Abstract
The incidence of esophageal adenocarcinoma (EAC) is rising in the United States. An important risk factor for EAC is the presence of Barrett esophagus (BE). BE is the replacement of normal squamous esophageal epithelium with a specialized columnar epithelium in response to chronic acid and bile reflux. However, the emergence of BE from squamous keratinocytes has not yet been demonstrated. Our research has focused on this. Wnt and cyclooxygenase 2 (Cox2) are two pathways whose activation has been associated with BE and progression to EAC, but their role has not been tested experimentally. To explore their contribution, we engineered a human esophageal keratinocyte cell line to express either a dominant-active Wnt effector CatCLef or a Cox2 complementary DNA. In a two-dimensional culture environment, Cox2 expression increases cell proliferation and migration, but neither transgene induces known BE markers. In contrast, when these cells were placed into three-dimensional organotypic culture conditions, we observed more profound effects. CatCLef-expressing cells were more proliferative, developed a thicker epithelium, and upregulated Notch signaling and several BE markers including NHE2. Cox2 expression also increased cell proliferation and induced a thicker epithelium. More importantly, we observed cysts form within the epithelium, filled with intestinal mucins including Muc5B and Muc17. This suggests that Cox2 expression in a three-dimensional culture environment induces a lineage of mucin-secreting cells and supports an important causal role for Cox2 in BE pathogenesis. We conclude that in vitro modeling of BE pathogenesis can be improved by enhancing Wnt signaling and Cox2 activity and using three-dimensional organotypic culture conditions.
Introduction
Esophageal adenocarcinoma (EAC) is a leading cause of death in the United States [1]. For the past 30 years, EAC incidence has risen at an alarming rate, making it an increasingly important human cancer. Typically, EAC does not arise in normal squamous esophageal mucosa but usually is seen developing in an altered, premalignant mucosa known as Barrett esophagus (BE) [2,3]. BE is characterized by the replacement of normal, squamous epithelium with a metaplasia consisting of columnar mucosa with a morphology and gene expression pattern more typical of intestinal epithelium. Intestinal proteins like villin, sucrase isomaltase, acidic mucins/MUC2, and the intestine-specific transcription factor Cdx2 are commonly detected in BE [2]. Whereas Cdx2, as the master regulator of the intestinal cell phenotype [4,5], undoubtedly plays an important role in the pathogenesis of BE, its expression alone has not been sufficient to induce BE metaplasia in esophageal keratinocytes [6–9]. Other developmental and signaling pathways seem to be necessary to induce the emergence of an intestinalized epithelium from keratinocyte precursors.
The current paradigm for the induction of BE requires gastric acid and bile reflux to induce tissue injury, resulting in the release of proinflammatory cytokines and other inflammatory mediators including eicosanoids like prostaglandin E2 (PGE2) [10–14]. PGE2 is the most abundant prostanoid in BE. It stimulates angiogenesis, increases cell proliferation and migration, and inhibits apoptosis [15–18]. Cyclooxygenases (Cox1 and Cox2) are the rate-limiting enzymes in prostaglandin biosynthesis [19–21]. Cox2 expression is induced in the esophagus by acid reflux, and its expression levels directly correlate with the degree of acid reflux injury [14,22]. Moreover, Cox2 is a known oncogene [23]. It is therefore hypothesized that Cox2 expression may promote esophageal adenocarcinogenesis, as Cox2 levels are increased in BE, and increase further with the onset of dysplasia and cancer [24–26]. Despite the supportive clinical data, a role for Cox2 in the pathogenesis of BE and EAC has not been formally tested.
The Wingless (Wnt) pathway is another signaling mechanism thought to contribute to BE pathogenesis. In the intestine, Wnt signaling, mediated by β-catenin intracellularly, is required for intestinal secretory cell fates, cell proliferation, and stem cell maintenance [27–29]. Moreover, it has been shown that active Wnt signaling during development can induce intestinal gene expression and morphology in lung endoderm, including the development of intestinal mucin-secreting goblet cells [30]. Several clinical observations suggest that activation of the Wnt signaling may contribute to the development of BE. The progression of BE to adenocarcinoma was marked by loss of several negative regulators of Wnt signaling as well and induction of Wnt-2 expression [31–35]. These changes would be expected to increase signaling along the Wnt axis. However, despite these findings, little is known about how Wnt signaling may promote intestinalization and the onset of BE.
Using a novel human esophageal keratinocyte cell line immortalized only by ectopic telomerase expression (EPC-hTERT, known here as STR cells) [36,37], we previously determined that ectopic expression of Cdx2, when combined with cyclin D1 overexpression and chromatin remodeling drugs or with c-Myc expression, induces a more BE-like gene expression pattern [6–8]. In the present study, we advance these observations by exploring the effects of ectopic activation of Cox2 or Wnt signaling on the STR cell function and differentiation in two-dimensional and three-dimensional culture systems.
Materials and Methods
Cell Culture and Transections
Immortalized human primary esophageal epithelial cells STR (EPC-hTERT) were developed and maintained as previously described [8,36,37]. Stable transduction of esophageal cells with retroviral vectors has been described previously [6,8]. The polymerase chain reaction (PCR) primers sequences are as the following: forward, 5-GCGCCTCGAGGCTCCACAGCC AGACGCC-3′; reverse, 5-GCCGGAATTCCATTAGACTTCTACAGTTC-3′. The MSCV vector includes an internal ribosomal entry site (IRES) and a 3′ neomycin resistance complementary DNA (cDNA). The Cox2 cDNA was subcloned into the multicloning site 5′ to the IRES. The inserted region of the constructs was verified by DNA sequencing. They were transfected into a Phoenix-Ampho packaging cell line (a gift from Dr Garry Nolan, Stanford University, Palo Alto, CA) with Lipofectamine 2000 reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions.
Culture supernatants from individual Phoenix-Ampho cells were purified as described elsewhere [38] and were used to infect STR cells. Cells were passaged 48 hours after infection and selected with 300 µg/ml neomycin (Invitrogen) for 5 days. The CatCLef1 cDNA fragment (kindly provided by Dr Brigid LM Hogan, Duke University, Durham, NC) was cloned into the lentiviral expression vector pLEX-MCS (Open Biosystems Products, Huntsville, AL) and lentivirus particles assembled in HEK293T. The PCR primer sequences are as follows: forward, 5′-CGTTCTTTTCACTCTGGTGGA-3′; reverse, 5′-GTTAACCAAAGATGACTTGATG-3′. Lentiviral particles were then used for polybrene-mediated transduction using standard protocols. Cells were selected with 1 µg/ml puromycin (Invitrogen) for 7 days. The MIGR1 and MIGR-Cdx2 retroviral vectors were described previously [39–41]. Prostaglandin E2 was purchased from a commercial supplier (14010; Cayman Chemical, Ann Arbor, MI).
Cell Proliferation and Migration Assays
Cell proliferation was quantified by colorimetry based on the metabolic cleavage of the tetrazolium salt WST-1 in viable cells as recommended by the manufacturer (Roche Applied Science, Mannheim, Germany). As a second approach, bromodeoxyuridine (BrdU) incorporation was measured in Cox2- and CatCLef1-expressing and control cells. Cells were incubated with BrdU (Zymed, Invitrogen Corp, Carlsbad, CA) for 1 hour before fixation. BrdU staining was conducted via standard methods. 4′, 6-Diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St Louis, MO) was used at a concentration of 1 µg/ml to highlight all cells. Fluorescent samples were visualized and imaged using software (IPLab; Scanalytics, Fairfax, VA). Cells stained for BrdU were scored by counting five high-power fields.
Migration assays were carried out as described [40]. In brief, after culturing cells without epidermal growth factor (EGF) for 48 hours, single-cell suspensions containing 4 x 104 cells/well in 0.5 ml of K-SFM (without EGF) were plated in triplicate into 24-well inserts (8 µm pore size; Falcon Cell Culture Inserts; BD Bioscience, Bedford, MA). The lower chamber was filled with 0.5 ml of K-SFM containing 0.5 µg/ml hydrocortisone (Sigma) and 10 ng/ml EGF (BD Biosciences, Rockville, MD). After incubating for 16 hours at 37°C, the cells on the upper side of the Transwell membrane (BD Bioscience) were removed by cotton swab and rinsed with phosphate-buffered saline. Cells migrating to the lower side of membrane were fixed in 4% paraformaldehyde for 20 minutes at room temperature, stained with crystal violet (Sigma), and counted. Means and SDs were calculated and compared using Student's t test.
