Abstract
Single-nucleotide polymorphisms (SNPs) around IL28B are associated with spontaneous hepatitis C virus (HCV) clearance of genotypes 1 and 3 in white and African-American populations. This study investigated whether the IL28B SNP (rs12979860) is associated with spontaneous clearance of HCV, principally genotype 4, in 162 Egyptians (80 with clearance). The protective C allele was more common in those with spontaneous clearance (76.3% vs 57.9%; P = .0006). Individuals with clearance were 3.4 (95% confidence interval, 1.8–6.5) times more likely to have C/C genotype. Thus, IL28B plays a role in spontaneous clearance of HCV genotype 4 in North Africa.
Spontaneous clearance of hepatitis C virus (HCV) occurs in ∼30% of acute infections [1]. Several host factors are determinants of spontaneous clearance including race, sex, human immunodeficiency virus (HIV) and hepatitis B virus coinfection status, and host genetics [1, 2]. The strongest genetic factor associated with spontaneous clearance described to date is single-nucleotide polymorphisms (SNPs) around IL28B, in particular rs8099917 and rs12979860 (reviewed in [3]). These SNPs have been studied largely in African American and white populations, which are primarily infected with HCV genotypes 1, 2, and 3 [4, 5]. It is not known whether the SNPs in this genomic region are also important in spontaneous clearance of other genotypes from other areas of the world. Such information may be helpful in understanding the mechanism of potential involvement of interleukin 28B (IL28B), a type III interferon (IFN), also called IFN-λ3, on HCV clearance.
Egypt has the highest HCV prevalence in the world. An estimated 10%–20% of the Egyptian population is chronically HCV infected, a seroprevalence that is ∼10- to 20-fold higher than in the United States. More than 90% of HCV infections in Egypt are genotype 4, which is found primarily in the Middle East and Central Africa and has a worldwide prevalence of 2% [6, 7].
The C allele of rs12979860, which is ∼3 kb upstream of IL28B, is associated with a favorable response to HCV treatment in patients with genotype 4 infection [8, 9]; however, the effects of this SNP in spontaneous clearance of genotype 4 is unknown. In addition, the prevalence of the favorable C allele in North Africa has never been investigated even though its prevalence in African Americans and in Africans from southern Africa is significantly lower than in whites [4].
In this study, we tested the hypothesis that rs12979860 is associated with spontaneous clearance of genotype 4 HCV in a North African population. To test this hypothesis, we used a prospectively followed up Egyptian cohort that was well characterized in terms of the natural outcome of HCV infection (clearance vs persistence).
Methods.
In 1997, half of the households of a village in the Nile Delta, Aghour El Soughra, were systematically selected and interviewed with a structured questionnaire to identify exposures potentially related to HCV acquisition. Adults and children >10 years old were interviewed themselves, with the head of the household providing information on children <10 years old. The subjects underwent annual interviews and serosurveys for 3 years. Household members who did not participate in the initial survey were invited to join during subsequent surveys. Of the 894 subjects examined, 366 were HCV antibody negative; 259 had inconsistent HCV antibody and/or HCV RNA results so were excluded from this study. The remaining 279 subjects (31.2%) had ≥2 positive HCV antibody tests. Of these, 130 (46.6%) had ≥2 negative HCV RNA tests and were classified as having HCV clearance, and 149 had ≥2 positive HCV RNA tests and were classified as persistently HCV infected. Of the 279 subjects with confidently determined HCV infection outcome, 169 were available for genotyping. There were no significant differences in median age (43 vs 38 years, respectively) or sex (41% vs 45% male) between the 169 subjects who were genotyped and the 110 with identified outcomes who were not genotyped. Based on prior studies in Egypt, including the region where this village is located, >90% of HCV-infected individuals carried HCV genotype 4 [6]. The presumed HCV transmission mode was nonsterile injections. All study participants were HIV negative. Only 2.4% of the subjects (4/169) tested positive for hepatitis B surface antigen, with no difference between HCV clearance and persistently infected groups. Informed consent was obtained from participants, or, in the case of minors, their parents. The study was approved by the Egyptian Ministry of Health and Population and the institutional review boards of the University of Maryland and Johns Hopkins University.
The initial HCV antibody at recruitment was determined with the Abbott HCV EIA 2.0 (Abbott Laboratories). Samples from subsequent years were tested with the third-generation EIA from the same manufacturer. Genomic DNA was isolated from peripheral blood mononuclear cells using a Qiagen DNA extraction kit (Qiagen). Genotyping of the rs12979860 SNP was performed using the ABI TaqMan allelic discrimination kit and the ABI7900HT sequence Detection System (Applied Biosystems) [4].
Hardy–Weinberg equilibrium was determined by χ2 test with 1 degree of freedom. Allele frequencies for rs12979860 were calculated and compared between those with HCV clearance and those with HCV persistence, using logistic regression (SAS, version 10). Odds ratios of >1 were associated with viral clearance, and odds ratios of <1 with persistence. Differences were considered statistically significant at P < .05 (2 sided).
