3.1830 interacts with TbTOPO3α.
(A) Tb927.3.1830-HA co-immunoprecipitates with TOPO3α-MYC. (B) TOPO3α-MYC co-immunoprecipitates with Tb927.3.1830-HA. TOPO3α was endogenously tagged with 3xMYC, and Tb927.8.2040 and Tb927.3.1830 with 3xHA. Cell lysates were immunoprecipitated either with anti-HA or anti-MYC antibodies and analyzed by western blot. (*) and (**) indicate antibody heavy and light chains. Arrow heads indicate break-down products of 1830-HA.
TbRMI1 deficiency increases VSG switching frequency
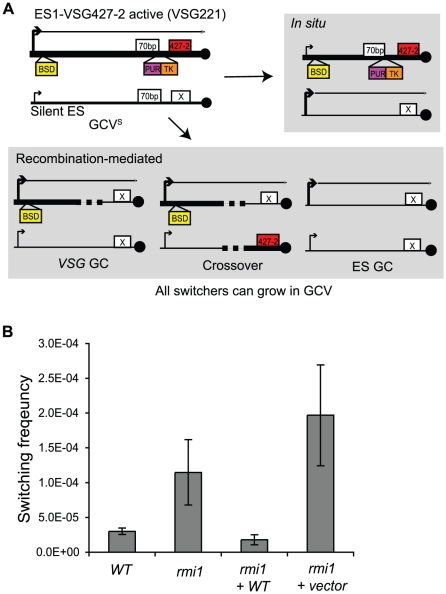
Deletion of TbTOPO3α caused significant increase (10–40-fold) in VSG switching frequency [39]. Switching frequency should increase in the absence of TbRMI1, if TbRMI1 works together with TbTOPO3α in antigenic switching. We measured the VSG switching frequency in Tbrmi1 null mutant, using the assay developed previously [39]. The active expression site, ES1-VSG 427-2 (VSG 221), was doubly marked with a blasticidin-resistance gene (BSD) and a PUR-TK, immediately downstream of the promoter and at the 3′ end of the 70-bp repeats, respectively (Figure 3A). VSG switching concurs with the loss or transcriptional repression of PUR-TK marker, so switched variants can be counter-selected in media containing ganciclovir (GCV), a nucleoside analogue that kills TK-expressing cells.
Figure 3. TbRMI1 deficiency increases VSG switching frequency.
(A) VSG switching reporter cell line and switching mechanisms. The active expression site, ES1-VSG 427-2, was doubly marked with BSD at the promoter and PUR-TK immediately at the 3′ end of the 70-bp repeats. VSG switching events will accompany either loss or repression of the PUR-TK marker, which allows only the switchers to grow in the presence of GCV [39]. Non-switchers can be eliminated as described in the text. Black circles are telomere repeats. (B) TbRMI1 deficiency increases VSG switching frequency. Switching frequency was measured in wild type, rmi1, and rmi1 cells transfected either with an empty vector or the wild-type RMI1.
Wild type and rmi1 strains were grown in media containing blasticidin and puromycin to maintain the homogeneity of the expressed VSG. The cells were then allowed to switch in the absence of selection for 3 days. 5×105 or 1×106 cells were suspended in media containing 5 µg/ml GCV and distributed in 96-well plates. Genuine switchers will be sensitive to puromycin. GCV-resistant clones that were not switched but carried mutations in TK were ruled out by examining puromycin sensitivity. GCVR PURS clones that were not switched and carried mutation(s) in PUR-TK should still express VSG 427-2. These were excluded by western blot using antibody against VSG 427-2. Five and three independent cultures from wild-type and rmi1 mutants, respectively, were examined and the ratio of GCVR switchers to the total number of cells plated was plotted in Figure 3B. The results are also summarized in Table 1. Switching frequency increased ∼ 4 fold in rmi1 (11.0±3.4×10−5), compared to wild type (2.8±0.5×10−5). To confirm that the switching phenotype was caused by the absence of RMI1, an empty vector or a vector containing wild type RMI1 were introduced into the rmi1 null mutant, and three independent clones from each transfection were examined for VSG switching frequency. As shown in Figure 3B, reintroduction of wild-type RMI1 complemented the increased-switching phenotype of rmi1 (1.8±0.7×10−5), while empty-vector transfected rmi1 cells still switched at a higher frequency (20.0±7.2×10−5), indicating that increased-switching phenotype was due to RMI1 deficiency.
Table 1. VSG-switching events in wild type, rmi1, rad51, rmi1 rad51, and rmi1 topo3α cultures.