PGE2 ELISA
Supernatant culture media were collected from overnight cultures of 1 x 104 cells in 96-well plates. Fifty microliters of culture supernatant was used per sample. All samples were run in duplicate. PGE2 levels in supernatants from STR.M.COX2 and control cells were quantified with enzyme immunoassay (EIA) system PGE2 assay kit (R&D Systems, Inc, Minneapolis, MN) following the manufacturer's protocol.
TOPflash Luciferase Assay
The EPC2-hTERT cells were transfected using Lipofectamine LTX reagent (Invitrogen). Cells were grown to 50% to 70% confluence and were transfected with 100 ng of pRC expression vectors, 100 ng of TOPflash (kindly provided by Dr Ken Kinzler, The Johns Hopkins University), 200 ng of CatCLef1, 1 ng of pRL-CMV Renilla control reporter (Promega, Madison, WI), and 700 µg of pCR2.1 (Stratagene, La Jolla, CA) as carrier. Expression plasmids for pCGCatCLef1 and Plex-MCS-CatCLef1 were described previously [30]. Media were changed at 24 hours, and at 48 hours, the cells were harvested. Luciferase assays were performed using the dual-luciferase assay kit (Promega). Luciferase activity was normalized to both total protein concentration and the transfection control Renilla luciferase levels. Statistical testing was by Student's t test.
RNA Isolation and Real-time Quantitative PCR Analysis
Total RNA was isolated from Neo- or Puro-selected and control cells using the RNAeasy kit (Qiagen, Germantown, MD). Five micrograms of total RNA was used for cDNA synthesis using the First-Strand cDNA synthesis Kit (Invitrogen). Reverse transcription (RT)-negative controls were included. Primer sequences for PCR are available in Table W1. For the RT-PCR, cDNA and primers were mixed with SYBR Green RT-PCR Master Mix (Applied Biosystems, Life Technologies Corp, Carlsbad, CA) and then assayed in an ABI Prism 7000 sequence detection system as directed by the manufacturer. A ribosomal phosphoprotein, 36B4, was used as the normalization control. Fold change in RNA levels was calculated from the Ct values using the formula previously described [39,42]. The ΔΔCt values for each gene were averaged across the RNA pools, SDs were calculated, and statistical comparisons were performed using analysis of variance and Tukey rank mean testing. These values were then converted to fold change to graphically report the findings.
Quantitative RT-PCR-Based TaqMan Low-Density Arrays
TaqMan Array 96-well fast plate (custom format 48; Applied Biosystems) included 45 specific genes and 3 housekeeping genes as internal controls. Total cellular RNA (2.5 µg) was reverse-transcribed using SuperScript VILO cDNA Synthesis Kit (Invitrogen). cDNA, 1.25 ng, was obtained for real-time PCR using ABI StepOnePlus Real-Time PCR System (Applied Biosystems). The comparative Ct method was applied for quantification. The amount of messenger RNA (mRNA) of a specific gene was measured by its threshold cycle (Ct) and normalized to that of 36B4, GAPDH, and 18S as n-fold difference (Ct), respectively. The expression of genes of interest in each sample was then compared with the amount of a calibrator sample—that is, the expression in the corresponding empty vector control (Ct). Customized 96-well low-density arrays for PCR amplification were designed using individual primers for genes of interest, chosen and purchased from the assays on demand gene expression products (Applied Biosystems) listed in Table W1. The probes were labeled with 5′-FAM and 3′-minor groove binder/nonfluorescent quencher. To control for the specificity of the assay, genomic DNA and total cellular RNA extracted from biopsies were also tested. Master Mix (20 µl) containing cDNA was loaded into each well. Each low-density array containing the genes of interest was loaded with cDNA from STR-MSCV-Cox2, STR-Plex-MCS-CatCLef1, and corresponding control sample for quantification of mRNA expression of all the selected genes in one experiment by the same array.
Organotypic Three-dimensional Culture
Organotypic culture was performed as previously described [6]. In brief, 0.5 x 106 of STR.M.COX2, STR.L.CatCLef, or their corresponding controls were seeded on top of the collagen/Matrigel matrices containing FEF3 human fetal esophageal fibroblasts and grown in submerged conditions for 4 days. Cultures were then raised to the air-liquid interface for an additional 4 days and harvested. The cultures were split, with half fixed and embedded for histochemical studies and the remainder processed for RNA extraction. Each organotypic culture experiment was performed in triplicate.
Protein Extraction and Western Blot Analysis
Whole cell extract was prepared as described [43]. Protein concentration of samples and bovine serum albumin standard was determined using the bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL). The blotted membranes were blocked with phosphate-buffered saline with 0.2% Tween containing 5% skim milk, followed by overnight incubation with the primary antisera or monoclonal antibodies including mouse anti-COX-2 (SC-1745, 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-LEF1 antibody (1:1000; Millipore Chemicon, Billerica, MA), anti-cyclin D1 (p16; Santa Cruz Biotechnology), mouse antitubulin (1:2000; Sigma), and antiactin (1:2000; Sigma) at 4°C. The secondary antibodies used were all from Sigma-Aldrich and used at 1:3000. Targeted proteins were visualized using an enhanced chemiluminescence detection system (ECL Plus; GE Healthcare Bio-Sciences Corp, Piscataway, NJ) and exposed to Blue Lite Autorad film (ISC-BioExpress, Kaysville, UT).
Alcian Blue Staining and Immunostaining
Harvested organotypic cultures were fixed in 1.5% paraformaldehyde and embedded in paraffin. For histology and immunohistochemistry, 5- to 11-µm-thick sections were cut from the paraffin blocks and stained with hematoxylin and eosin or Alcian blue by standard procedures. Immunohistochemical staining was performed with standard techniques. Primary antibodies used include BrdU antibody (1:2500; Upstate, Charlottesville, VA), chicken anti-NHE2 (1:500; Chemicon), rabbit anti-Muc5B (1:50; Sigma-Aldrich), rabbit anti-Muc17 (1:20; Sigma-Aldrich), rabbit anti-Filaggrin (1:1500, no. 24584; Abcam, Cambridge, MA), rat anti-Notch1 (1:10; gift from Dr Spyros Artavanis-Tsakonas, Harvard Medical School, Boston, MA), and Ki67 (1:3000, VP-RM04; Vector Laboratories, Burlingame, CA). Sections were incubated with primary and biotinylated secondary antibodies and an avidin-horseradish peroxidase conjugate (Vectastain Elite ABC Kit; Vector Laboratories) following the manufacturer's protocol. The signal was developed using the 3,3-diaminobenzidine substrate kit (Vector Laboratories). Sections were counterstained with hematoxylin. Images were obtained at 40x using a Leica microscope (Leica Microsystems, Inc, Buffalo Grove, IL) with the Retiga 2000R digital camera (QImaging, Surrey, British Columbia, United Kingdom) and the IP Lab imaging software application (BD Biosciences). Exposure times were kept constant for all samples. Muc5B antibody and Cy3-labeled antirabbit secondary antibody were used for immunofluorescence. For Alcian blue staining, slides were deparaffinized. After application of 3% aqueous acetic acid to the slides, 1% Alcian blue in 3% acetic acid, pH 2.5, was applied. Sections were washed and counterstained with 0.1% nuclear fast red, dehydrated, and mounted.
Results
Expression of a Protein Chimera Containing LEF1 and the β-Catenin C-terminus Activates Wnt Signaling in Human Keratinocytes
To provoke a direct, unopposed Wnt signal in our human keratinocyte cell line, we obtained a dominant active chimera in which an amino-terminally truncated β-catenin is fused to a full-length LEF1 (Figure 1A). The chimera is called CatCLef1 (kindly provided by Dr Brigid Hogan, Duke University) [44]. This chimera induces intestinalization when ectopically expressed in lung endoderm during embryogenesis [30]. We subcloned this chimera into a lentiviral vector with a puromycin-selectable marker, generated infectious lentiviral particles for the CatCLef1 and control vectors, and infected STR cells with the viral supernatant. As a control for these studies, STR cells also were infected with the original lentivirus that lacked the CatCLef cDNA, we will refer to as the empty-cassette control. STR cells have a normal cell morphology, wild-type p53, and form morphologically normal-appearing squamous epithelium when cultured using three-dimensional techniques [36,37].