Results and Discussion.
Of the 169 subjects available for genotyping, the mean age of the HCV clearance and persistence groups was similar: 44 years and 41 years, respectively. Female sex was more common in the clearance (62.7%) than in the persistently infected group (55.8%). The rs12979860 SNP was successfully typed in 162 of the 169 subjects (95.9%), including 80 in the HCV clearance group and 82 in the HCV persistence group. The SNP was in Hardy–Weinberg equilibrium (P = .67).
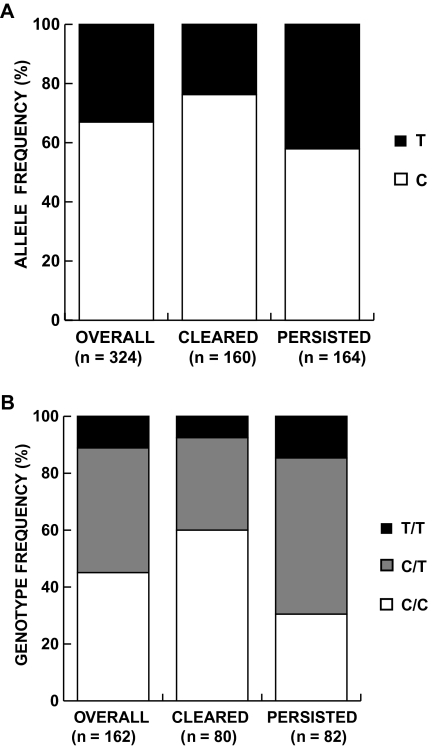
The overall frequency of the protective C allele was 67%, similar to that in European populations and higher than in southern African populations [4]. The C allele frequency was higher in the clearance than in the persistence group (76.3% vs 57.9%; P = .0006) (Figure 1). Individuals who carried the C allele were 2.3 (95% confidence interval, 1.4–3.8) times more likely to have HCV clearance than those who carried the T allele (P = .0005) (Table 1). When the diploid genotype was considered, the overall frequency of the protective C/C genotype was 45%, and this was also more common in subjects with HCV clearance than in those with persistence (66% vs 34%, respectively) (Figure 1). Subjects with HCV clearance were 3.4 times more likely to have C/C than they were to have T/T or C/T combined (95% confidence interval, 1.8–6.5; P = .0002) (Table 1). The C allele has a recessive protective effect, because C/C was similarly protective when compared with T/T or C/T separately (Table 1). The addition of sex to these models did not change the magnitude or strength of the associations.
Figure 1.
The distribution of IL28B (rs12979860) variants (y-axis) among hepatitis C virus (HCV)–infected Egyptian subjects overall and stratified by HCV clearance and persistence status (x-axis) is shown for chromosomes with C or T alleles (A) and diploid genotype (C/C, C/T, or T/T) (B). The number of observations in each group is shown in parentheses.
Table 1.
Predictive Value of IL28B rs12979860 Genotype for Natural Clearance of Hepatitis C Virus Genotype 4
| rs12979860 Genotype | Allele frequency in those with spontaneous clearance (%) | Allele frequency in those with persistence (%) | Comparison | OR (95% CI) | P |
| C (n = 217) | 122 (76.3) | 95 (57.9) | C vs T | 2.3 (1.4–3.8) | .0005 |
| T (n = 107) | 38 (23.8) | 69 (42.1) | … | … | … |
| C/C (n = 73) | 48 (60.0) | 25 (30.5) | C/C vs T/T | 3.8 (1.3–11.5) | .0126 |
| C/T (n = 71) | 26 (32.5) | 45 (54.9) | C/C vs C/T | 3.3 (1.7–6.6) | .0005 |
| T/T (n = 18) | 6 (7.5) | 12 (14.6) | C/C vs C/T and T/T | 3.4 (1.8–6.5) | .0002 |
Of the 162 subjects in this study for whom the rs12979860 SNP was successfully typed, 80 of the subjects were associated with spontaneous clearance and 82 with persistence.
Abbreviations: CI, confidence interval; OR, odds ratio.
This study, the first to examine genetic variation in the IL28B genomic region and the natural history of principally HCV genotype 4 infections, demonstrates that the rs12979860 C/C genotype is strongly associated with spontaneous clearance. Notably, the magnitude of this association is similar to that seen in HCV genotype 1 infections [4, 5] and the association was found in an Egyptian population, a group that has not previously been studied. Thus, the mechanism by which IL28B affects spontaneous HCV clearance operates across HCV genotypes and across ethnic populations.
