Abstract
A hallmark of malignant gliomas is their ability to disperse through neural tissue, leading to long-term failure of all known therapies. Identifying new antimigratory targets could reduce glioma recurrence and improve therapeutic efficacy, but screens based on conventional migration assays are hampered by the limited ability of these assays to reproduce native cell motility. Here, we have analyzed the motility, gene expression, and sensitivity to migration inhibitors of glioma cells cultured on scaffolds formed by submicron-sized fibers (nanofibers) mimicking the neural topography. Glioma cells cultured on aligned nanofiber scaffolds reproduced the elongated morphology of cells migrating in white matter tissue and were highly sensitive to myosin II inhibition but only moderately affected by stress fiber disruption. In contrast, the same cells displayed a flat morphology and opposite sensitivity to myosin II and actin inhibition when cultured on conventional tissue culture polystyrene. Gene expression analysis indicated a correlation between migration on aligned nanofibers and increased STAT3 signaling, a known driver of glioma progression. Accordingly, cell migration out of glioblastoma-derived neurospheres and tumor explants was reduced by STAT3 inhibitors at subtoxic concentrations. Remarkably, these inhibitors were ineffective when tested at the same concentrations in a conventional two-dimensional migration assay. We conclude that migration of glioma cells is regulated by topographical cues that affect cell adhesion and gene expression. Cell migration analysis using nanofiber scaffolds could be used to reproduce native mechanisms of migration and to identify antimigratory strategies not disclosed by other in vitro models.
Introduction
Cell migration away from the site of the primary tumor is a hallmark of malignant cancers often leading to recurrence and the failure of existing therapies. This is particularly evident in malignant gliomas, the most challenging tumor of the central nervous system characterized by its ability to disperse through normal neural tissue and recur after initial treatment [1].
Histologic evidence has shown that glioma cell dispersion in the brain occurs along preferential patterns, in many cases following the orientation of thin, elongated anatomic structures such as capillaries, white matter fibers, and unmyelinated axons [2]. Unfortunately, standard assays devised to study glioma cell motility do not incorporate such topographical cues guiding cell adhesion and traction in vivo, focusing instead on cell motility on either rigid (i.e., polystyrene or glass) surfaces or invasion through a homogeneous, collagen-based matrix that is absent in neural tissue [3,4].
Motile glioma cells are more resistant than nonmotile cells to apoptotic stimuli [5,6], and current evidence suggests that conventional therapies may in fact trigger glioma cell dispersion [7,8]. Thus, understanding the mechanisms of glioma cell migration is critical to the development of more efficient targeting strategies as part of adjuvant therapy. Antimigratory approaches against gliomas have targeted cell adhesion molecules or tumor-associated proteases, following antimetastatic strategies used in other solid tumors [9]. However, these approaches have been largely ineffective in the clinical setting, partly because of the ability of brain tumor cells to shift between different mechanisms of cell adhesion as well as proteolytic and nonproteolytic modes of migration [3,10]. This underscores the need for additional studies to identify antimigratory compounds capable of targeting the master regulators of tumor cell locomotion [3].
In a recent study, we demonstrated that glioma cells can be cultured on scaffolds made of poly-ε-caprolactone nanofibers produced by electrospinning [11]. Fiber density, alignment, and stiffness can be controlled in these scaffolds, thus providing the cells with a topographically complex substrate. Glioma cells were able to grow on nanofibers of different alignment and accurately reproduced the morphologies described for these cells migrating through neural tissue [10,12]. Here, we show that migration of glioma cells on nanofiber scaffolds reproduces not only the morphology but also characteristic molecular features of three-dimensional migration and results in a pattern of gene expression dependent on fiber alignment. Moreover, we show that active cell migration on aligned nanofibers correlates with activation of the transcription factor STAT3, a central regulator of tumor progression and metastasis in solid cancers [13]. Accordingly, subtoxic inhibition of STAT3 specifically reduced glioma cell migration on nanofibers, suggesting that this novel culture technology could be used for screening of antimigratory compounds.
Materials and Methods
Preparation of Nanofiber-Coated Culture Plates
Poly-ε-caprolactone nanofibers were prepared as previously described [11] with minimal modifications. Briefly, optically clear polystyrene film (thickness, 190 µm; MultiPlastics, Inc, Lewis Center, OH) was cut to the desired final size and attached to the side of a rotating drum (Nanofiber Solutions, Columbus, OH). Nanofibers were deposited by electrospinning, using a syringe perpendicular to the polystyrene film as described [11]. Fiber alignment was controlled by the rotational speed of the drum and scaffold thickness by the amount of time used to deposit the nanofibers. Films covered with multilayered nanofiber scaffolds were trimmed, attached to bottomless culture plates (Figure 1A), and sterilized with UV radiation (350 mJ/cm2) before use.
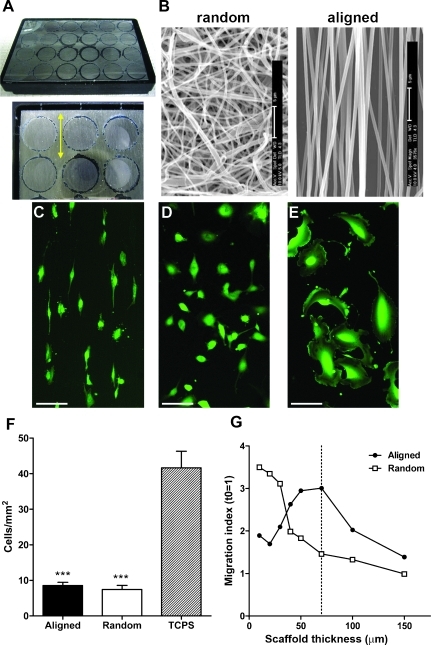
Figure 1.