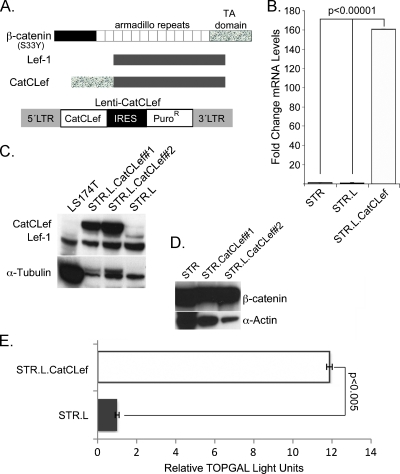
Figure 1.
Development of a human esophageal keratinocyte cell line with enhanced Wnt/β-catenin signaling. (A) Map of β-catenin, Lef1, and the CatCLef chimeric construct, as well as the lentiviral vector into which CatCLef is subcloned. (B) Fold change in CatCLef mRNA levels after Lenti-CatCLef infection and selection (STR.L.CatCLef), compared with control lentivirus-infected cells (STR.L), n = 3. (C) Western blot analysis for CatCLef protein levels in control (STR.L) and CatCLef-expressing cell lines (STR.L.CatCLef#1 and #2). (D) Western blot analysis for endogenous β-catenin protein levels after CatCLef expression. (E) Wnt/β-catenin transcriptional activity on a canonical Wnt reporter construct TOPflash in CatCLef-expressing (STR.L.CatCLef) and control cells (STR.L), n = 3.
We generated two distinct cell lines (STR.L.CatCLef#1 and STR. L.CatCLef#2) and a control cell line (STR.L) and can demonstrate a significant increase in CatCLef mRNA and protein levels by quantitative PCR and Western blot analysis in both when compared with STR.L control cells (Figure 1, B and C). CatCLef expression did not seem to alter β-catenin protein levels (Figure 1D) but did increase transcriptional activity from a canonical Wnt reporter TOPflash by 12-fold (Figure 1E). We have therefore established human esophageal keratinocyte cell lines with ectopic β-catenin/T-cell factor transcriptional activity.
Development of Human Esophageal Keratinocytes with Ectopic Cox2 Expression
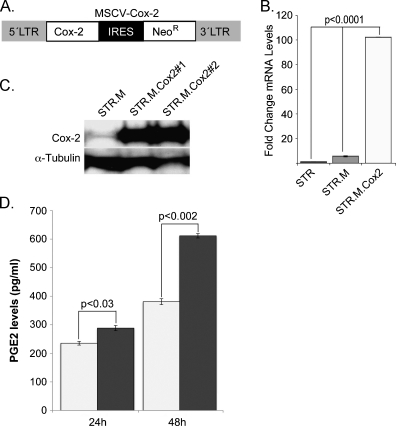
We subcloned a Cox2 cDNA into an MSCV-based retroviral vector to drive expression of Cox2 in human esophageal keratinocytes. This vector has a neomycin resistance cDNA downstream from an IRES as the selectable marker (Figure 2A). Thus, cells resistant to neomycin selection should also express Cox2. We generated active retroviral particles using this vector and subsequently infected the human esophageal keratinocyte STR cells as before. After selecting for neomycin resistance, we demonstrated Cox2 expression and normal enzyme function in the STR-Cox2 cells. Cox2 mRNA was increased 100-fold over controls that received no virus or the control MSCV-neo virus with an empty cassette upstream of the IRES (Figure 2B). As before, we isolated two distinct Cox2-expressing cell lines and a single control cell line after separate infections and puromycin selections and can demonstrate abundant Cox2 protein by Western blot analysis in the STR.M.Cox2 cells but not the control STR.M cells (Figure 2C). Moreover, Cox2 expression was associated with significantly increased production of prostaglandin PGE2 detected in the cell culture media of STR-Cox2 cells compared with control infected cells (Figure 2D). PGE2 levels are increased by 20% at 24 hours after plating cells and by 60% at 48 hours. These findings confirm ectopic Cox2 expression in STR keratinocytes is associated with increased eicosanoid PGE2 biosynthesis.
Figure 2.
Establishing active Cox2 expression in human esophageal keratinocytes. Human STR keratinocytes were infected with a retroviral vector to induce Cox2 expression (MSCV-Cox2) or the control empty viral vector MSCV-Neo. Neomycin resistance serves as a marker for viral infection and gene expression. (A) Map of MSCV-Cox2 retroviral construct. (B) Fold change in Cox2 mRNA levels after MSCV-Cox2 infection and selection (STR.M.Cox2), compared with control MSCV virus-infected cells (STR.M), n = 3. (C) Western blot analysis for Cox2 protein levels in control (STR.M) and Cox2-expressing cell lines (STR.M.Cox2#1 and #1). (D) PGE2 levels in the cell culture media of control STR.M cells (white bar) and Cox2-expressing cells (gray bar) at 24 and 48 hours, n = 4.
Enhanced Wnt Signaling or Cox2 Activity Increased Cell Proliferation and Migration in Cox2 Expressing Cells Only
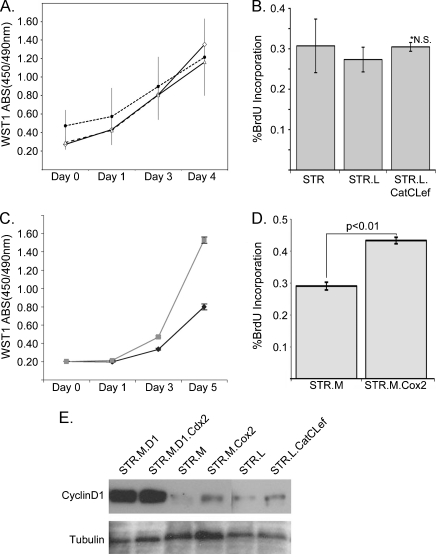
Despite the significant induction of effective “Wnt” signaling by the chimeric CatCLef, as evidenced by the increased luciferase activity from the TOPflash reporter, there was no effect on keratinocyte proliferation, as determined either by WST-1 assays (Roche Applied Science), or BrdU incorporation (Figure 3, A and B). In contrast, Cox2 expression was associated with a robust increase in cell proliferation (Figure 3, C and D). In a previous study, we demonstrated that the expression of the intestine-specific transcription factor Cdx2 was maintained in human STR cells only if cell proliferation was enhanced by ectopic cyclin D1 expression [8]. Whereas both CatCLef and Cox2 expression did modestly increase cyclin D1 protein levels, neither reached the levels achieved in the STR.D1 cells (Figure 3E) and they were not enough to sustain Cdx2 expression in STR keratinocytes (data not shown).
Figure 3.
Increased cell proliferation was observed with Cox2 expression but not CatCLef. (A) WST-1 cell accumulation studies (Roche) performed on STR (solid line) parental cells as well as STR.L (short dash line) and STR.L.CatCLef cells (long dash line), n = 8, each time point. (B) BrdU incorporation assay performed on performed on STR, STR.L, and STR.L.CatCLef cells, n = 3. (C) WST-1 cell accumulation study of STR.M (solid black line) and STR.M.Cox2 cells (solid gray line), n = 8. (D) BrdU incorporation assay for STR.M and STR.M.Cox2 cells, n = 3. (E) Western blot analysis for cyclin D1 protein levels in STR cells overexpressing cyclin D1, cyclin D1 and Cdx2, CatCLef, Cox2, and control cells. Tubulin served as loading control.
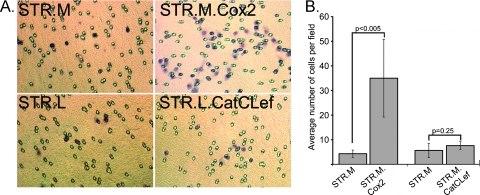
Wnt signaling and Cox2 activity have both previously been associated with increased cell migration [21,45]. We therefore tested the cell migratory activity of our STR.L.CatCLef and STR.M.Cox2 cells using Boyden chambers. Cells were plated in the upper chamber, and growth media were placed in the lower chamber. The number of cells that passed through the 8.0-µm pores was determined at 24 hours. Whereas CatCLef-expressing cells had no significant change in cell migration activity when compared with controls, the Cox2-expressing cells had a seven-fold increase in cell migration (Figure 4). In summary, Cox2 expression, but not CatCLef, was associated with significantly enhanced cell proliferation and migration in human STR keratinocytes.