| Genotype | Total # cells plated | Number of switchers | |||||
| Total | VSG GC | XO | ‘ES GC or loss’ | In situ | ES+XO | ||
| WT | 500,000 | 17 | 3 | 3 | 11 | 0 | 0 |
| WT | 500,000 | 11 | 5 | 0 | 6 | 0 | 0 |
| WT | 1,000,000 | 26 | 4 | 0 | 20 | 1 | 1 |
| WT | 1,000,000 | 26 | 9 | 0 | 16 | 1 | 0 |
| WT | 1,000,000 | 31 | 6 | 2 | 23 | 0 | 0 |
| rmi1 | 500,000 | 74 | 56 | 16 | 2 | 0 | 0 |
| rmi1 | 478,000 | 39 | 29 | 10 | 0 | 0 | 0 |
| rmi1 | 500,000 | 50 | 34 | 14 | 2 | 0 | 0 |
| rad51 | 2,000,000 | 39 | 30 | 4 | 2 | 3 | 0 |
| rad51 | 2,000,000 | 26 | 9 | 4 | 1 | 9 | 3 |
| rad51 | 2,000,000 | 10 | 7 | 1 | 1 | 0 | 1 |
| rmi1 rad51 | 2,000,000 | 4 | 0 | 2 | 1 | 1 | 0 |
| rmi1 rad51 | 2,000,000 | 2 | 0 | 1 | 0 | 1 | 0 |
| rmi1 rad51 | 2,000,000 | 3 | 2 | 1 | 0 | 0 | 0 |
| rmi1 topo3α | 500,000 | 28 | 22 | 6 | 0 | 0 | 0 |
| rmi1 topo3α | 500,000 | 37 | 21 | 13 | 2 | 0 | 1 |
TbRMI1 monitors VSG recombination, similarly to TbTOPO3α
Hyper-recombination and elevated crossover are prominent phenotypes caused by TOPO3α deficiency in T. brucei [39] and in other organisms [26], [30], [31], [43], [44]. To determine whether TbRMI1 is required to suppress VSG GC and crossover, 275 cloned switchers (112 from five wild type cultures and 163 from three rmi1 cultures) were examined. The diagram in Figure 3A illustrates four major switching mechanisms characterized. Each event can be distinguished by assessing blasticidin sensitivity, and BSD and VSG 427-2 presence by PCR. As summarized in Figure 4 and Table 1, VSG GC and crossover rates increased ∼ 13- and ∼ 14-fold in the absence of RMI1, compared to wild type.
Figure 4. TbRMI1 monitors VSG recombination.
TbRMI1 deficiency increases VSG GC and crossover (XO) but decreases ES GC. 275 cloned switchers were examined and the frequency of each switching mechanism was plotted. White bars are wild type and dark grey bars are rmi1 mutant. Error bars indicate standard deviation. The results are also summarized in Table 1. Switched variants were assigned as follows (Figure 3A diagram); VSG-GC switchers, BSD+ VSG 427-2-; ‘ES GC or ES loss’ switchers, BSD- VSG 427-2-; crossover switchers, BSD+ VSG 427-2+; in situ, BSD+ VSG 427-2+.
In our assay, switchers that arose by ES GC and ES loss cannot be distinguished, because these switchers will be negative for BSD and VSG 427-2 when examined by PCR [39]. Frequent loss of entire active ES was observed when TK was targeted next to the active ES promoter [45]. Therefore, BSD- VSG 427-2- switchers should represent either ES GC or ES loss (could be associated with multiple events). Deletion of RMI1 decreased the rate of ‘ES GC or ES loss’, indicating that RMI1 is required for ES GC, a gene conversion event that could span ∼50 kb. Collectively, we conclude that RMI1 also controls the outcome of recombinational switching near VSGs, similarly to TOPO3α, and that RMI1 functions together with TOPO3α in T. brucei antigenic variation. Our data suggest that the RMI1-TOPO3α pathway has two distinct effects on VSG switching, suppressing recombination near the VSG but promoting recombination near the ES promoter.
Genetic interaction between TbRMI1 and TbTOPO3α
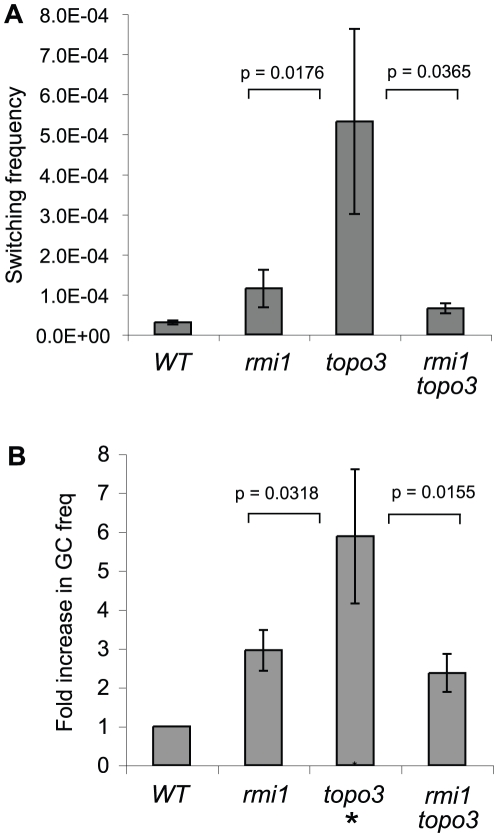
Although TbRMI1 interacts with TbTOPO3α and the rmi1 null mutation caused similar outcomes of VSG switching as the topo3α, RMI1 deficiency increased VSG switching frequency by only ∼ 4-fold, which is less than previously reported for TOPO3α deficiency [39]. If TOPO3α-RMI1 works in the same pathway, the rmi1 topo3α double mutant should exhibit the same phenotype as one of the single mutants. If rmi1 topo3α exhibits an additive phenotype, it is possible that they play additional separate roles. To determine the genetic relationship between RMI1 and TOPO3α, we compared the switching frequencies of wild type, rmi1, topo3α, and rmi1 topo3α double mutant. Switching was significantly increased in the topo3α single mutant (53.2±23.1×10−5), ∼ 10–30-fold higher than wild type (2.8±0.5×10−5), and this was ∼ 5-fold higher than Tbrmi1 (11.0±3.4×10−5) (Figure 5A). The double mutant behaved more like rmi1 single mutant, exhibiting only ∼ 2–3-fold increase (6.5±1.3×10−5) relative to wild type, suggesting that RMI1 is epistatic to TOPO3α in antigenic switching. In the rmi1 topo3α double mutant, as summarized in Table 1, VSG switching also occurred mainly by VSG GC and crossover.