Glioma cell morphology and migration are directed by substrate topography. (A) Representative photography of a multiwell culture plate with a nanofiber-coated backing film. The enlarged image shows the translucent coating of nanofibers, and the double-headed arrow indicates the direction of fiber alignment. (B) Scanning electron microscope images of randomly oriented and highly aligned nanofibers. Bars, 5 µm. (C–E) Images of calcein-stained U251 glioma cells cultured on highly aligned nanofibers (C), randomly oriented nanofibers (D), or conventional TCPS (E). Bars, 100 µm. (F) Quantification of cell adhesion (U251 cells/mm2) showed significantly lower adhesion of cells to nanofibers compared with TCPS (***P < .001), but no differences between randomly oriented and aligned nanofibers (analysis by one-way ANOVA and Bonferroni post hoc tests). (G) Radial dispersion of U251 cells from cell aggregates was measured using nanofiber scaffolds of different thickness. Cells gained motility much faster on aligned than on randomly oriented nanofibers as the fibers became sparser. Use of 70-µm-thick scaffolds (dashed line) resulted in the highest cell motility on aligned nanofibers but little motility of the same cells on randomly oriented nanofibers.
Cell Cultures and Reagents
The human glioma cell lines U87 and U251 (American Type Culture Collection, Manassas, VA) were grown at 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. The identity of these cells was confirmed through “Cell Check” authentication service provided by the Research Animal Diagnostic Laboratory (Columbia, MO). Two cultures of glioblastoma-derived tumor initiating cells (G8 and G9) were prepared from freshly resected tumors and cultured as neurospheres in serum-free medium as described [14]. These cells have been characterized as tumor stem cells elsewhere [15], display self-renewal in vitro, and are highly tumorigenic in vivo, replicating the phenotype of the original tumors. Only low passages of G8 and G9 cells (up to five passages) were used.
To prepare tumor xenografts, G8 and G9 cells were implanted in the striatum of athymic mice as described [16]. After 2 weeks, animals were perfused with saline, and tumors were resected and minced in Hank's balanced salt solution. Tissue explants were cultured in suspension in neurosphere medium for 72 hours, with daily changes of medium to remove debris. Explants containing viable tumor cells were detected by uptake of the fluorescent dye calcein-AM (5 µM; Invitrogen, Carlsbad, CA) and deposited on nanofiber scaffolds. As an alternative strategy, cells were first stably transduced with the lentiviral vector pCDH-EF1-coGFP (System Biosciences, Mountain View, CA) and used to generate tumors as above. Tissue pieces containing GFP-expressing tumor were processed under a fluorescence dissection microscope, cleaned of debris in Hank's balanced salt solution, and deposited on nanofiber scaffolds. Patterns of cell migration out of tissue explants were undistinguishable using either approach.
To inhibit cell migration, glioma cells were treated with the myosin II inhibitor blebbistatin (Sigma-Aldrich, St Louis, MO) and the actin polymerization inhibitor cytochalasin D (Sigma-Aldrich). To test the involvement of STAT3 on migration, cells were treated with the STAT3 inhibitors stattic (Tocris Bioscience, Ellisville, MO) [17] and LLL12 (Division of Medicinal Chemistry, Ohio State University, Columbus, OH) [18]. Cell viability was determined using an assay for reduction of soluble tetrazolium (CellTiter kit; Promega, Madison, WI).
Adhesion and Migration Assays
To quantify cell adhesion to nanofibers, glioma cells were dissociated and plated (30,000 cells/200 µl) in triplicate on nanofiber-coated or tissue culture polystyrene (TCPS) plates. After 30 minutes at 37°C, cells were washed, fixed, and quantified as described [19].
To analyze cell migration on nanofibers, 50,000 to 75,000 glioma cells were plated on 35-mm agar plates to form spheroids [16]. After 48 hours, glioma spheroids 200 to 250 µm in diameter were stained with 5 µM CellTracker CMFDA (Invitrogen) and manually placed within nanofiber-coated wells. To analyze the migration of glioblastoma-derived initiating cells, either from neurospheres or from tumor explants, nanofibers were first precoated with 5 µg/ml fibronectin in phosphate-buffered saline for 2 hours. Migration index was calculated as the ratio of maximum dispersion (Figure W1A) divided by the original diameter of the spheroids.
To analyze cell migration on TCPS plates, glioma cells were tested using conventional wound healing and radial dispersion assays as previously described [20]. To analyze cell translocation (“Transwell assay”), 30,000 cells were applied to uncoated cell culture inserts with 8-µm pores (Greiner Bio-One, Monroe, NC). Migration in response to a chemoattractant gradient (0%–10% serum) was measured after 8 hours by counting the number of transmigrated cells.
To analyze cell migration using an organotypic culture model, cultures of mouse neonatal brain slices were prepared as we have previously described [16]. Aggregates of GFP-expressing glioma cells were pretreated overnight with STAT3 inhibitors, deposited on the tissue slices, and followed by fluorescence microscopy for up to 96 hours. Dispersion was quantified by analyzing the total area and perimeter covered by the migratory cells [16].
All experiments of cell migration were performed three times with at least three independent replicates per condition except brain slice experiments, which used eight replicates.
Microarray Experiments
To compare gene expression of glioma cells migrating on aligned versus randomly oriented nanofibers 5 x 105 U251 cells were cultured in nanofiber-coated 60-mm culture plates, collected after 48 hours, and processed with TRIzol (Invitrogen) according to standard protocols. Total RNA from independent triplicates was processed for hybridization to Affymetrix U133+ 2.0 microarrays at The Ohio State University Microarray Shared Resource. Data were deposited in the National Center for Biotechnology Information's Gene Expression Omnibus Repository for Functional Genomic Studies (data set GSE28167). Significantly upregulated genes (>1.5-fold; Table W1) in cells cultured on aligned nanofibers were analyzed using Ingenuity Pathway Analysis to identify top functional networks and DAVID bioinformatics resources to identify top functional clusters (Table W2). Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed in parallel samples to verify the expression level of selected messenger RNA (primers listed in Table W3).
Western Blot Analysis
Dissociated U251 and G9 cells cultured in nanofiber-coated 60-mm culture plates were lysed in situ in 20 mM Tris-HCl buffer, pH 7.6, containing 150 mM NaCl, 1% vol/vol NP-40, 0.5% vol/vol sodium deoxycholate, and protease plus phosphatase inhibitors (Complete and PhosSTOP cocktails; Roche Applied Science, Indianapolis, IN). Proteins were processed for Western blot analysis with antibodies against STAT3 and phospho-Tyr705 STAT3 (Cell Signaling Technology, Danvers, MA), myosin light chain 2 (MLC2) and phospho-Ser19 MLC2 (Cell Signaling), and β-tubulin (Sigma-Aldrich).