Figure 4.
Cox2 expression but not CatCLef increases STR human keratinocyte cell migration in Boyden chamber assays. Cell migration was evaluated in Boyden chambers for STR.L.CatCLef and STR.M.Cox2 cells compared with control STR.L and STR.M, respectively. (A) Example view of filter from each evaluated cell line. (B) Quantitation of cell counts from filters; three fields/filter were evaluated. Experiment was performed in triplicate, n = 9.
CatCLef and Cox2 Expressing Cells Cultured under Three-dimensional Organotypic Conditions Demonstrate Altered Cell Proliferation and Morphology
We examined the STR.L.CatCLef and STR.M.Cox2 cells for changes in cell morphology but observed none (data not shown). We next examined for changes in mRNA levels for a small panel of genes associated with intestinal epithelium and BE, including Alkaline Phosphatase, NHE2, Muc2, Villin, and Keratin20, but there were no significant differences observed between expressing and control cells (data not shown). However, in a previous study, we noted that STR cells expressing Barrett associated genes Cdx1 and cMyc demonstrated significant intestinalization, including the production of Muc5AC, only when cultured under three-dimensional organotypic conditions that provokes STR cell differentiation [6]. We therefore cultured our STR.L.CatCLef and STR.M.Cox2 cells similarly to determine whether CatCLef or Cox2 expression could alter STR cell differentiation.
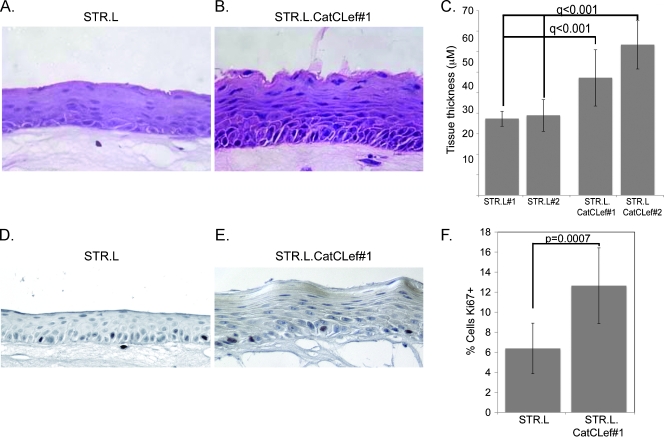
One finding that was immediately apparent was that the stratified epithelium that developed from the STR.L.CatCLef cells was noticeably thicker than the control STR.L cells (Figure 5, A and B), nearly double the thickness, on average (Figure 5C). Moreover, the basal cell layer contained more cells and appeared crowded and more disorganized than is observed in the control STR.L cells. Staining for Ki67+ cells demonstrated a two-fold increase in cell proliferation in the STR.L.CatCLef organotypic epithelium when compared with controls.
Figure 5.
CatCLef increases epithelial thickness and cell proliferation when cells cultured under organotypic conditions. Hematoxylin and eosin stain of epithelial tissues sections from STR.L control (A) and STR.L.CatCLef cells (B) after cultured under organotypic conditions. (C) Quantification of epithelial thickness, measured in three regions from three different tissue sections per culture, n = 9. (D and E) Ki67 staining in tissue sections from STR.L (D) and STR.L.CatCLef cells (E). (F) Quantitation of Ki67-positive staining. The number of Ki67+ cells was expressed as a percentage of the total number of basal keratinocytes in a counted field, n = 6.
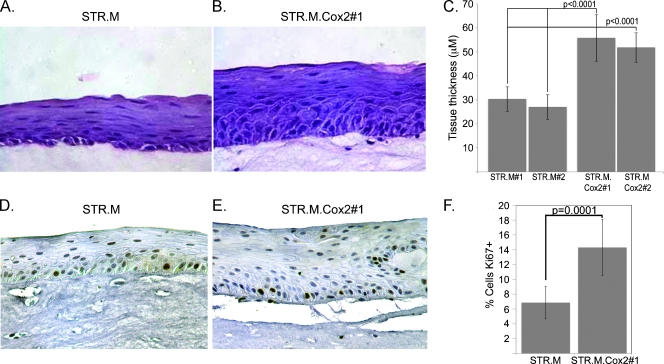
Likewise, the STR.M.Cox2 cells also yielded a thicker epithelial layer and increased Ki67+ cells compared with control STR.M cells when both are cultured under organotypic conditions (Figure 6). These differences were statistically significant and were in excess of two-fold greater than controls. Moreover, they exceeded levels seen in the STR. L.CatCLef cells, although this difference was not statistically significant. Together, these findings support the conclusion that excess WNT signaling in the form of CatCLef expression or Cox2 activity enhances human keratinocyte STR cell proliferation and tissue thickness when the cells are cultured under organotypic conditions.
Figure 6.
Cox2 expression increases epithelial thickness and cell proliferation of STR cells grown under organotypic conditions as in Figure 5. Hematoxylin and eosin stain of epithelial tissues sections from STR.M control (A) and STR.M.Cox2 cells (B) cultured under organotypic conditions. (C) Quantification of epithelial thickness measured as before, n = 9. (D and E) Ki67 staining in tissue sections from STR.M (D) and STR.M.Cox2 cells (E). (F) Quantitation of Ki67-positive staining determined as before, n = 6.
The STR Keratinocyte Differentiation Program Is Altered by CatCLef or Cox2 Expression
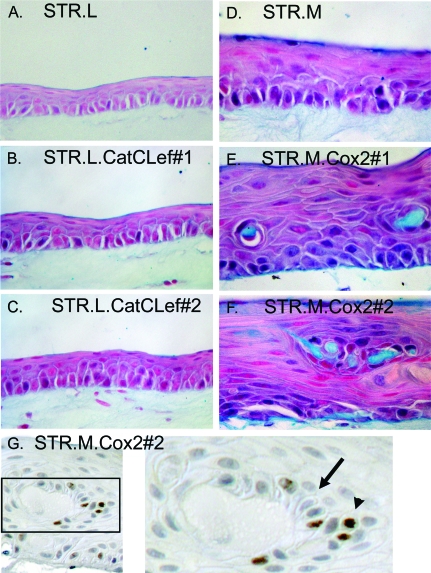
We next evaluated the organotypic cultures for evidence of altered cell differentiation and the promotion of a more intestinalized epithelium morphologically or by gene expression patterns. Alcian blue is frequently used to identify the production of intestinal mucins and goblet cells in Barrett epithelial biopsies. Staining of the CatCLef or control STR.L cultures did not find a significant production of intestinal mucins (Figure 7, A, B, or C). However, Cox2 expression did give rise to cells producing and secreting intestinal mucins, whereas the control culture did not (Figure 7, D, E, and F). Small pools of Alcian blue-positive, acellular material are observed in cultures derived from both STR.M.Cox2 cell lines. These Alcian blue-positive pools resemble mucinous cysts, with a border of cells surrounding them. There are no visible goblet-like cells in any of the sections studied. However, a closer examination of sections stained for other markers revealed a well-defined cyst with polarized, columnar cells in one section stained for Ki67 (Figure 7G). Of note, associated with this cystic structure, there are proliferating Ki67+ cells. These proliferating cells, which seem to be contributing cells to the cyst, are out of their usual position, which should be along the basal layer of the epithelium. Together, these findings suggest that Cox2 expression in human esophageal keratinocytes can elicit morphologic alterations and intestinal mucin production in a subset of cells.
Figure 7.
Cox2 expression induces intestinal mucin secretion in a subset of STR cells grown under organotypic conditions. Sections from the organotypic cultures of established cell lines expressing CatCLef, Cox2, or their controls were stained with Alcian blue to identify intestinal mucins. (A) STR.L control cells in culture. (B) STR.L.CatCLef cell line #1. (C) STR.L.CatCLef cell line #2. (D) STR.M control cells in culture. (E) STR.M.Cox2 cell line #1. (F) STR.M.Cox2 cell line #2. (G) Cystic structure noted in section from STR.M.Cox2 cells stained for Ki67. Note the cells with a polarized morphology along the cyst border (black arrow) and the proliferating Ki67+ cells (brown nuclei, black arrowhead) that have migrated above the basal cell layer.