Figure 5. Genetic interaction between T. brucei RMI1 and TOPO3α.
(A) TbRMI1 is epistatic to TbTOPO3α in VSG switching. (B) RMI1 absence increases gene-conversion frequency at URA3 locus and TbRMI1 is epistatic to TbTOPO3α in gene conversion at this locus. Two counter-selectable markers, TK and URA3, were used to measure the GC rates [39]. GCVR and FOAR clones were counted and fold increase relative to wild type was plotted. Error bars indicate standard deviation. (*) in Figure 5B indicates topo3α data taken from the previous report [39]. Unpaired T tests were done and means of switching and GC frequencies were significantly different in the mutants.
Next, we examined whether Tbrmi1 causes hyper-recombination elsewhere and whether RMI1 is also epistatic to TOPO3α in this context. Previously, by replacing one allele of TbURA3 with HYG-TK, we devised a recombination assay [39] in which GC at a non-ES locus can be measured using two counter-selecting drugs: GCV that kills TK+ cells and 5-FOA that kills URA3+. The frequency of GCVR and FOAR represents the GC rate at the URA3 locus. Using this assay, GC increased by 5.9-fold in the absence of TOPO3α [39]. One allele of URA3 was replaced with HYG-TK in wild type, rmi1 or rmi1 topo3α strains. Three independent HYGR clones each were examined and fold increase relative to wild type was plotted (Figure 5B). RMI1 deficiency increased GC rate (2.9-fold), and the double mutant behaved more like rmi1 single mutant, exhibiting 2.4-fold increase of GC. The data show that RMI1 is epistatic to TOPO3α in gene conversion at theURA3 locus and in VSG switching.
TbRMI1 is required for both RAD51-dependent and RAD51-independent VSG switching
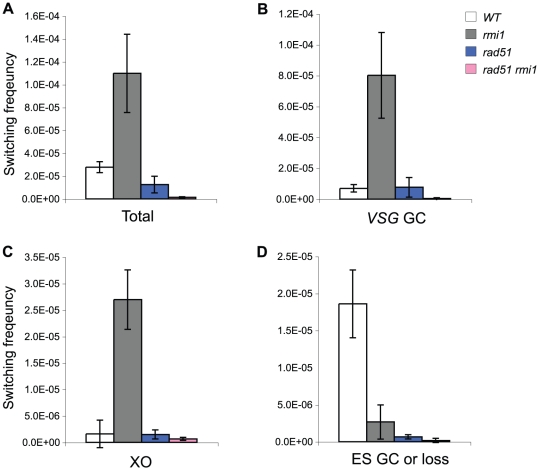
The increased-switching phenotype of Tbtopo3α was dependent on TbRAD51 [39]. Therefore, we asked whether RAD51 is required for the increased switching observed in the rmi1 mutant. Both alleles of RAD51 were deleted in wild type and rmi1 mutant. rad51 single and rmi1 rad51 double mutants were examined for VSG switching frequency and mechanisms. 2×106 cells from three independent cultures of each mutant were plated, and cloned GCVR switchers were analyzed. Deletion of RAD51 caused only 2-fold reduction in overall VSG-switching in the presence of RMI1. On the other hand, it caused ∼ 19-fold reduction in the absence of RMI1, compared to wild type (Figure 6A and Table 1). This suggests that there are at least two recombination pathways working in antigenic switching mechanisms: RAD51-dependent and -independent.
Figure 6. TbRMI1 is required for both RAD51-dependent and RAD51-independent VSG switching.
(A) Simultaneous deletion of RMI1 and RAD51 severely impairs VSG switching. Overall VSG switching frequencies of wild type, rmi1, rad51, and rmi1 rad51 were plotted. (B) Increased VSG GC rate in rmi1 was dependent on RAD51, but RMI1 also functions in VSG GC switching independently of RAD51. (C) Increased crossover in rmi1 was dependent on RAD51. (D) ‘ES GC or ES loss’ switching was impaired in a rad51 or rmi1 single mutant. The results are also summarized in Table 1.