Statistical Analysis
Cell adhesion and translocation experiments were analyzed by one-way analysis of variance (ANOVA) followed by post hoc Bonferroni tests. Cell migration on nanofibers, TCPS, or brain slices was analyzed by two-way ANOVA for repeated measures. Results of STAT3 inhibition on nanofibers were analyzed by one-way ANOVA for each inhibitor and cell type. P < .05 indicates significant differences between treatments. Bar graphs in the figures represent mean ± SD. Microarray data were analyzed by robust multichip average procedure using quantile normalization. Differentially expressed genes between two groups were identified with a modified t test [21] using BRB-Array Tools software.
Results
Glioma Cell Morphology and Migration Depend on Fiber Alignment
To better understand the mechanisms underlying glioma cell migration in response to variable topographical cues, we first analyzed the morphology and behavior of glioma cells cultured on three-dimensional nanofiber scaffolds versus conventional two-dimensional surfaces. Dissociated U251 glioblastoma cells were plated on conventional TCPS plates and compared against cells cultured on two distinct types of nanofiber scaffolds (50 µm thick; Figure 1, A and B). Cells cultured on aligned nanofibers adopted a fusiform morphology, usually with a leading process following an individual fiber (Figure 1C). In contrast, cells on randomly oriented fibers remained relatively rounded (Figure 1D). In neither case did we see obvious lamellipodia or fan-shaped morphologies that were typical of these cells cultured on TCPS (Figure 1E). Despite their different morphologies, initial cell adhesion was similar on both types of nanofiber substrates, although considerably lower than adhesion to TCPS (Figure 1F).
Next, we quantified cell migration on nanofibers using a radial migration assay to measure cell dispersion out of a tumor aggregate or “core” [11,20] (Figure W1A). Glioma cell spheroids were plated on nanofiber scaffolds of different thickness (10–200 µm) to determine the effect of fiber density on cell migration. We observed that cell migration was very restricted at the highest fiber densities and, as expected, increased as the nanofibers became sparser (Figures 1G and W1B). Interestingly, migration on highly aligned nanofibers peaked on relatively thick scaffolds (60–80 µm), whereas migration on randomly oriented nanofibers remained low until the fibers were very sparse (<30 µm thick), which likely allowed the cells to contact the underlying substrate [11]. Therefore, we chose 70-µm-thick nanofiber scaffolds for our subsequent experiments to provide maximum differences in total cell motility between the two different types of fiber orientations.
Glioma Cell Migration on Aligned Nanofibers Is Myosin II Dependent
Recent work has shown that cell motility in a three-dimensional environment is a substantially different process from migration on rigid two-dimensional surfaces, being less dependent on focal adhesions and long, anchored, stress fibers and more on the local contraction of actomyosin complexes to squeeze the tail-end of the cell through intercellular spaces [10,22]. To determine whether migration of glioma cells on nanofiber scaffolds reproduced this key molecular feature of three-dimensional migration, we assessed the effect of inhibitors targeting myosin II and actin polymerization on cell migration.
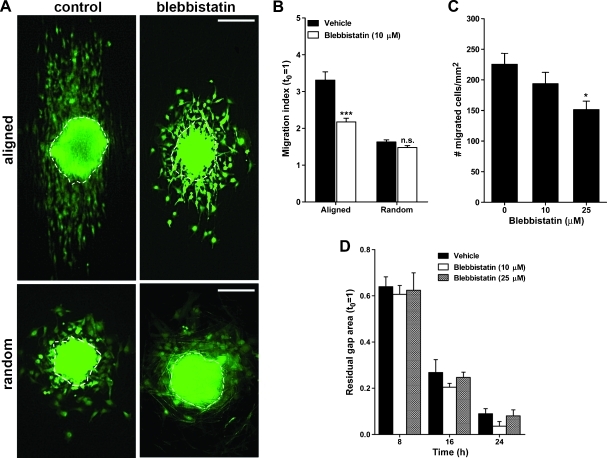
Migration of U251 glioma cells out of aggregates seeded on aligned nanofibers was significantly inhibited by the myosin II inhibitor blebbistatin (Figure 2, A and B). However, blebbistatin did not affect glioma cells on randomly oriented nanofibers, where motility was already restricted. When we compared these results with a conventional cell translocation assay where the cell body must be squeezed through the pores of culture inserts, we observed that blebbistatin partially inhibited the translocation of glioma cells but at a considerably higher concentration than that needed to inhibit cell migration on nanofibers (Figure 2C). In contrast, in a conventional wound healing assay (Figure 2D), glioma cell migration was not affected by blebbistatin, in agreement with the literature [10]. Overall, these results suggested that cell motility on aligned nanofibers was highly dependent on myosin II activity as in other three-dimensional models.
Figure 2.
Glioma cell migration on aligned nanofibers is myosin II-dependent. (A) Representative images of U251 glioma cell aggregates after being cultured for 24 hours on aligned or randomly oriented nanofibers, in the presence of 10 µM blebbistatin (blebbistatin) or its vehicle (control). Notice the parallel and elongated migration profile of cells on aligned nanofibers. Dashed outlines indicate the size and shape of the same aggregates at t = 0 h. Bars, 200 µm. (B) Effect of blebbistatin on cell dispersion on nanofibers. Results indicate a significant inhibition of cell migration on aligned but not on randomly oriented nanofibers. ***P < .001 by two-way ANOVA. (C) Effect of blebbistatin on cell translocation through cell culture inserts. Results indicate a significant effect only at 25 µM blebbistatin. *P < .05 by one-way ANOVA and Bonferroni post hoc test. (D) Effect of blebbistatin on two-dimensional cell migration measured with a wound healing assay. Migration of U251 cells in this assay was not affected by the myosin II inhibitor.