Only a few studies have examined IL28B and spontaneous HCV clearance, and these study populations were primarily infected with HCV genotypes 1 and 3 [4, 5, 10]; thus, it was not known whether this genomic region would be equally important in the clearance of other less common HCV genotypes. It is plausible that the association would not be present in all HCV genotypes, because in studies of IL28B SNPs and HCV treatment response, associations are variable based on HCV genotype. In genotype 1, there is a strong association of IL28B SNPs, including rs12979860, with treatment-induced clearance [3, 5]. However, in studies of individuals infected with genotypes 2 and 3, the association of IL28B genotype on the outcome is less pronounced [5, 11], which may be related to the higher susceptibility of these genotypes to IFN.
Patients with HCV genotype 4 have been included in only 4 studies of IL28B and HCV treatment outcomes. The first was a genome-wide association study performed in Swiss cohorts [5], wherein HCV genotype 1 and 4 patients were combined and an association was found between a SNP linked to rs12979860 and treatment-induced clearance. However, the association between IL28B and HCV genotype 4 was not addressed specifically, so it is not known whether the association with genotype 4 was weaker than with genotype 1. In a group of Austrian patients, those infected with HCV genotype 4 were evaluated separately and the C/C at rs12979860 was associated with sustained virologic response [8]. Two studies in HIV-HCV–coinfected patients found an association with rs12979860 and treatment response in patients infected with HCV genotype 4 [9, 12]. Taken together, these studies suggest that IL28B genotype is a determinant of treatment-induced clearance in HCV genotype 4 infections.
This study is the first to genotype an IL28B SNP in a North African population. Interestingly, the prevalence of the C allele in the Egyptian population is more similar to that in populations from European ancestry than that in populations from African ancestry [4]. Another unique aspect of this study is the high homogeneity in both the study population and the HCV genotype. The only other study of a homogeneous population and HCV genotype was in German women in the anti-D cohort who were infected with genotype 1b, which also showed a strong association of C/C genotype with rs12979860 and HCV clearance [13].
One limitation of this study is that HCV sequence data could not be obtained from those with viral clearance; thus, we were unable to determine whether the association of IL28B genotype with outcome varies between different strains of HCV genotype 4. However, phylogenetic analyses of HCV strains in prior studies from Egypt showed geographic differences in subgenotype distribution but not region-specific phylogenetic clustering [14, 15]. This strongly suggests that the observed association in this study of IL28B genotype with outcome is not likely to be attributable to a particular phylogenetic cluster of HCV strains circulating in the village.
In conclusion, this study clearly demonstrates that the rs12979860 IL28B SNP has an important role in spontaneous clearance of principally HCV genotype 4 infections in an Egyptian population, confirming the importance of IL28B in the natural history of acute HCV infections. It is a priority to determine the mechanisms through which IL28B promotes defense against HCV and to determine the full range of viruses affected by these mechanisms.
Notes
Acknowledgements.
The authors thank G. Thomas Strickland for his contributions to the Egyptian cohort used in this study and to the subjects who participated in this study.
Funding.
This work was supported in part by the Egyptian Ministry of Health United States Agency for International Development (USAID)–funded Schistosomiasis Research Project (263-014.2, grant 02-04-22); and the National Institutes of Health (R01-Da13324).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Thomas DL, Astemborski J, Rai RM, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–6. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 2.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 3.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–76. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45. doi: 10.1053/j.gastro.2009.12.056. 1345 e1–7. [DOI] [PubMed] [Google Scholar]
- 6.Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis. 2000;182:698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen MH, Keeffe EB. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005;3:S97–S101. doi: 10.1016/s1542-3565(05)00711-1. [DOI] [PubMed] [Google Scholar]
- 8.Stattermayer AF, Stauber R, Hofer H, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naive patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–350.e2. doi: 10.1016/j.cgh.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 9.Pineda JA, Caruz A, Rivero A, et al. Prediction of response to pegylated interferon plus ribavirin by IL28B gene variation in patients coinfected with HIV and hepatitis C virus. Clin Infect Dis. 2010;51:788–95. doi: 10.1086/656235. [DOI] [PubMed] [Google Scholar]
- 10.Clausen LN, Weis N, Astvad K, et al. Interleukin-28B polymorphisms are associated with hepatitis C virus clearance and viral load in a HIV-1-infected cohort. J Viral Hepat. 2011;18:e66–74. doi: 10.1111/j.1365-2893.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- 11.Scherzer TM, Hofer H, Staettermayer AF, et al. Early virologic response and IL28B polymorphisms in patients with chronic hepatitis C genotype 3 treated with peginterferon alfa-2a and ribavirin. J Hepatol. 2011;54:866–71. doi: 10.1016/j.jhep.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Rallon NI, Naggie S, Benito JM, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–9. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tillmann HL, Thompson AJ, Patel K, et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139:1586–92. doi: 10.1053/j.gastro.2010.07.005. 1592 e1. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Hamid M, El-Daly M, Molnegren V, et al. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88:1526–31. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 15.Elkady A, Tanaka Y, Kurbanov F, et al. Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication. J Med Virol. 2009;81:1015–23. doi: 10.1002/jmv.21492. [DOI] [PubMed] [Google Scholar]