75 and 10 cloned switchers were analyzed from rad51 and rmi1 rad51 mutants, respectively (Figure 6B, C, and D and Table 1). VSG GC was not affected in rad51, but it was significantly reduced in rad51 rmi1 double mutants, indicating that at least two pathways are operating for VSG GC, one of which requires RMI1 independently of RAD51. RAD51 deletion eliminated the increased VSG GC and crossover phenotype of rmi1, suggesting that RMI1 may be required to suppress RAD51-dependent recombinogenic structures arising between the active VSG and VSG donors, similar to the genetic interaction between TOPO3α and RAD51. Unlike VSG GC, ‘ES GC or ES loss’ was reduced in an rmi1 or rad51 single mutant, suggesting that RAD51-dependent recombinogenic structure may not be the cause of this switching mechanism. Therefore, recombination near ES promoter is fundamentally different from that near VSG. Collectively, we propose that RMI1 is required to suppress RAD51-dependent recombination intermediates near VSG, but it is required to promote recombination near ES promoter.
Discussion
Extensive genetic and biochemical studies from yeasts have established the mechanisms of several key recombination pathways. These pathways serve multiple purposes, depending on cellular needs. Choice of recombination pathways determines cell fate. Failure to make the right choices can result in chromosome instability and/or in incompetence in responding to the environment, both eventually leading to high chance of lethality.
T. brucei expresses one type of VSG gene at a time, with the potential to express countless different VSGs. Switching to a different VSG allows parasites to escape a host's immune responses. As illustrated in Figure 3A, recombination-mediated VSG switching can be initiated near or downstream of the ES promoter. An entire silent ES or a relatively smaller region containing a silent VSG can be duplicated and transposed into the active ES to replace a previously active ES-associated VSG. This appears to occur through break-induced replication (BIR) [46], which is at least partly due to the 70-bp repeats that are present upstream of VSG genes, but limited homologies are present within and downstream of VSG genes. Telomere repeats may facilitate VSG switching, providing a second region of homology, but this possibility has not been extensively examined. How the recombination-mediated VSG switching occurs is still not clear.
Similar to mitotic recombination, crossover is relatively infrequent in recombination-mediated VSG switching [13]-[16], [39]. The RTR complex is one of the major factors that control mitotic crossover in other organisms. The RTR complex consists of a RecQ-family helicase (BLM/SGS1), a Topoisomerase III α, and RMI1/2 (BLAP75/18) [17]-[23]. In vitro studies showed that human hRMI1 stimulates the dHJ dissolution activity of TOPO3α and the dHJ unwinding activity of BLM-TOPO3α [35], [36].
We have previously shown that TOPO3α deficiency is associated with hyper-recombination, consistent with studies in other organisms, and with hyper-VSG switching phenotypes in T. brucei [39]. TOPO3α is required to suppress crossover VSG switching in T. brucei. In this study, we characterized a T. brucei RMI1 ortholog that physically interacts with TbTOPO3α. Similar to TOPO3α, deficiency of TbRMI1 increased the rate of VSG crossover, a hallmark of defects in RTR complex function. In addition, TbRMI1 deficiency increased VSG GC switching to a similar extent as TbTOPO3α deficiency, which caused dramatic increase in VSG GC rate. VSG GC was around 70% in both wild type and Tbtopo3α mutant, indicating that VSG GC frequency was 10-40 fold higher than wild type, as the overall VSG switching frequency was 10-40 fold higher in the Tbtopo3α mutant [39]. One of major functions of the RTR complex is to control recombination intermediates accumulating during replication stress. Genetic studies from budding yeast demonstrated that the growth defect of top3 is caused by the accumulation of recombination intermediates and that the defect could be relieved by mutations in SGS1 and in the RAD51-pathway [28], [32], [33]. Recombination intermediates accumulated when TOP3 enzyme activity was compromised [34]. Our data from this study suggests that RMI1 works together with TOPO3α during VSG switching. We propose that accumulation of recombination intermediates near the active VSG causes hyper-VSG GC and hyper-crossover phenotypes when TOPO3α-RMI1 function is compromised (Figure 7 and [39]).
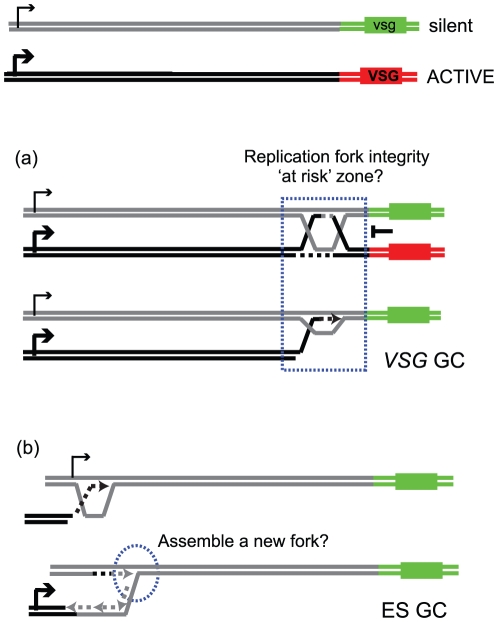
Figure 7. Multiple genetic networks in recombination-mediated VSG variation.