To explore this response further, we screened mRNA expression levels for a custom panel of 45 genes using a quantitative PCR array (Applied Biosystems), planning to carefully validate any gene found different in the screening study. The genes were selected as markers associated with keratinocyte differentiation, BE, or intestinal epithelial cells and included a number of keratins as well as mucins. Total RNA isolated from the epithelium of the organotypic cultures of STR.L.CatCLef and STR.M.Cox2 cells, and their control STR.L and STR.M cells, were subjected to this quantitative PCR array screen. We found from this screen that 19 genes in the STR.L.CatCLef were different by two-fold or more when compared with the control STR.L cells similarly cultured (Table W2). Many of the differences were with keratins, which were largely induced. Some of these keratins are associated with multilayered squamous epithelium (K4, K13, K15) and others expressed more typically with simple epithelia (K10 and K19) [46]. Comparing STR.M and STR.M.Cox2 cells identified 21 genes that were two-fold or greater different between the cell lines (Table W3). Four of the eight genes induced were mucins, supporting the Alcian blue staining results, and two were keratins 7 and 19, associated with simple epithelium. Moreover, we observed reductions in mRNA levels of stratified squamous epithelial factors including keratins 1, 4, 5, 13, and 14, Filaggrin, and Loricrin. Together, these findings suggest that the ectopic expression of CatCLef or Cox2, in conjunction with culture under differentiation-producing organotypic conditions, can induce significant alterations in the normal keratinocyte differentiation patterns.
CatCLef Expression Increases Notch Signaling and Alters Keratinocyte Differentiation
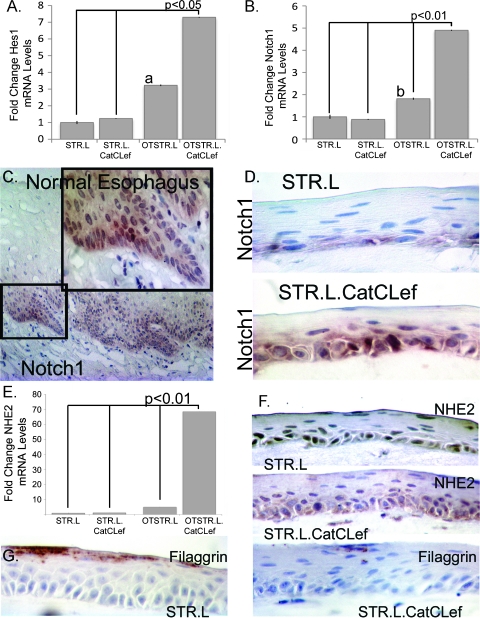
We next performed validation studies on selected genes from the screening studies. We noticed the induction of two Notch pathway factors, Notch1 and Hes1, in our screening PCR array for STR.L.CatCLef cells. Given the association of Notch signaling with esophageal squamous cell differentiation [47] and the requirement of Notch and β-catenin for cell proliferation in the intestine [48], we selected these as two mRNA products for additional validation. As expected, when control cells are cultured under organotypic conditions, levels of these two mRNA species are increased two- to three-fold, whereas in cells expressing CatCLef, a significant five- to seven-fold increase was observed (Figure 8, A and B). Notch1 protein expression in normal human esophagus localizes to the lower layers of the epithelium but is not exclusive to the basal cell layer (Figure 8C). In our organotypic cultures, we see this pattern recapitulated; however, CatCLef tissues express a considerably greater level of Notch1 protein, supporting the quantitative PCR findings (Figure 8D).
Figure 8.
CatCLef expression enhances Notch signaling and alters STR cell differentiation. (A and B) Quantitative SYBR Green RT-PCR analysis of Hes1 (A) and Notch1 (B) mRNA expression in STR.L or STR.L.CatCLef cells cultured under normal two-dimensional conditions or under three-dimensional organotypic conditions (OTSTR.L and OTSTR.L.CatCLef). ΔCt values were calculated after duplicate PCRs for each sample and statistical analysis was performed (analysis of variance and Tukey rank mean). ΔΔCt values were then calculated and used to determine fold change in expression. n = 4. aSignificantly differs from STR.L cells, P < .05. bSignificantly differs from STR.L.CatCLef cells, P < .05. (C) Immunohistochemistry for Notch1 in normal human esophagus. (D) Notch1 immunohistochemistry in organotypic cultures of STR.L and STR.L.CatCLef cells. (E) Quantitative SYBR Green RT-PCR for NHE2 mRNA levels in STR.L or STR.L.CatCLef cells cultured under normal two-dimensional conditions or under three-dimensional organotypic conditions, n = 4. (F) NHE2 immunohistochemistry in organotypic cultures of STR.L and STR.L.CatCLef cells. (G) Filaggrin staining in organotypic cultures of STR.L and STR.L.CatCLef cells.
Similarly, we noted an increased expression of NHE2 mRNA, a marker we previously identified as increased in BE [8], and reductions in Filaggrin mRNA levels, a keratin-binding protein expressed in terminally differentiated keratinocytes [49] in CatCLef-expressing cells, but only under organotypic culture conditions. NHE2 mRNA levels are increased 70-fold, and cytoplasmic staining for NHE2 localizes to the basal and suprabasal layers and compares well with staining patterns in normal human colon (Figure 8, E and F; data not shown). In contrast, there was a significant reduction in Filaggrin protein levels (Figure 8G), suggesting CatCLef expression interfered with keratinocyte terminal differentiation. In summary, ectopic Wnt signaling mediated by CatCLef enhanced Notch signaling and induced a more intestinalized gene expression pattern in human esophageal keratinocytes cultured under organotypic conditions.
Cox2 Expression Induces the Expression of Intestinal Mucins in Human Esophageal Keratinocytes
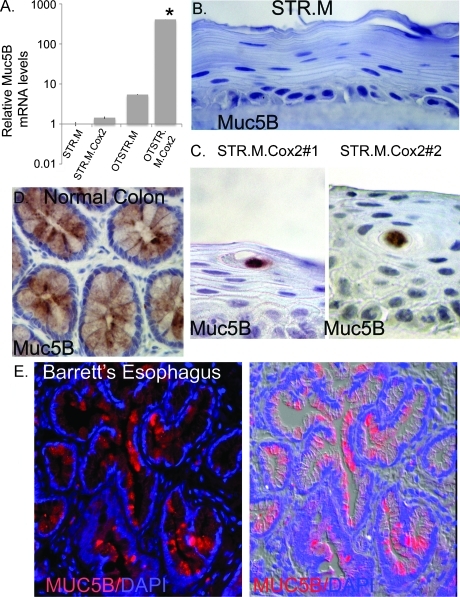
Given the findings of mucin-filled cystic structures in the organotypic STR.M.Cox2 cultures, we next verified the enhanced expression of some of the mucins observed in our PCR array screen. Muc1, although strongly induced, is normally expressed in keratinocytes and was therefore less interesting. The three remaining mucins are all normally expressed in intestinal epithelium and were therefore of interest [50]. Muc5B is a secreted gel-forming mucin previously reported in esophageal submucosal gland cells but not BE [51]. We determined that its expression was significantly induced 400-fold over control cells cultured under two-dimensional conditions and 75-fold over organotypic-cultured control STR.M cells (Figure 9A). This could be confirmed immunohistochemically, with the control STR.M cells not staining for Muc5B but the mucin-filled cystic structures strongly positive for Muc5B in the organotypic STR.M.Cox2 cultures (Figure 9, B and C). Staining with this antibody is highly specific for colonic goblet cells (Figure 9D). Moreover, using this antibody, we were able to detect Muc5B histologically in human Barrett specimens (Figure 9E).
Figure 9.
