Abstract
Background. Many studies of sexual behavior have shown that individuals vary greatly in their number of sexual partners over time, but it has proved difficult to obtain parameter estimates relating to the dynamics of human immunodeficiency virus (HIV) transmission except in small-scale contact tracing studies. Recent developments in molecular phylodynamics have provided new routes to obtain these parameter estimates, and current clinical practice provides suitable data for entire infected populations.
Methods. A phylodynamic analysis was performed on partial pol gene sequences obtained for routine clinical care from 14 560 individuals, representing approximately 60% of the HIV-positive men who have sex with men (MSM) under care in the United Kingdom.
Results. Among individuals linked to others in the data set, 29% are linked to only 1 individual, 41% are linked to 2–10 individuals, and 29% are linked to ≥10 individuals. The right-skewed degree distribution can be approximated by a power law, but the data are best fitted by a Waring distribution for all time depths. For time depths of 5–7 years, the distribution parameter ρ lies within the range that indicates infinite variance.
Conclusions. The transmission network among UK MSM is characterized by preferential association such that a randomly distributed intervention would not be expected to stop the epidemic.
Human immunodeficiency virus (HIV) infection, like all sexually transmitted infections, spreads along the sexual contact network, and contact tracing played a key role in establishing the etiology of AIDS [1]. However, more recent studies have struggled to achieve any reconciliation between contact networks revealed by interview data and transmission networks reconstructed from the viral phylogeny [2, 3]. There are several possible reasons for this but 2 important factors are the long period of infectiousness (in the absence of therapy) and the low average risk of transmission per sexual contact [4]. The consequence of the first factor is that although sexual contact networks constructed from interview data usually work within a time frame of 1 year [5, 6], the HIV infection of the subject may have occurred at any time over a period of several years. The consequence of the second factor is reduction of the confidence that any particular contact was associated with infection. Other possible reasons for the lack of agreement that may be associated with particular risk groups include network complexity and anonymous sex. In addition, earlier studies of HIV-infected communities using sequence data were generally based on a small sample of the infected population, reducing the probability of including individuals from the same transmission network [7]. More recently, often working within defined risk groups, a number of studies have identified transmission networks by means of sequence analysis [8–10]. However, this static picture alone does not permit the quantitative description of the network through which HIV has been transmitted [11].
We have introduced an approach to estimating the HIV transmission network structure of a population based on the analysis of HIV sequences generated in the course of routine clinical care to test for antiretroviral resistance [12, 13]. Under UK guidelines from 2003, such tests are now performed for all patients entering therapy, which is recommended for patients with a CD4 cell count of ≤350 cells/μL [14]. Central collection of the assay results from the majority of clinics performing the assays in the United Kingdom allowed a data set of sequences from >26 000 patients to be constructed, including approximately two-thirds of the total number of men who have sex with men (MSM) receiving care for HIV infection in the United Kingdom [15]. We have already demonstrated that phylogenetic analysis of large population-based data sets reveals much more of the transmission pattern than has been seen before [12] and that in combination with relaxed clock models it has shed new light on the dynamics of the HIV infection epidemic in both the MSM and heterosexual contact risk groups.
Here we extend our earlier analysis [12] to the MSM population of the entire United Kingdom. Using viral sequences from almost 60% of MSM diagnosed with HIV infection in 2009, we infer the degree distribution of the transmission networks, based on the time-resolved viral phylogenies, for a range of time depths. We have already shown that that the degree distribution inferred in this way can be approximated by a power law [13], as shown earlier for sexual contact networks inferred from interview data [5]. We here test a series of distributions for fit to the data to determine whether a preferential attachment model is, as has been suggested, the most appropriate model for transmission in this group.