The diagram shows the active ES (ES1) expressing VSG (red) and a silent ES containing vsg (green). RMI1 deficiency exhibited opposite phenotype in VSG-GC and ES GC switching, suggesting that molecular mechanisms of these events are distinct. We propose that there may be ‘replication fork instability zone(s)’ near VSG, potentially the 70-bp repeats, but not in ES promoter region. The RMI1-TOPO3α is required to dissolve recombination intermediates arising in this zone during replication. When their function is compromised, recombination intermediates accumulate and result in VSG-GC and crossover switches (a). RMI1 is required to promote ES GC. As ES GC requires extension of much longer regions compared to VSG GC, having a stable replication fork must be advantageous than having a migrating D-loop. Budding yeast SGS1 is required for replisome stability. Therefore, we propose that T. brucei RMI1-TOPO3α might stabilize a newly assembled replication fork between the active and a silent ES near promoter region. Formation of a stable replication fork may be the key step to generate ES GC switching (b).
What could cause recombination intermediates to accumulate in the active VSG locus? Genome-wide analysis of ORC1 association showed that ORC1 was enriched at subtelomeric regions (Kinetoplastid Molecular Cell Biology meeting April 2011, Tiengwe CM et al.). Some of these ORC1 binding sites may initiate replication. VSGs are located close to telomere repeats. It was shown that the telomere repeats are fragile in mammalian cells, dependent on replication and telomere binding factor TRF1 [47]. In the absence of TRF1, telomere repeats break more frequently and replication was frequently initiated within the TTAGGG repeats, increasing telomere instability and sister-chromatid bridging. Interestingly, BLM and RTEL helicases were required to suppress the telomere fragility. Thus, it is possible that fragility of the active ES could be due to a combination of telomere instability and replication defects. In the absence of TOPO3α or RMI1, these could cause the accumulation of recombination intermediates.
Due to lack of information on DNA replication in T. brucei, it is impossible to determine the identities of recombination intermediates that might accumulate in topo3α or rmi1 trypanosome mutants. Recombination intermediates could arise when replication forks encounter DNA damage, during replication progression and repair. Intermediates could form between sister-chromatids with entangled regions, called hemicatenanes, which can be processed to ‘rec-X’ structures, whose resolution requires SGS1-TOP3 in budding yeast [34], [48], [49]. These structures are different in regard to base pairing and presence of single stranded regions and they can be visualized by DNA 2D gel analysis by Southern blot, a technique unavailable in T. brucei. We showed that VSG GC and crossover are suppressed by TOPO3α and RMI1 in a RAD51-dependent manner. Therefore, we think that TOPO3α-RMI1 may play roles in maintaining ES integrity near VSGs by removing these ‘unwanted’ recombination intermediates. Without TOPO3α-RMI1, they might be resolved by other nucleases (crossover), or a silent VSG locus might be used as a new template during hemicatenane-mediated template switching using its homology sequences and complete replication (VSG GC). It is possible that TOPO3α-RMI1 deficiency could be associated with location specific VSG switching phenotypes due to replication stability ‘at risk’ zone(s) near VSG genes. In the normal situation, TOPO3α-RMI1 may be responsible for protecting the integrity of that region(s) (Figure 7(a)).
When double strand breaks (DSBs) occur, broken DNA ends are resected, leaving a 3′ overhang, and a RAD51 filament on single stranded DNA invades a template strand to form a D-loop. How this structure is processed can decide how the breaks are repaired. End resection requires SGS1-DNA2-RPA, and EXO1, and SAE2-MRE11 complex and the end resection activity of SGS1 is stimulated by TOP3-RMI1 in budding yeast [50], [51]. BIR frequency increases 39-fold in sgs1Δ exo1Δ double mutants [52]. Interestingly, in combination with exo1Δ, BIR was increased in a separation-of-function allele of SGS1, sgs1-D664Δ, which is proficient for gene conversion but defective for resolution of recombinogenic X-structures formed by replication fork stalling [52], [53]. The residue D664 is located at the second acidic region (AR2) in the N-terminus of SGS1 and the sgs1-AR2Δ shows similar phenotype as the sgs1-D664Δ. There are two recQ family helicases in T. brucei, Tb927.8.6690 and Tb927.6.3580, and Tb927.8.6690 appears to be a BLM/SGS1 ortholog. The region encompassing D664 of SGS1 seems to be conserved in Tb927.8.6690; YPWSD 664E in SGS1 and YPWSE 449E in Tb927.8.6690. Tb927.8.6690 and mutations at this particular region would be useful tools to study molecular mechanism of VSG switching more in detail in the future.
It is notable that VSG GC frequency decreased further in rmi1 rad51 double mutants compared to wild type, suggesting that there are pathways that require RMI1, independently of RAD51. Studies in yeasts and in T. brucei have shown that recombination still occurs in rad51Δ cells [54]-[56]. In the absence of telomerase, telomeres are maintained by a RAD51-dependent and independent pathways [57]-[59]. It is possible that RMI1 may play distinct roles in VSG switching in a RAD51-independent manner.