Intestinal and subepithelial gland mucin Muc5B induced by Cox2 expression in STR cells. (A) Quantitative SYBR Green RT-PCR analysis of Muc5B mRNA expression in STR.M or STR.M.Cox2 cells cultured under normal two-dimensional conditions or under three-dimensional organotypic conditions (OTSTR.M and OTSTR.M.Cox2). ΔCt values and statistical analysis were calculated as before after duplicate PCRs for each sample, n = 4. *Significantly differs from all other cells, P < .01. NS indicates nonsignificant difference with STR.M or STR.M.Cox2. (B) Negative immunohistochemical staining for Muc5B in STR.M control cells in organotypic cultures. (C) Muc5B immunohistochemical staining localized to the mucin-containing cysts in organotypic cultures of both STR.M.Cox2 cell lines (#1 and #2). (D) Pattern for Muc5B expression in normal human colon by immunohistochemical staining. (E) Epifluorescent staining for Muc5B (red) in human Barrett epithelium biopsy section. Nuclei counterstained with DAPI (blue). Shown on the right is a merged image of the fluorescent channels with a differential interference contrast image of the tissue.
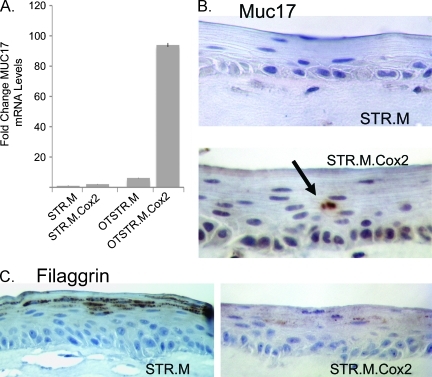
We further extended this work by examining the expression of Muc17 and Filaggrin in the organotypically cultured STR.M.Cox2 cells. Muc17 is a membrane-bound mucin thought to be expressed exclusively in the intestine [50]. We found levels of Muc17 mRNA were increased 90-fold over control cultured cells, including 18-fold over organotypically cultured STR.M control cells (Figure 10A). This was supported by immunohistochemical studies demonstrating Muc17 expression specifically in the mucin-cystic structures of the STR.M. Cox2 cells but not control STR.M cells (Figure 10B). Lastly, Cox2 expression reduced keratinocyte differentiation, as evidenced by the significant reduction in Filaggrin protein levels (Figure 10C). In summary, Cox2 expression in human keratinocytes promoted the expression and secretion of intestinal mucins when the cells were cultured under organotypic conditions that promote cell differentiation.
Figure 10.
Altered keratinocyte differentiation and induction of Muc17 in organotypically cultured Cox2-expressing keratinocytes. (A) Quantitative SYBR Green RT-PCR analysis of Muc17 mRNA expression in STR.M or STR.M.Cox2 cells cultured under normal two-dimensional conditions or under three-dimensional organotypic conditions (OTSTR.M and OTSTR.M.Cox2). ΔCt values and statistical analysis were calculated as before after duplicate PCRs for each sample, n = 4. *Significantly differs from all other cells, P < .01. (B) Negative immunohistochemical staining was observed for Muc17 in STR.M control cells but Muc17 staining localized to the mucin-containing cysts in organotypic cultures of STR.M.Cox2 cells. (C) Filaggrin protein expression by immunohistochemistry in organotypic cultures of STR.M and STR.M.Cox2 cells.
Discussion
BE is an important clinical condition that increases one's risk for developing EAC. One critical limitation in our understanding of the pathogenesis for BE has been the dearth of research models. It has hampered our ability to identify the BE progenitor cell, test hypotheses regarding causation, evaluate preventative and therapeutic strategies, and determine which genes and signaling pathways are critical for BE development. This study was initiated to extend earlier work by us seeking to model BE pathogenesis in an in vitro cell culture system [6,8]. In these studies, we observed that ectopic Cdx1 or Cdx2 expression could promote a more intestinalized pattern of gene expression in human esophageal STR keratinocyte. However, in both studies, it was determined that ectopic expression of these factors alone would not be enough to induce Barrett epithelial cells from the human keratinocytes; other agents or gene products would be needed. In the present study, we identified two likely cofactors in the pathogenesis of BE: enhanced Wnt signaling and Cox2 enzymatic activity.
Increased Wnt Activity Significantly Alters Human Keratinocyte Proliferation and Differentiation When Cultured under Organotypic Conditions
The importance of Wnt signaling for proliferation and differentiation of the normal intestinal epithelium cannot be overstated [52]. This argues for a similarly important role for Wnt in the pathogenesis of BE. Although there are some data in support of this hypothesis [31–35], more work needs to be done to clarify the role for Wnt in this disease process. Our findings here do support this hypothesis because enhanced Wnt signaling (mediated by the chimeric protein CatCLef) did increase the expression of keratins associated with simple epithelium, as well as NHE2, an exchange protein highly expressed in intestinal epithelium and BE.
In addition, we unexpectedly found that Notch signaling was also increased with Wnt signaling. Notch and Wnt/β-catenin are well-known collaborators in the intestine, where, together, they support intestinal stem cell maintenance and normal epithelial cell proliferation as well as drive cell proliferation in colon neoplasias [48]. It is interesting to speculate that the observed increase in cell proliferation noted in the three-dimensional culture system is due to an interaction between Wnt and Notch that does not occur in the two-dimensional system because the cells are maintained in sparse, noncontacting conditions to prevent differentiation. Moreover, the observed diminished keratinocyte differentiation associated with increased Notch activity (failed expression of Filaggrin) runs contrary to a recently published study demonstrating a role for Notch in keratinocyte differentiation [47]. It is not clear whether this failure is due to the Notch signaling being too high or if it is an effect of the Wnt pathway in conjunction with Notch. The elevated Notch levels also may be the reason we failed to see induction of intestinal secretory gene products because, in the intestine, Notch is known to prevent goblet cell development [48]. These remain open questions and important areas for future investigation.
Cox2 Expression Can Induce a Cell Morphology and Gene Expression Shift in Human Esophageal Keratinocytes
The fact that Cox2 expression yielded highly significant findings in this study are in some respects not surprising. It is consistent with clinical studies of BE pathogenesis, in which Cox2 expression is induced in the esophagus by acid reflux and further increased in BE and EAC [24–26]. Moreover, Cox2 inhibition in a rat surgical model of Barrett and EAC decreased tumor formation [53]. Despite these supportive data, a role for Cox2 in the pathogenesis of BE and EAC has not been formally tested until our studies reported here.
The induction of intestinal mucin-containing cysts within the developing multilayered epithelium of STR.M.Cox2 cells is an important finding. It suggests that the mucin secretion is polarized to collect in the discreet cysts, and it therefore establishes in principle that polarized mucin-secreting cells can emerge from the squamous cells and therefore suggests the possibility that the Barrett specialized columnar epithelial cells likewise can be derived from a keratinocyte precursor. It is important to note that not all cells expressing Cox2 adopted the mucin-secreting phenotype. We believe this is not a limitation of our approach; instead, it better reflects the in vivo emergence of BE cells under the conditions of gastroesophageal reflux. Current thinking on BE pathogenesis suggests that BE progression is due to clonal progression [54]. This in vitro system may phenocopy this, as we observe a phenotypic shift in a limited subset of cells. Under the selective pressure of gastroesophageal reflux, these limited numbers of cells may have a growth advantage. This is a hypothesis we are trying to test in future studies.
We also found it interesting that Muc5B was one of the mucins produced by these novel cell types because it is a mucin produced by esophageal subepithelial gland cells [51,55]. We speculate that epithelial stem cells may retain a capacity to give rise to alternative cell lineages, particularly of nearby epithelial structures. There is some support for this plasticity in the literature; hair follicle stem cells can contribute to the epidermis in the setting of wound healing [51,55]. The long-term effects of a chronic reflux injury may provoke these cells to adopt the related epithelial lineage of subepithelial glandular cells in response.
One important question left unanswered is how Cox2 expression leads to the observed morphologic and gene expression changes. It may simply be the effects of prostaglandins on cell differentiation patterns, mediated by their cell receptors. Alternatively, prostaglandin production is associated with significant oxidative stress for the cells, which can damage DNA and alter epigenetic configurations and gene expression patterns [56]. We favor the latter hypothesis and plan to investigate this possibility in subsequent experiments.