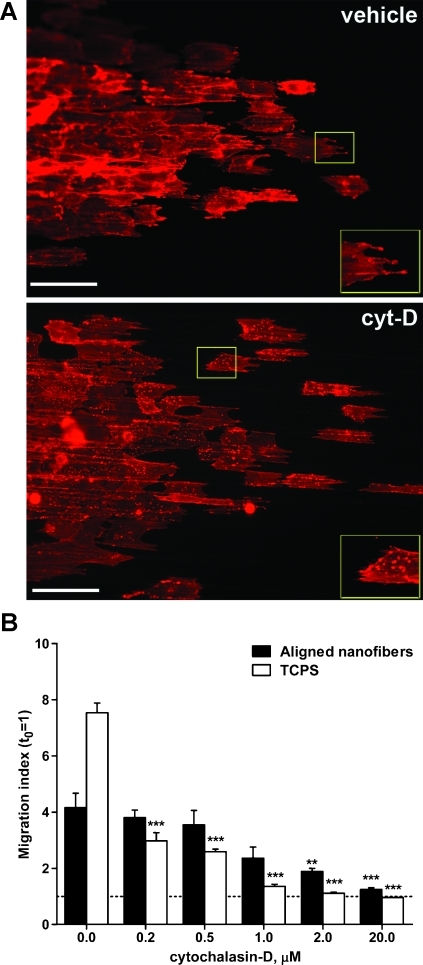
On the other hand, the inhibitor of actin polymerization cytochalasin D was much less effective in inhibiting cell motility on nanofibers compared with the same cells plated on TCPS. Cytochalasin D treatment of U251 cells caused some disruption of cortical F-actin, visualized as appearance of punctuate actin staining (Figures 3A and W2). However, cell dispersion on aligned nanofibers was only reduced significantly (P < .01) when U251 cells were exposed to toxic concentrations of this inhibitor (>2 µM). In comparison, dispersion of the same glioma cells on TCPS, measured with a radial migration assay (Figure 3B) or a wound healing assay (Figure W2), was significantly reduced (P < .001) at the lowest concentration of cytochalasin D tested (0.2 µM), and cells not only stopped migrating but also detached from the plates at a 2-µM concentration of this inhibitor. These results suggested that, while actin polymerization was still required for cell migration on nanofibers, the formation of long stress fibers was less critical than for motility of a rigid surface [23]. Together, the results from acto-myosin II disruption suggested that cell migration on nanofibers reproduced more closely the molecular features observed on three-dimensional migration rather than those observed for migration on rigid two-dimensional surfaces.
Figure 3.
Cell migration on nanofibers is less sensitive to stress fiber disruption than two-dimensional migration. (A) U251 glioma cell aggregates were cultured on aligned nanofibers, fixed after 24 hours, and stained with phalloidin. Alexa 594 to label actin F. Results show diffuse cortical staining in vehicle-treated cells (vehicle) versus punctuate staining (insets) along actin filaments in cells treated with the actin polymerization inhibitor cytochalasin D (cyt-D, 2 µM). Cell morphology was little or not affected in these conditions. Bars, 100 µm. (B) Quantification of radial dispersion of U251 cells shows that two-dimensional cell dispersion on TCPS was significantly reduced even at the lowest concentration of cytochalasin D tested (0.2 µM), whereas dispersion on nanofibers required 10 to 100x higher concentrations to be significantly inhibited. **P < .01, ***P < .001 by one-way ANOVA for each treatment.
Cell Migration on Aligned Nanofibers Results in Elevated JAK/STAT Signature
Both here and in a previous study [11], we have observed a substantial difference in the behavior of glioma cells cultured on aligned nanofibers, where the cells migrate efficiently, versus randomly oriented nanofibers, where migration is highly restricted without evident effects on viability. Therefore, we investigated whether the different migratory behavior of glioma cells that were otherwise equivalent for viability [11] or adhesion (Figure 1F) was reflected in differential gene expression.
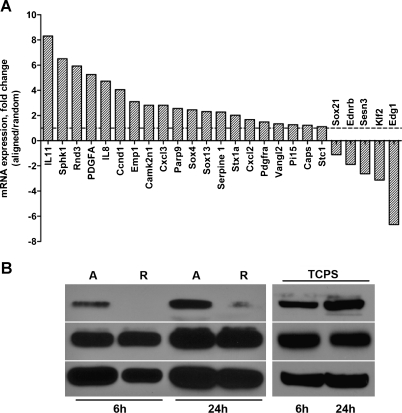
Using microarray analysis to compare U251 glioma cells cultured on aligned versus randomly oriented nanofibers, we observed significant differences in gene expression (summarized in Table W1). Pathway and gene ontology analysis suggested a strong association between the genes upregulated in cells cultured on aligned nanofibers and functional clusters involved in positive regulation of cell motility (Table W2). Strikingly, there was a remarkable up-regulation of genes that are known activators or targets of JAK/STAT signaling [24–26], including IL8, IL11, TLP, CXCL2, CCND1, PIK3CD, SPHK1, PIK3CD, and SERPINE1. Up-regulation of these genes in cells cultured on aligned nanofibers was subsequently validated by quantitative RT-PCR (Figure 4A).
Figure 4.
Glioma cell migration on aligned nanofibers correlates with activation of STAT3 signaling. (A) Quantitative RT-PCR results showing differential gene expression levels in glioma cells cultured on aligned versus randomly oriented nanofibers (selected genes are a subset of those listed in Table W1). Many of the upregulated genes are known modulators or targets of JAK/STAT and are involved in positive regulation of cell migration (Table W2). (B) Dissociated U251 glioma cells (5 x 105 cells/ml) were plated on aligned (A) or randomly oriented (R) nanofibers and collected 6 or 24 hours after attachment. Cells recovered from aligned nanofibers showed a substantial increase in Y705-phosphorylated STAT3 (p-STAT3) compared with cells recovered from randomly oriented nanofibers. As a control, U251 cells were plated on conventional TCPS and processed in the same manner, showing considerable expression of total and active STAT3 at all times tested.