The frequency of ‘ES GC or ES loss’ decreased in the absence of RMI1 or RAD51, unlike VSG GC, which was significantly increased in the absence of RMI1 and was not affected by RAD51. This is interesting but puzzling, considering that hyper-recombination is the hallmark phenotype of RMI1 deficiency. ‘ES GC or ES loss’ switching seems to occur in a RAD51 and/or RMI1 dependent manner, but their genetic relations cannot be deduced from the current data. However, it is clear that ES GC is not triggered by the same source as the VSG GC. Random breaks may occur at the active promoter due to high level of transcription and these breaks would normally be repaired by homologous recombination, using sister-chromatids, or by NHEJ. However, infrequently, template switching (to a silent ES promoter region) could occur and be repaired by BIR to generate ES GC switchers. When BIR was assayed in a budding yeast cell line with two template chromosomes available, template switching occurred about 20 percent of the time, usually near the sites of strand invasion [60]. The current hypothesis is that repair could be completed by D-loop migration coupled with lagging strand DNA synthesis moving along the template or strand invasion intermediates can be cleaved by endonuclease(s) [60], [61]. This cleavage would allow a new replication fork to establish between two chromosomes (the active and a silent ES in the case of VSG switching). As ES GC switching requires duplication of much larger fragment (∼ 50 kb) than the VSG GC (∼ 4 kb), establishing a stable replication fork could be more advantageous than a migrating D-loop for ES GC, while it would not matter for VSG GC events that require duplication of shorter regions. The repair can be completed by a newly assembled replication fork between the active and silent ES, giving rise to an ES GC switcher (Figure 7 (b)).
In yeast, SGS1 is required for replisome stability, affecting the assembly of replication factors at the fork, in collaboration with checkpoint kinase MEC1 [62]. Therefore, TbRMI1-TOPO3α might be required for the stability of a newly formed replication fork between the active and a silent ES promoter region. If this is the case, generation of ES GC switching should require RAD51 and RMI1-TOPO3α functions. Alternatively, RMI1 might have novel function in the context of ES promoter, which may indirectly affect RAD51-dependent BIR. Transcription of ES occurs polycistronically with a VSG located ∼ 50 kb downstream of the promoter, the only site where transcription machinery assembles within the unit. ES GC and VSG GC occur in different chromatin environments, so the mechanisms of these gene conversions could be fundamentally distinct.
The rmi1 mutant was epistatic to topo3α in VSG switching phenotype. This genetic interaction is interesting because one would expect the opposite scenario considering that TOPO3α possesses enzymatic activity for resolution of recombination intermediates, which is stimulated by RMI1 [35], [36]. The epistatic interaction of TbRMI1 and TbTOPO3α in VSG switching suggests that TbRMI1 may have a role, potentially with BLM helicase, in a process upstream of TbTOPO3α.
Given high level of sequence similarities between ESs, it is likely that silent ESs can recombine at a certain rate and yet the ES structures seem to be well maintained, suggesting that silent ES recombination may also be suppressed by protectors of genome integrity, and recombination of silent ESs might be elevated in topo3α or rmi1 trypanosome mutants. It is difficult to examine recombination rates at or between silent ESs quantitatively, due to technical difficulties, such as strong gene silencing and sequence similarities.
It appears that trypanosomes suppress VSG GC effectively, although VSG switching is essential for parasite survival in the host and VSG GC is a dominant immune evasion mechanism. During host infection, parasite numbers rise and fall periodically. We think that recombination-mediated VSG switching is generally suppressed but one or two silent VSG loci may have a higher chance to recombine with the active ES, potentially due to their proximity. VSG loci might be highly recombinogenic without this suppression, which could increase the possibility for parasites to use up their expressible VSGs at the early stage of infection, consequently resulting in eradication by host immunity. To prevent this, parasites may expose their ES-linked VSG repertoire gradually to preserve the population survival by VSG switching and to prolong their infection, while having some time to accumulate novel VSGs at transcribable loci.
VSG switching requires key factors in recombination and replication and their interplay controls the outcome of VSG switches. Although it is beginning to reveal its mystery, the control mechanism of antigenic switching must involve various cellular processes, whose contributions are largely unknown. Additional factors as well as specific alleles of the RTR complex members should be studied thoroughly to understand mechanism of antigenic variation in depth.
Materials and Methods
Trypanosome strains and plasmids
Trypanosoma brucei bloodstream forms (strain Lister 427 antigenic type MITat1.2 clone 221a (expressing VSG 427-2)) were cultured in HMI-9 at 37°C. The cell lines constructed for this study are listed in Table 2. They are of ‘single marker (SM)’ background that expresses T7 RNA polymerase and Tet repressor [63]. Stable clones were obtained and maintained in HMI-9 media containing necessary antibiotics at the following concentrations, unless otherwise stated: 2.5 µg/ml, G418 (Sigma); 5 µg/ml, blasticidin (Invivogen); 5 µg/ml, hygromycin (Sigma); 0.1 µg/ml, puromycin (Sigma); 1 µg/ml, phleomycin (Invivogen). Plasmids used for this study are listed in Table 3.