Three-dimensional Culture Conditions Are a Critical Component to In Vitro Modeling of BE
The importance of culture conditions on the behavior and differentiation of human esophageal STR cells is well established by our studies. Cell culture studies using primary esophageal keratinocytes have explored the effects of various Barrett etiologic agents, including chronic acid and bile acid treatment [57,58], bone morphogenetic protein 4 [59], retinoic acid [60,61], and the intestine-specific transcription factors Cdx1 and Cdx2 [6,8,62]. Although these treatments did yield increased expression of intestine-specific genes and some changes in cell morphology, none succeeded at inducing a mucin-secreting cell type from the keratinocytes. Early studies of gene expression patterns in the STR.L.CatCLef and STR.M.Cox22 cells cultured under traditional two-dimensional conditions were not encouraging. Only when the cells were placed in conditions favoring cell differentiation did we observe the significant changes, including greater intestinalization and, in the case of Cox2, the emergence of polarized mucin-secreting cells.
The studies with the STR.M.Cox2 cells were particularly striking. To our knowledge, the induction of polarized mucin-secreting cells from human esophageal keratinocytes exceeds those of any previously published study modeling BE in vitro. In addition, the presence of Ki67+ cells associated with the large cyst suggests that a stem cell with altered niche requirements is giving rise to the polarized secretory cells. More work is needed to understand specifically what is happening with these cells and how it might relate to the pathogenesis of BE. However, based on these observations, all our future studies examining candidate genes for contributions to BE will include culturing cells under organotypic conditions as a critical modeling approach.
Finally, it is not clear what it is about the organotypic culture condition that enables Cox2 or Wnt signals to provoke intestinalized gene expression patterns. We suspect the close cell-cell contact provides a permissive environment for these alterations. Moreover, with the cells cultured densely, there will naturally be higher local levels of prostaglandins produced by Cox2 activity. This alone may be enough to induce the observed changes. Alternatively, fibroblasts in the organotypic culture may respond to the prostaglandins by secreting other growth factors or cytokines that influence epithelial cell differentiation. We have plans to test these possibilities in future experiments.
In conclusion, in vitro modeling of BE pathogenesis can be improved by the inclusion of strategies to enhance Wnt signaling and Cox2 activity and, most importantly, using three-dimensional organotypic culture conditions. Combining these strategies with others to enhance goblet cell differentiation or the expression of Muc2 (such as expressing the transcription factor Math1/HATH1) should be very effective at improving the in vitro models. Once refined, these cell culture-based models would serve as the rational basis to pursue the development of appropriate transgenic mouse models and provide a platform for testing novel diagnostic and therapeutic agents.
Supplementary Material
Acknowledgments
The authors thank Gary Swain for his immunohistochemical and microscopy expertise.
Footnotes
This work was supported by the National Cancer Institute (Program Project PO1 CA098101 to Anil Rustgi and K99 CA138498 to D.B.S.) and by the Morphology, Cell Culture, and Molecular Biology Core Facilities of the Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania (P30-DK50306 and PO1 CA098101).
This article refers to supplementary materials, which are designated by Tables W1 to W3 and are available online at www.neoplasia.com.
References
- 1.American Cancer Society, author. Cancer Facts and Figures 2010. Atlanta, GA: American Cancer Society; 2010. [Google Scholar]
- 2.Morales CP, Souza RF, Spechler SJ. Hallmarks of cancer progression in Barrett's oesophagus. Lancet. 2002;360:1587–1589. doi: 10.1016/S0140-6736(02)11569-8. [DOI] [PubMed] [Google Scholar]
- 3.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6:11–16. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 6.Stairs DB, Nakagawa H, Klein-Szanto A, Mitchell SD, Silberg DG, Tobias JW, Lynch JP, Rustgi AK. Cdx1 and c-Myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward Barrett's esophagus. PLoS One. 2008;3:e3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong J, Stairs DB, Lynch JP. Modeling Barrett's esophagus. Biochem Soc Trans. 2010;38:321–326. doi: 10.1042/BST0380321. [DOI] [PubMed] [Google Scholar]
- 8.Kong J, Nakagawa H, Isariyawongse B, Funakoshi S, Silberg D, Rustgi AK, Lynch JP. Induction of intestinalization in human esophageal keratinocytes is a multi-step process. Carcinogenesis. 2009;30:122–130. doi: 10.1093/carcin/bgn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong J, Crissey MA, Funakoshi S, Kreindler JL, Lynch JP. Ectopic Cdx2 expression in murine esophagus models an intermediate stage in the emergence of Barrett's esophagus. PLoS One. 2011;6:e18280. doi: 10.1371/journal.pone.0018280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rieder F, Cheng L, Harnett KM, Chak A, Cooper GS, Isenberg G, Ray M, Katz JA, Catanzaro A, O'Shea R, et al. Gastroesophageal reflux disease-associated esophagitis induces endogenous cytokine production leading to motor abnormalities. Gastroenterology. 2007;132:154–165. doi: 10.1053/j.gastro.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Corrado G, Zicari A, Cavaliere M, Rea P, Pacchiarotti C, Cerroni F, Pontieri G, Cardi E. Increased release of interleukin-6 by oesophageal mucosa in children with reflux oesophagitis. Eur J Gastroenterol Hepatol. 1999;11:839–843. doi: 10.1097/00042737-199908000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Hamaguchi M, Fujiwara Y, Takashima T, Hayakawa T, Sasaki E, Shiba M, Watanabe T, Tominaga K, Oshitani N, Matsumoto T, et al. Increased expression of cytokines and adhesion molecules in rat chronic esophagitis. Digestion. 2003;68:189–197. doi: 10.1159/000075698. [DOI] [PubMed] [Google Scholar]
- 13.Kuramochi H, Vallbohmer D, Uchida K, Schneider S, Hamoui N, Shimizu D, Chandrasoma PT, DeMeester TR, Danenberg KD, Danenberg PV, et al. Quantitative, tissue-specific analysis of cyclooxygenase gene expression in the pathogenesis of Barrett's adenocarcinoma. J Gastrointest Surg. 2004;8:1007–1016. doi: 10.1016/j.gassur.2004.09.025. discussion 1016–1007. [DOI] [PubMed] [Google Scholar]
- 14.Hamoui N, Peters JH, Schneider S, Uchida K, Yang D, Vallbohmer D, Hagen JA, DeMeester SR, DeMeester TR, Danenberg K, et al. Increased acid exposure in patients with gastroesophageal reflux disease influences cyclooxygenase-2 gene expression in the squamous epithelium of the lower esophagus. Arch Surg. 2004;139:712–716. doi: 10.1001/archsurg.139.7.712. discussion 716–717. [DOI] [PubMed] [Google Scholar]
- 15.Pai R, Soreghan B, Szabo IL, Pavelka M, Baatar D, Tarnawski AS. Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med. 2002;8:289–293. doi: 10.1038/nm0302-289. [DOI] [PubMed] [Google Scholar]
- 16.Leahy KM, Ornberg RL, Wang Y, Zweifel BS, Koki AT, Masferrer JL. Cyclooxygenase-2 inhibition by celecoxib reduces proliferation and induces apoptosis in angiogenic endothelial cells in vivo. Cancer Res. 2002;62:625–631. [PubMed] [Google Scholar]
- 17.Husain SS, Szabo IL, Tamawski AS. NSAID inhibition of GI cancer growth: clinical implications and molecular mechanisms of action. Am J Gastroenterol. 2002;97:542–553. doi: 10.1111/j.1572-0241.2002.05528.x. [DOI] [PubMed] [Google Scholar]
- 18.Fosslien E. Review: molecular pathology of cyclooxygenase-2 in cancer-induced angiogenesis. Ann Clin Lab Sci. 2001;31:325–348. [PubMed] [Google Scholar]
- 19.Crofford LJ. Prostaglandin biology. Gastroenterol Clin North Am. 2001;30:863–876. doi: 10.1016/s0889-8553(05)70217-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang MT, Honn KV, Nie D. Cyclooxygenases, prostanoids, and tumor progression. Cancer Metastasis Rev. 2007;26:525–534. doi: 10.1007/s10555-007-9096-5. [DOI] [PubMed] [Google Scholar]