Because these results suggested a possible involvement of STAT signaling in glioma cell migration, we focused on the transcription factor STAT3, which has been recently highlighted as a central regulator of malignant progression in high-grade gliomas [27]. In agreement with our results of gene expression, Western blot analysis results showed that the active phosphorylated form of STAT3 was markedly increased in cells cultured on aligned nanofibers shortly after the cells attached to the substrate, whereas it was barely detectable in cells on randomly oriented nanofibers, even after 24 hours in culture (Figure 4B). In agreement with previous literature [28–30], active phosphorylated STAT3 was also highly expressed in glioma cells cultured on TCPS, where cell motility is unimpeded by the substrate.
Inhibition of STAT3 Reduces the Migration of Glioma Cells on Nanofiber Scaffolds
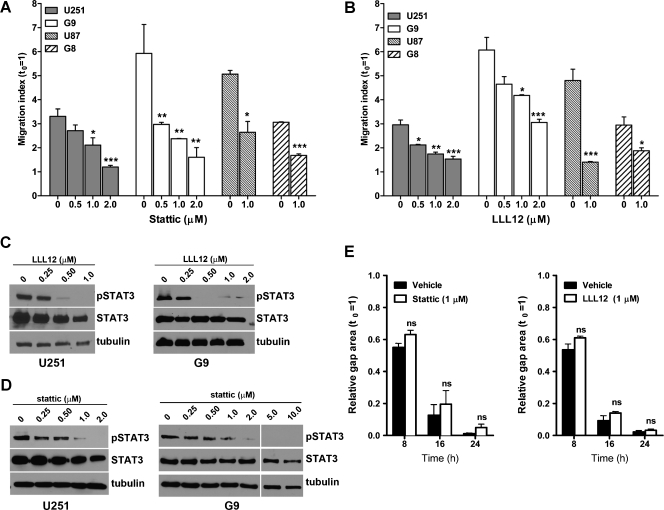
To determine whether cell migration on nanofibers could be used as a model to analyze the role of STAT3 on glioma cell migration, we tested two STAT3 inhibitors that specifically prevent STAT3 phosphorylation of Tyr705, a critical residue necessary for STAT3 dimerization and transcriptional activity. Both inhibitors, stattic and LLL12, significantly inhibited the migration of U251 cells cultured on aligned nanofibers (Figures 5, A and B, and W3), at concentrations (0.5–2.0 µM) that did not affect cell viability during these short assays (Figure W4). These results were reproduced with an additional glioma cell line (U87) and two preparations of primary glioblastoma-derived initiating cells (G8 and G9); all of them tested on aligned nanofibers. Moreover, the inhibition of cell migration matched the reduction or complete inhibition of STAT3 phosphorylation by these compounds (Figure 5, C and D).
Figure 5.
STAT3 inhibition reduces glioma cell migration on aligned nanofibers. (A and B) Cell dispersion was measured for two glioma cell lines (U251 and U87) and neurospheres from two glioblastoma-derived initiating cells (G8 and G9), in the presence of the STAT3 inhibitors stattic (A) and LLL12 (B). Both inhibitors significantly reduced cell migration at concentrations between 0.5 and 2 µM. *P < .05, **P < .01, ***P < .001 by one-way ANOVA for each cell type and inhibitor. (C and D) Western blot analysis of U251 and G9 cells collected from aligned nanofibers confirmed the inhibition of STAT3 phosphorylation (pSTAT3) using LLL12 (C) and stattic (D) at the same concentrations used to inhibit cell dispersion. STAT3 inhibition in G9 cells—which form large compact tumorspheres—was sometimes incomplete at low concentrations of LLL12 or stattic, but cell motility was, nevertheless, reduced. Complete STAT3 inhibition was achieved at higher concentrations of these inhibitors. (E) Migration of U251 cells using a wound healing assay was not affected by low concentrations of STAT3 inhibitors. NS indicates nonsignificant.
Interestingly, these subtoxic concentrations of stattic or LLL12 were unable to inhibit cell motility when tested in a conventional wound healing assay (Figure 5E), suggesting that the inhibitors had disrupted a sensitive mechanism necessary for cell migration in three-dimensional nanofiber scaffolds but not on rigid two-dimensional surfaces. In agreement, the phosphorylation of STAT3 and MLC2, a regulatory chain of myosin II necessary for myosin activity, were significantly reduced in cells treated with a low concentration of stattic on aligned nanofibers but not on polystyrene dishes (Figure W5). Together with our results showing that myosin II activity was critical for cell migration on aligned nanofibers (Figure 2), these results suggested that actomyosin activity on nanofibers, but not on TCPS, was finely regulated by STAT3 and highly sensitive to partial STAT3 inhibition. The sensitivity of glioma cell migration to low concentrations of STAT3 inhibitors was also observed in cells dispersing in cultured brain slices, a complex three-dimensional environment reproducing the natural cytoarchitecture and natural barriers to cell movement in the brain (Figure W6), suggesting that cell migration on nanofibers was supported by similar or the same mechanisms as in complex three-dimensional organotypic cultures. In contrast, inhibition of STAT3 did not reduce cell translocation in a Transwell migration assay at low concentrations of inhibitors (not shown) and had only a partial effect at high concentrations (LLL12 > 10 µM) that likely affected cell viability to some extent [18].
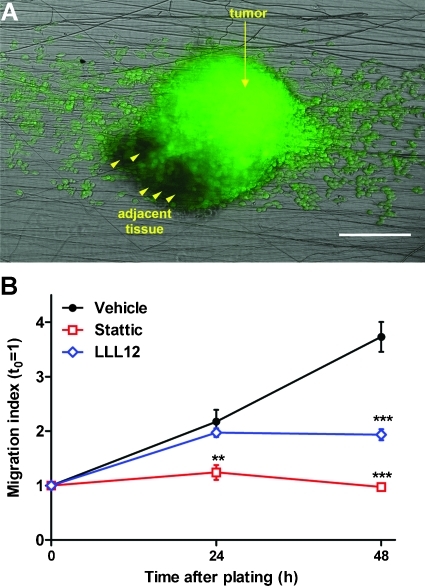
Because culture of tumor-derived neurospheres on nanofiber scaffolds required minimal processing steps and was highly reproducible, we finally asked if this culture model could be used to analyze cell migration directly out of fresh, biopsy-like tissue explants. We generated intracranial tumor xenografts using primary glioblastoma cells and recovered the tumors as well as adjacent brain tissue after 2 weeks. Viable tumor pieces, identified by calcein uptake or GFP fluorescence as indicated in the methods section, were minced, cleared of debris, and plated on fibronectin-coated aligned nanofibers. Cell migration took longer to be detected than migration out of homogeneous tumor neurospheres, but there was consistent glioma cell migration out of the tissue explants and along aligned nanofibers within 48 to 72 hours (Figure 6A). More importantly, treatment of the tumor explants with the two STAT3 inhibitors reduced significantly or abolished the outward migration of glioma cells (Figure 6B).