Table 2. Strains used in this study.
| Names (sources) | Genotypes | Manipulations |
| SM | WT | T7 RNA polymerase and Tet repressor (TetR):: NEO |
| HSTB-188 [39] | WT | ES1 promoter::BSD |
| HSTB-261 [39] | WT | ES1 promoter::BSD, 70-bp repeats-PUR-TK |
| HSTB-229 | rmi1-/+ | ES1 promoter::BSD, rmi1ΔloxP-HYG-TK-loxP/+ |
| HSTB-286 | rmi1-/- | ES1 promoter::BSD, rmi1ΔloxP-HYG-TK-loxP/ΔloxP-PUR-loxP |
| HSTB-298 | rmi1-/- | ES1 promoter::BSD, rmi1ΔloxP/ΔloxP |
| HSTB-336 | rmi1-/- | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rmi1ΔloxP/ΔloxP |
| HSTB-411, 412, 413 | rmi1-/- + Vector | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rmi1ΔloxP/ΔloxP + pHD309 (empty vector) |
| HSTB-414, 415, 416 | rmi1-/- + RMI1-wt | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rmi1ΔloxP/ΔloxP + pSY38 (RMI1-wt in pHD309) |
| HSTB-344 [39] | topo3α-/- | ES1 promoter::BSD, 70-bp repeats-PUR-TK, topo3αΔloxP/ΔloxP |
| HSTB-365 [39] | rad51-/- | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rad51ΔHYG/rad51ΔPHELO |
| HSTB-384, 385, 386 | rmi1-/- rad51-/- | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rmi1ΔloxP/ΔloxP, rad51ΔHYG/rad51ΔPHELO |
| HSTB-456 | rmi1-/- topo3α-/- | ES1 promoter::BSD, 70-bp repeats-PUR-TK, rmi1ΔloxP/ΔloxP, topo3αΔloxP/ΔloxP |
| HSTB-201 | 2040-3HA (*) | 2040-3HA::HYG |
| HSTB-210 | RMI1-3HA | RMI1-3HA::PUR |
| HSTB-213 | TOPO3α-3MYC | TOPO3α-3MYC::PHLEO |
| HSTB-218 | 2040-3HA( * ) TOPO3α-3MYC | 2040-3HA::HYG, TOPO3α-3MYC::PHLEO |
| HSTB-222 | RMI1-3HA TOPO3α-3MYC | RMI1-3HA::PUR, TOPO3α-3MYC::PHLEO |
(*) The full annotation is Tb927.8.2040.
Table 3. Plasmids used in this study.
| Names | Inserts, targeting loci, and markers | Sources |
| pHD309 | Vector to target at TUB array, HYG | |
| pLHTL-pyrFE | TbURA3::HYG-TK | [64] |
| pLEW100-Cre | Cre-recombinase in expression vector, PHLEO | [64] |
| pHJ17 | loxP-HYG-TK-loxP | [39] |
| pHJ18 | loxP-PUR-TK-loxP | [39] |
| pHJ23 | To target BSD downstream of ES promoter | [39] |
| pHJ63 | TOPO3α Δ::loxP-PUR-TK-loxP | [39] |
| pHJ64 | TOPO3α Δ::loxP-HYG-TK-loxP | [39] |
| pSY1 | RAD51Δ::HYG | [39] |
| pSY23 | RAD51Δ::PHLEO | [39] |
| pHJ71 | RMI1Δ::loxP-HYG-TK-loxP | This study |
| pHJ77 | RMI1Δ:: loxP-PUR-loxP | This study |
| pSY38 | To insert wild-type RMI1 at TUB array, HYG | This study |
| pMOTag53M | One-step PCR-3xMYC tagging construct, PHLEO | [42] |
| pMOTag4H | One-step PCR-3xHA tagging construct, HYG | [42] |
| pMOTag2H | One-step PCR-3xHA tagging construct, PUR | [42] |
Construction of Tbrmi1 knock-out lines
To generate T. brucei rmi1 mutants, HSTB-188, a wild-type ‘single marker’ line with the active ES marked with a blasticidin-resistance gene (BSD), was sequentially transfected with deletion cassettes. The cassettes contain either a hygromycin-resistance gene conjugated with a Herpes simplex virus thymidine kinase (HYG-TK) or a puromycin-resistance gene (PUR) flanked by homology sequences to upstream and downstream of Tb927.3.1830. Deletions were confirmed by PCR analyses. Single knock-out (sKO), rmi1-/+, and double KO, rmi1-/- were used to assess growth phenotypes, as described previously [39]. Selection markers flanked by loxP sites were removed by transiently expressing Cre-recombinase (pLew100-Cre). The cells that lost both HYG-TK and PUR were selected in 50 µg/ml ganciclovir (GCV). Loss of markers was confirmed by resistance to puromycin and hygromycin, and by PCR analysis. The sequences of primers used here are available upon request. These cell lines were used for recombination and VSG switching assays.
Co-immunoprecipitation and western blot
About 108 cells were lysed in lysis buffer (25 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM phenylmethylsulfonylfloride (PMSF), 1 mM dithiothreitol (DTT), protease inhibitor cocktails (Sigma)). Whole cell lysates were immunoprecipitated with rabbit anti-HA or anti-MYC antibodies. Immunoprecipitates were analyzed by western blot using mouse anti-HA or anti-MYC antibodies.
VSG switching assay and analyses of switchers
To create switching reporter strains, the promoter and 70-bp repeats of the active ES were marked with BSD and PUR-TK, respectively, as described previously [39]. To determine VSG switching frequency and mechanisms, cells were maintained in the presence of blasticidin and puromycin to exclude switchers from the starting population. Cells were then allowed to switch in the absence of drug selection for 3 days. Indicated numbers of cells (Table 1) were suspended in HMI-9 media containing 5–10 µg/ml GCV and distributed in 96-well plates. GCV-resistant clones were examined for sensitivity to 2 µg/ml puromycin, to exclude non-switchers that carry spontaneous mutation(s) in TK but not in PUR gene. Non-switchers that carry mutations both in PUR and TK were ruled out by western blot analysis using anti-VSG 427-2 antibodies. Cloned switchers were analyzed for blasticidin sensitivity. Genomic DNA was analyzed by PCR for the presence of BSD and VSG 427-2. The sequences of primers used here are available upon request.