- 21.Lynch JP, Lichtenstein GR. Cyclooxygenase activity in gastrointestinal cancer development and progression—prospects as a therapeutic target. In: Morgan D, Fossman U, Nakaqda M, editors. Cancer and Inflammation. Basel, Switzerland: Birkhauser Verlag Basel; 2004. pp. 147–176. [Google Scholar]
- 22.Lurje G, Vallbohmer D, Collet PH, Xi H, Baldus SE, Brabender J, Metzger R, Heitmann M, Neiss S, Drebber U, et al. COX-2 mRNA expression is significantly increased in acid-exposed compared to nonexposed squamous epithelium in gastroesophageal reflux disease. J Gastrointest Surg. 2007;11:1105–1111. doi: 10.1007/s11605-007-0210-3. [DOI] [PubMed] [Google Scholar]
- 23.Muller-Decker K, Furstenberger G. The cyclooxygenase-2-mediated prostaglandin signaling is causally related to epithelial carcinogenesis. Mol Carcinog. 2007;46:705–710. doi: 10.1002/mc.20326. [DOI] [PubMed] [Google Scholar]
- 24.Morris CD, Armstrong GR, Bigley G, Green H, Attwood SE. Cyclooxygenase-2 expression in the Barrett's metaplasia-dysplasia-adenocarcinoma sequence. Am J Gastroenterol. 2001;96:990–996. doi: 10.1111/j.1572-0241.2001.03599.x. [DOI] [PubMed] [Google Scholar]
- 25.Majka J, Rembiasz K, Migaczewski M, Budzynski A, Ptak-Belowska A, Pabianczyk R, Urbanczyk K, Zub-Pokrowiecka A, Matlok M, Brzozowski T. Cyclooxygenase-2 (COX-2) is the key event in pathophysiology of Barrett's esophagus. Lesson from experimental animal model and human subjects. J Physiol Pharmacol. 2010;61:409–418. [PubMed] [Google Scholar]
- 26.Sonoda R, Naomoto Y, Shirakawa Y, Fujiwara Y, Yamatsuji T, Noma K, Tanabe S, Takaoka M, Gunduz M, Tsujigiwa H, et al. Preferential up-regulation of heparanase and cyclooxygenase-2 in carcinogenesis of Barrett's oesophagus and intestinal-type gastric carcinoma. Histopathology. 2010;57:90–100. doi: 10.1111/j.1365-2559.2010.03594.x. [DOI] [PubMed] [Google Scholar]
- 27.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 29.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okubo T, Hogan BL. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett's esophagus. Oncogene. 2006;25(21):3084–3092. doi: 10.1038/sj.onc.1209338. [DOI] [PubMed] [Google Scholar]
- 32.Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett's oesophagus patients at risk for malignant transformation. J Pathol. 2006;208:100–107. doi: 10.1002/path.1884. [DOI] [PubMed] [Google Scholar]
- 33.Clement G, Guilleret I, He B, Yagui-Beltran A, Lin YC, You L, Xu Z, Shi Y, Okamoto J, Benhattar J, et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter occurs early in the carcinogenesis of Barrett's esophagus. Cancer Sci. 2008;99:46–53. doi: 10.1111/j.1349-7006.2007.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584–591. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 35.Darlavoix T, Seelentag W, Yan P, Bachmann A, Bosman FT. Altered expression of CD44 and DKK1 in the progression of Barrett's esophagus to esophageal adenocarcinoma. Virchows Arch. 2009;454:629–637. doi: 10.1007/s00428-009-0769-z. [DOI] [PubMed] [Google Scholar]
- 36.Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B, von Werder A, Enders GH, Opitz OG, Rustgi AK. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–738. [PubMed] [Google Scholar]
- 37.Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry WS, Cukierman E, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller MS, Ezaki T, Guo RJ, Lynch JP. Cdx1 or Cdx2 expression activates E-cadherin-mediated cell-cell adhesion and compaction in human Colo 205 cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G104–G114. doi: 10.1152/ajpgi.00484.2003. [DOI] [PubMed] [Google Scholar]
- 40.Ezaki T, Guo RJ, Li H, Reynolds AB, Lynch JP. The homeodomain transcription factors Cdx1 and Cdx2 induce E-cadherin adhesion activity by reducing beta- and p120-catenin tyrosine phosphorylation. Am J Physiol Gastrointest Liver Physiol. 2007;293:G54–G65. doi: 10.1152/ajpgi.00533.2006. [DOI] [PubMed] [Google Scholar]
- 41.Opitz OG, Suliman Y, Hahn WC, Harada H, Blum HE, Rustgi AK. Cyclin D1 overexpression and p53 inactivation immortalize primary oral keratinocytes by a telomerase-independent mechanism. J Clin Invest. 2001;108:725–732. doi: 10.1172/JCI11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 43.Lynch J, Suh ER, Silberg DG, Rulyak S, Blanchard N, Traber PG. The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by down-regulation of D-type cyclins. J Biol Chem. 2000;275:4499–4506. doi: 10.1074/jbc.275.6.4499. [DOI] [PubMed] [Google Scholar]
- 44.Galceran J, Hsu SC, Grosschedl R. Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proc Natl Acad Sci USA. 2001;98:8668–8673. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller T, Bain G, Wang X, Papkoff J. Regulation of epithelial cell migration and tumor formation by beta-catenin signaling. Exp Cell Res. 2002;280:119–133. doi: 10.1006/excr.2002.5630. [DOI] [PubMed] [Google Scholar]
- 46.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, Klein-Szanto AJ, Herlyn M, Diehl JA, Katz JP, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–2123. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 49.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 50.Byrd JC, Bresalier RS. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004;23:77–99. doi: 10.1023/a:1025815113599. [DOI] [PubMed] [Google Scholar]
- 51.Guillem P, Billeret V, Buisine MP, Flejou JF, Lecomte-Houcke M, Degand P, Aubert JP, Triboulet JP, Porchet N. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int J Cancer. 2000;88:856–861. doi: 10.1002/1097-0215(20001215)88:6<856::aid-ijc3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 52.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 53.Oyama K, Fujimura T, Ninomiya I, Miyashita T, Kinami S, Fushida S, Ohta T. Cyclooxygenase (COX)-2 expression in a rat duodenoesophageal reflux model and chemoprevention of adenocarcinoma by the selective COX-2 inhibitor nimesulide [in Japanese] Nippon Shokakibyo Gakkai Zasshi. 2007;104:1183–1191. [PubMed] [Google Scholar]
- 54.Maley CC, Reid BJ. Natural selection in neoplastic progression of Barrett's esophagus. Semin Cancer Biol. 2005;15:474–483. doi: 10.1016/j.semcancer.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Arul GS, Moorghen M, Myerscough N, Alderson DA, Spicer RD, Corfield AP. Mucin gene expression in Barrett's oesophagus: an in situ hybridisation and immunohistochemical study. Gut. 2000;47:753–761. doi: 10.1136/gut.47.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stairs DB, Kong J, Lynch JP. Cdx genes, inflammation, and the pathogenesis of intestinal metaplasia. Prog Mol Biol Transl Sci. 2010;96:231–270. doi: 10.1016/B978-0-12-381280-3.00010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchetti M, Caliot E, Pringault E. Chronic acid exposure leads to activation of the cdx2 intestinal homeobox gene in a long-term culture of mouse esophageal keratinocytes. J Cell Sci. 2003;116:1429–1436. doi: 10.1242/jcs.00338. [DOI] [PubMed] [Google Scholar]
- 58.Kazumori H, Ishihara S, Rumi MA, Kadowaki Y, Kinoshita Y. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut. 2006;55:16–25. doi: 10.1136/gut.2005.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milano F, van Baal JW, Buttar NS, Rygiel AM, de Kort F, DeMars CJ, Rosmolen WD, Bergman JJ, Van Marle J, Wang KK, et al. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132:2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 60.Cooke G, Blanco-Fernandez A, Seery JP. The effect of retinoic acid and deoxycholic acid on the differentiation of primary human esophageal keratinocytes. Dig Dis Sci. 2008;53:2851–2857. doi: 10.1007/s10620-008-0240-z. [DOI] [PubMed] [Google Scholar]
- 61.Chang CL, Lao-Sirieix P, Save V, De La Cueva Mendez G, Laskey R, Fitzgerald RC. Retinoic acid-induced glandular differentiation of the oesophagus. Gut. 2007;56:906–917. doi: 10.1136/gut.2006.097915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu T, Zhang X, So CK, Wang S, Wang P, Yan L, Myers R, Chen Z, Patterson AP, Yang CS, et al. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007;28(2):488–496. doi: 10.1093/carcin/bgl176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.