Figure 6.
STAT3 inhibition reduces cell dispersion from tumor explants. (A) Representative image of a tissue explant from a GFP-expressing G9 glioma cultured on aligned nanofibers during 48 hours. Arrowheads indicate nonfluorescent areas of adjacent brain tissue. Bar, 200 µm. (B) Tumor explants were cultured in the presence of 1 µM stattic (stattic), 1 µM LLL12 (LLL12), or their vehicle (vehicle), and cell dispersion was measured as before. Both STAT3 inhibitors significantly reduced (LLL12) or abolished (stattic) outward cell migration. **P < .01, ***P < .001 by two-way ANOVA for repeated measures.
Taken together, our results revealed a possible role for STAT3 in the migration of glioma cells in response to topographical cues and demonstrated the advantages of three-dimensional nanofiber scaffolds as a culture model to investigate pathways involved in cancer cell migration.
Discussion
Malignant gliomas have a very poor prognosis owing to their extensive infiltration of the surrounding normal neural tissue [2]. This infiltration is triggered in part by chemotherapy and radiotherapy [7,8,31], and motile glioma cells are highly resistant to these treatments [5,32]. Thus, understanding the mechanisms that drive glioma cell motility may improve not only the development of anti-invasive strategies but also the efficacy of current adjuvant therapies.
In this context, a major problem in studying cell motility in vitro is the difficulty of reproducing the native behavior of these tumor cells. With few exceptions [9], assays to study glioma cell invasion have largely reproduced the models used to study motility of other epithelial solid tumors, such as the wound healing assay (two-dimensional motility) and invasion through collagen-based matrices (three-dimensional motility) [9]. Glioma cells in these assays are exposed to a uniform environment—either an infinite flat surface or a uniform matrix—that lacks directional mechanical cues relevant to native mechanisms of cell migration in the brain. In response to limitations of other models, we developed a topographically complex environment for cell culture, using biocompatible scaffolds formed by electrospun submicron-sized fibers [11]. These scaffolds have mechanical properties, such as a low-tensile modulus (∼7 MPa), comparable with those of biologic tissues [33] and are therefore highly compliant compared with tissue culture polystyrene (tensile modulus ∼3600 MPa). This has allowed us to challenge glioma cells with a deformable substrate containing variable topography and analyze the molecular mechanisms involved in cell migration under these conditions.
Glioma cells adhered to nanofibers with less efficiency than to conventional TCPS, possibly due to less resistance from the substrate for the formation of focal adhesions [34,35], but total adhesion was independent of substrate topography (Figure 1F). In contrast, the actual migration of the cells was tightly dependent on the properties of the substrate, including both nanofiber alignment and density. Although the cells were not embedded in a matrix, we have previously shown that they can crawl through or become entangled in several layers of fibers [11]. The substrate is therefore irregular enough for the cells to exhibit three-dimensional migratory patterns, such as the marked body alignment and formation of protrusions along fibers, mimicking the formation of protrusions through the pores of a matrix and the elongated appearance of glioma cells migrating in vivo [10,36]. Furthermore, our data suggest that cell motility in nanofibers reproduced, at least in part, molecular features of three-dimensional motility such as stringent myosin II dependence [10] and low sensitivity to disruption of stress fibers [23], which contrasted with the opposite features of the cells cultured on rigid two-dimensional surfaces.
Using an optimal combination of nanofiber density and alignment to promote (aligned nanofibers) or restrict (random nanofibers) cell dispersion, we demonstrated a substantial up-regulation of STAT3 signaling in migratory glioma cells on nanofibers. The transcription factor STAT3 is a key regulator of development and metastasis in solid tumors [13,37] and has been recently proposed as a major driver of glioblastoma progression [27]. STAT3 promotes glioma stem cell proliferation and pluripotency [38] and drives tumor development toward an aggressive mesenchymal phenotype [27], therefore being a target with significant clinical potential [39]. Indeed, down-regulation of STAT3 efficiently reduces glioma cell proliferation, induces apoptosis, and inhibits tumor growth in vivo [40–42]. This has prompted the recent development of novel small-molecule therapeutic agents targeting STAT3 in brain tumors [43,44].
Because the down-regulation of STAT3 in gliomas causes rapid cell death in vitro, the role of this transcription factor in glioma cell migration has not been extensively explored. de la Iglesia et al. [45] have reported that overexpression of constitutively activated STAT3 reduced glioma cell migration, possibly due to repression of interleukin 8 (IL-8) signaling. However, because STAT3 is known to activate IL-8 expression in other cell models [46,47] and is in turn regulated by IL-8 and other cytokines [48], this paradoxical effect of STAT3 could have been caused by an overexpressed construct lacking regulatory feedback in transfected cells. In contrast, recent studies have suggested that inhibition of STAT3 reduces glioma cell migration [18,30,49], although that effect was achieved in most cases using conditions that induced cell apoptosis at the same time.