It is noted that all VSG switching assays in this study were performed without the MACS-enrichment step. MACS step may have reduced the number of some switchers, as ‘ES GC or ES loss’ switchers appear to have growth disadvantages, compared to parental cells and VSG GC switchers.
Recombination assay
Gene-conversion frequency at TbURA3 locus was determined as described previously [39].
Acknowledgments
We thank Bibo Li and Nina Papavasiliou for comments on the manuscript, and members of Cross and Papavasiliou labs for useful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Rockefeller University Women in Science fellowship to HK and by grant no. R01AI021729 from the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dalgaard JZ, Vengrova S. Selective gene expression in multigene families from yeast to mammals. Sci STKE 2004. 2004:re17. doi: 10.1126/stke.2562004re17. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annual review of immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chrom Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- 4.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. In: Baker JR, Muller R, Rollinson D, editors. Advances in Parasitology. Vol. 49. London: Academic Press Ltd.; 2001. pp. 1–70. [DOI] [PubMed] [Google Scholar]
- 5.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium faiciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Kratz E, Dugas JC, Ngai J. Odorant receptor gene regulation: implications from genomic organization. Trends Genet. 2002;18:29–34. doi: 10.1016/s0168-9525(01)02579-3. [DOI] [PubMed] [Google Scholar]
- 8.Palenchar J, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, et al. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machado C, Augusto-Pinto L, McCulloch R, Teixeira S. DNA metabolism and genetic diversity in Trypanosomes. Mutat Res. 2006;612:40–57. doi: 10.1016/j.mrrev.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Cross GAM. Marr J, Koumuniecki R, Nilsen TW, editors. Antigenic variation in african trypanosomes and malaria. Mol Med Parasitol: Academic Press. 2002. pp. 89–110.
- 13.Pays E, Guyaux M, Aerts D, vanMeirvenne N, Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- 14.Aitcheson N, Talbot S, Shapiro J, Hughes K, Adkin C, et al. VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection. Mol Microbiol. 2005;57:1608–1622. doi: 10.1111/j.1365-2958.2005.04795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudenko G, McCulloch R, Dirksmulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- 16.Bernards A, van der Ploeg LHT, Gibson WC, Leegwater P, Eijgenraam F, et al. Rapid change of the repertoire of variant surface glycoprotein genes in trypanosomes by gene duplication and deletion. J Mol Biol. 1986;190:1–10. doi: 10.1016/0022-2836(86)90070-7. [DOI] [PubMed] [Google Scholar]
- 17.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, et al. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M. The Arabidopsis BLAP75/Rmi1 Homologue Plays Crucial Roles in Meiotic Double-Strand Break Repair. PLoS Genet. 2008;4:e1000309. doi: 10.1371/journal.pgen.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neff NF, Ellis NA, Ye TZ, Noonan J, Huang K, et al. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol Biol Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 27.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 30.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki M, Nakagawa T, Seki T, Kato G, Tada S, et al. Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol. 2006;26:6299–6307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shor E, Gangloff S, Wagner M, Weinstein J, Price G, et al. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mankouri HW, Hickson ID. Top3 processes recombination intermediates and modulates checkpoint activity after DNA damage. Mol Biol Cell. 2006;17:4473–4483. doi: 10.1091/mbc.E06-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 37.Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raynard S, Zhao W, Bussen W, Lu L, Ding YY, et al. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HS, Cross GA. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Yang Y, Singh TR, Busygina V, Guo R, et al. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure. 2010;18:1159–1170. doi: 10.1016/j.str.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoadley KA, Xu D, Xue Y, Satyshur KA, Wang W, et al. Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome. Structure. 2010;18:1149–1158. doi: 10.1016/j.str.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Seki M, Tada S, Enomoto T. Function of recQ family helicase in genome stability. Subcell Biochem. 2006;40:49–73. doi: 10.1007/978-1-4020-4896-8_5. [DOI] [PubMed] [Google Scholar]
- 44.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 45.Cross M, Taylor MC, Borst P. Frequent loss of the active site during variant surface glycoprotein expression site switching in vitro in Trypanosoma brucei. Mol Cell Biol. 1998;18:198–205. doi: 10.1128/mcb.18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, et al. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5:e160. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrero VA, Symington LS. Extensive DNA end processing by exo1 and sgs1 inhibits break-induced replication. PLoS Genet. 2010;6:e1001007. doi: 10.1371/journal.pgen.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, et al. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai Y, Symington L. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 55.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of rad51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conway C, Proudfoot C, Burton P, Barry JD, McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 57.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 59.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 60.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 61.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 62.Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 64.Scahill MD, Pastar I, Cross GA. CRE recombinase-based positive-negative selection systems for genetic manipulation in Trypanosoma brucei. Mol Biochem Parasitol. 2008;157:73–82. doi: 10.1016/j.molbiopara.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]