An important finding of our study is the observation that cell motility in nanofiber scaffolds was STAT3-dependent and could be specifically disrupted with low, subtoxic concentrations of STAT3 inhibitors. This effect was reproduced with glioma cells dispersing in cultured brain slices, suggesting that migration through both types of topographically complex environments was supported by similar molecular mechanisms. Remarkably, low concentrations of STAT3 inhibitors did not affect cell motility on two-dimensional surfaces. A possible explanation for these results is that STAT3 could regulate molecular mechanisms leading to actomyosin activity in glioma cells, which is critical for three-dimensional motility but can be compensated by alternative mechanisms in two-dimensional motility [10,23]. Accordingly, low concentrations of a STAT3 inhibitor reduced the activation of a myosin regulatory chain (MLC2) only in cells cultured on nanofibers, which could explain the significant effect of partial STAT3 inhibition on three-dimensional motility and lack of effect on two-dimensional motility. In agreement, STAT3 has been implicated as a mediator of Rho GTPase signaling [50], which is critical for actomyosin contraction and tail-end retraction necessary for glioma cell movement in three-dimensional matrices [51]. Interestingly, the expression of proinvasive metalloproteases that are STAT3 targets, such as MMP2 and MMP9 [30], did not change after STAT3 inhibition on nanofibers (not shown), suggesting that the role of STAT3 was specific to regulating motility rather than invasive mechanisms in a three-dimensional context. Overall, our results show that partial inhibition of STAT3 phosphorylation is sufficient to reduce glioma cell migration, underscoring the potential of this transcription factor as a novel target for combined anti-invasive and cytotoxic strategies in gliomas.
Although we have used the nanofiber scaffolds as a novel culture model for glioma cells, it should be possible to extend these studies to other tumor cell types that disperse in vivo along anatomic structures, such as pancreatic, prostate, or head and neck tumors that use perineural migration for metastasis [52]. In all these cases, the topographically complex nature of nanofiber scaffolds could provide considerable advantages over other models to study three-dimensional cell migration. Whereas organotypic models (i.e., tissue slices) represent a more accurate mimicry of the microenvironment, challenging cells to migrate and invade in a manner consistent with their behavior in vivo [53], nanofiber scaffolds offer several practical advantages such as a simpler setup, scaling-up capability, and easy cell recovery for downstream analysis. In addition, our results suggest that, for glioma cells, migration in nanofibers and brain slices is comparable and highly sensitive to subtoxic doses of antimigratory compounds that may lack effect on rigid two-dimensional surfaces or may require much higher concentrations to elicit a comparable effect.
Finally, the possibility of measuring cell migration out of tissue explants suggests that this model could potentially be used as a bioassay for drug testing in specimens and tumorspheres derived from individual patients.
Supplementary Material
Abbreviations
- TCPS
tissue culture polystyrene
Footnotes
This work was supported by grants from the National Institutes of Health (1R01CA152065-01 to M.S.V. and 1R43CA150763-01 to J.J.L. and J.K.J.) and the National Science Foundation (STTR 1010406 to J.K.J.). J.J.L. and J.K.J. hold Ohio State University-sponsored patent applications for the design of nanofiber-based scaffolds.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN. Molecular pathology of malignant gliomas. Ann Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 3.Decaestecker C, Debeir O, Van Ham P, Kiss R. Can anti-migratory drugs be screened in vitro? A review of 2D and 3D assays for the quantitative analysis of cell migration. Med Res Rev. 2007;27:149–176. doi: 10.1002/med.20078. [DOI] [PubMed] [Google Scholar]
- 4.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Joy AM, Beaudry CE, Tran NL, Ponce FA, Holz DR, Demuth T, Berens ME. Migrating glioma cells activate the PI3-K pathway and display decreased susceptibility to apoptosis. J Cell Sci. 2003;116:4409–4417. doi: 10.1242/jcs.00712. [DOI] [PubMed] [Google Scholar]
- 6.Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Demuth T, Ross KR, Berens T, Coons SW, Watts G, Trent JM, et al. Glioma cell motility is associated with reduced transcription of proapoptotic and proliferation genes: a cDNA microarray analysis. J Neurooncol. 2001;53:161–176. doi: 10.1023/a:1012253317934. [DOI] [PubMed] [Google Scholar]
- 7.Lamszus K, Kunkel P, Westphal M. Invasion as limitation to antiangiogenic glioma therapy. Acta Neurochir Suppl. 2003;88:169–177. doi: 10.1007/978-3-7091-6090-9_23. [DOI] [PubMed] [Google Scholar]
- 8.Zhai GG, Malhotra R, Delaney M, Latham D, Nestler U, Zhang M, Mukherjee N, Song Q, Robe P, Chakravarti A. Radiation enhances the invasive potential of primary glioblastoma cells via activation of the Rho signaling pathway. J Neurooncol. 2006;76:227–237. doi: 10.1007/s11060-005-6499-4. [DOI] [PubMed] [Google Scholar]
- 9.Viapiano MS, Lawler SE. Glioma invasion: mechanisms and therapeutic challenges. In: Van Meir E, editor. CNS Cancer: Models, Prognostic Factors and Targets. Totowa, NJ: Humana Press; 2009. pp. 1219–1252. [Google Scholar]
- 10.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson J, Nowicki OM, Lee CH, Chiocca AE, Viapiano MS, Lawler SE, Lannutti JJ. Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy. Tissue Eng Part C. 2009;15:531–540. doi: 10.1089/ten.tec.2008.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia. 2006;53:799–808. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- 13.Devarajan E, Huang S. STAT3 as a central regulator of tumor metastases. Curr Mol Med. 2009;9:626–633. doi: 10.2174/156652409788488720. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 16.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7:1756–1770. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a smallmolecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Hutzen B, Li PK, Ball S, Zuo M, DeAngelis S, Foust E, Sobo M, Friedman L, Bhasin D, et al. A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells. Neoplasia. 2010;12:39–50. doi: 10.1593/neo.91196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Kong LL, Matthews RT, Viapiano MS. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem. 2008;283:24848–24859. doi: 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim H, Hu B, Viapiano MS. Reduced expression of the hyaluronan and proteoglycan link proteins in malignant gliomas. J Biol Chem. 2009;284:26547–26556. doi: 10.1074/jbc.M109.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 22.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghibaudo M, Trichet L, Le Digabel J, Richert A, Hersen P, Ladoux B. Substrate topography induces a crossover from 2D to 3D behavior in fibroblast migration. Biophys J. 2009;97:357–368. doi: 10.1016/j.bpj.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh YM, Kim JK, Choi Y, Choi S, Yoo JY. Prediction and experimental validation of novel STAT3 target genes in human cancer cells. PLoS One. 2009;4:e6911. doi: 10.1371/journal.pone.0006911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang K, Krous LC, Knowlton N, Chen Y, Frank MB, Cadwell C, Centola M, Jarvis JN. Ablation of Stat3 by siRNA alters gene expression profiles in JEG-3 cells: a systems biology approach. Placenta. 2009;30:806–815. doi: 10.1016/j.placenta.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Merchant JL. What lurks beneath: IL-11, via Stat3, promotes inflammation-associated gastric tumorigenesis. J Clin Invest. 2008;118:1628–1631. doi: 10.1172/JCI35344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao RD, Mladek AC, Lamont JD, Goble JM, Erlichman C, James CD, Sarkaria JN. Disruption of parallel and converging signaling pathways contributes to the synergistic antitumor effects of simultaneous mTOR and EGFR inhibition in GBM cells. Neoplasia. 2005;7:921–929. doi: 10.1593/neo.05361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Brown C, Buettner R, Hedvat M, Starr R, Scuto A, Schroeder A, Jensen M, Jove R. Sorafenib induces growth arrest and apoptosis of human glioblastoma cells through the dephosphorylation of signal transducers and activators of transcription 3. Mol Cancer Ther. 2010;9:953–962. doi: 10.1158/1535-7163.MCT-09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senft C, Priester M, Polacin M, Schroder K, Seifert V, Kogel D, Weissenberger J. Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J Neurooncol. 2011;101:393–403. doi: 10.1007/s11060-010-0273-y. [DOI] [PubMed] [Google Scholar]
- 31.Narayana A, Kunnakkat SD, Medabalmi P, Golfinos J, Parker E, Knopp E, Zagzag D, Eagan P, Gruber D, Gruber ML. Change in pattern of relapse after antiangiogenic therapy in high-grade glioma. Int J Radiat Oncol Biol Phys. 2010 doi: 10.1016/j.ijrobp.2010.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 33.McKee CT, Last J, Russell P, Murphy CJ. Indentation versus tensile measurements of Young's modulus for soft biological tissues. Tissue Eng Part B Rev. 2011;17:155–164. doi: 10.1089/ten.teb.2010.0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 35.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki SO, Iwaki T. Dynamic analysis of glioma cells: looking into “movement phenotypes”. Neuropathology. 2005;25:254–262. doi: 10.1111/j.1440-1789.2005.00626.x. [DOI] [PubMed] [Google Scholar]
- 37.Blando JM, Carbajal S, Abel E, Beltran L, Conti C, Fischer S, DiGiovanni J. Cooperation between Stat3 and Akt signaling leads to prostate tumor development in transgenic mice. Neoplasia. 2011;13:254–265. doi: 10.1593/neo.101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atkinson GP, Nozell SE, Benveniste ET. NF-κB and STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother. 2010;10:575–586. doi: 10.1586/ern.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dasgupta A, Raychaudhuri B, Haqqi T, Prayson R, Van Meir EG, Vogelbaum M, Haque SJ. Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur J Cancer. 2009;45:677–684. doi: 10.1016/j.ejca.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li GH, Wei H, Lv SQ, Ji H, Wang DL. Knockdown of STAT3 expression by RNAi suppresses growth and induces apoptosis and differentiation in glioblastoma stem cells. Int J Oncol. 2010;37:103–110. [PubMed] [Google Scholar]
- 43.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 44.Fuh B, Sobo M, Cen L, Josiah D, Hutzen B, Cisek K, Bhasin D, Regan N, Lin L, Chan C, et al. LLL-3 inhibits STAT3 activity, suppresses glioblastoma cell growth and prolongs survival in a mouse glioblastoma model. Br J Cancer. 2009;100:106–112. doi: 10.1038/sj.bjc.6604793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA, Mischel PS, Louis DN, Bonni A. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci. 2008;28:5870–5878. doi: 10.1523/JNEUROSCI.5385-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gharavi NM, Alva JA, Mouillesseaux KP, Lai C, Yeh M, Yeung W, Johnson J, Szeto WL, Hong L, Fishbein M, et al. Role of the Jak/STAT pathway in the regulation of interleukin-8 transcription by oxidized phospholipids in vitro and in atherosclerosis in vivo. J Biol Chem. 2007;282:31460–31468. doi: 10.1074/jbc.M704267200. [DOI] [PubMed] [Google Scholar]
- 47.Tong KM, Shieh DC, Chen CP, Tzeng CY, Wang SP, Huang KC, Chiu YC, Fong YC, Tang CH. Leptin induces IL-8 expression via leptin receptor, IRS-1, PI3K, Akt cascade and promotion of NF-κB/p300 binding in human synovial fibroblasts. Cell Signal. 2008;20:1478–1488. doi: 10.1016/j.cellsig.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senft C, Polacin M, Priester M, Seifert V, Kogel D, Weissenberger J. The nontoxic natural compound curcumin exerts anti-proliferative, anti-migratory, and anti-invasive properties against malignant gliomas. BMC Cancer. 2010;10:491. doi: 10.1186/1471-2407-10-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debidda M, Wang L, Zang H, Poli V, Zheng Y. A role of STAT3 in Rho GTPase-regulated cell migration and proliferation. J Biol Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 51.Salhia B, Hwang JH, Smith CA, Nakada M, Rutka F, Symons M, Rutka JT. Role of myosin II activity and the regulation of myosin light chain phosphorylation in astrocytomas. Cell Motil Cytoskeleton. 2008;65:12–24. doi: 10.1002/cm.20240. [DOI] [PubMed] [Google Scholar]
- 52.Marchesi F, Piemonti L, Mantovani A, Allavena P. Molecular mechanisms of perineural invasion, a forgotten pathway of dissemination and metastasis. Cytokine Growth Factor Rev. 2010;21:77–82. doi: 10.1016/j.cytogfr.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Palfi S, Swanson KR, De BS, Chretien F, Oliveira R, Gherardi RK, Kros JM, Peschanski M, Christov C. Correlation of in vitro infiltration with glioma histological type in organotypic brain slices. Br J Cancer. 2004;91:745–752. doi: 10.1038/sj.bjc.6602048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.