METHODS
Data
HIV sequence data from the protease and partial reverse transcriptase coding regions were obtained from the UK HIV Drug Resistance Database (UKHIVRDB; http://www.hivrdb.org; 2007 download). Before the data set was released for analysis, the records were fully anonymized and delinked from clinical identifiers. In most cases, up to 1200 nucleotides from the start of the protease coding region are available, according to the method used for genotyping. For this study, sequences <850 nucleotides in length were removed, as were sequences with a large number of ambiguities. A check for identical or near-identical sequences (>99.9% identity) was applied to the entire data set. When these were associated with different patient identifications, the sequence groups affected were examined in detail and individual decisions were made whether to discard them on the basis of the available background information. The first available sequence, which was usually obtained before antiretroviral treatment was initiated, was selected from patients with multiple sequences in the database, giving a total of 25 136 sequences each from different patients. In a current substudy with access to sequential viral load data, approximately 12% of the first sequences reported as being obtained before antiretroviral therapy (ART) was initiated appear from analysis of the viral loads to have actually been taken after ART initiation. HIV-1 subtyping was then applied by means of a combination of the Rega [16] method and nucleotide distance comparisons within the data set, which identified 14 560 sequences belonging to subtype B (Figure 1). Because submission of the entire sequence data set to public databases would permit transmission networks to be identified and thus risk breaching patient confidentiality, following Kouyos et al [10] we have submitted a random sample of 10% to GenBank under accession numbers JN100661–JN101948.
Figure 1.
Flowchart of data manipulation and processing. NJ, neighbor-joining; UK HIVRDB, United Kingdom Human Immunodeficiency Virus Drug Resistance Database.
Ethical approval for the use of anonymized data in this work was given by the London Multicentre Research Ethics Committee (MREC/01/2/10; 5 April 2001). Data held in the UKHIVRDB can be accessed for collaborative projects if a project proposal is approved by the steering committee. The proposal form can be downloaded from the database Web site at http://www.hivrdb.org.
Phylogenetic Analysis
The size of the data set required a hierarchical approach to define clusters. For the initial characterization, a genetic distance threshold was calculated following stripping of antiretroviral resistance-associated sites from the sequence data as previously described [13] to avoid clustering artifacts among those sequences that might be affected by ART. The genetic distances were estimated for all pairwise comparisons by means of the HKY+ γ model [17]), and bootstrap testing with PAUP* software [18] was used. PAUP* (version 4.0) output was analyzed using a combination of Python and R scripts. Thresholds of 3.5%, 4.0%, 4.5%, 5%, and 5.5% were examined using a range of bootstrap percentage support values (Supplementary Table 1); 4.5% genetic distance plus 95% bootstrap support was adopted to identify sequences for further analysis as previously described [12]. A total of 5809 sequences with phylogenetic associations that could be resolved using the neighbor-joining method [19] were analyzed using BEAST (version 1.6) software as previously described [13], but with the use of random collections of clusters in 30 pools of approximately 200 sequences per pool for computational feasibility (Figure 1).
Network Creation
A network representing possible HIV transmissions was inferred from the BEAST consensus trees from all the pooled sequence data. The nodes of the network are the HIV sequences, but they represent individuals because only 1 sequence for each individual was used. Two nodes are linked by an edge if the highest posterior density time to the most recent common ancestor (MRCA) of the respective viral sequences in the BEAST consensus tree is less than or equal to some network depth cutoff value. For example, 2 individuals are linked if their viral sequences are predicted to have diverged (ie, the transmission event) <5 years before the most recent sampled sequence. The tree data were processed using a custom R [20] script utilizing the mrca function in the R package ape [21].
Networks of varying time depths (1–16 years) were created by concatenating the subnetworks from the 30 individual consensus trees from the pooled data sets. A network with a time depth of 31 years, representing the maximum depth and maximum number of connections between the sequences, was also created for comparison. Clusters were identified using the clusters function within the R package igraph.
Network Shape Analysis
The degree of a node (number of links per node) approximates the number of possible infected contacts an individual has had within the period of the network. We used the statnet package [22] within R to fit several models to the degree distribution of the nodes within our networks, in addition to fitting a simple power law (not shown). Maximum likelihood parameter estimates, log likelihood values, Akaike information criterion for small sample sizes (AICC) values, and Bayesian information criterion (not shown) values were obtained for the discrete Pareto, negative binomial, Yule, and Waring distributions. For network depths of 3–7 years we also performed 1000 bootstrap replicates to obtain the 95% confidence intervals on the parameters and AICC scores.
RESULTS
Cluster Size Distribution
The UKHIVRDB holds HIV sequence data from the protease and partial reverse transcriptase coding regions obtained in the course of routine clinical care, submitted by collaborating clinical virology laboratories across the entire United Kingdom. We analyzed the 2007 download containing sequences from 25 136 infected individuals, of which 14 560 sequences were classified as subtype B. Direct information on risk activity associated with transmission was not available for this data set, but in an earlier data set from the UKHIVRDB from 2004 for which 1288 individuals infected with subtype B virus had the associated risk group data, the proportions were recorded as follows: MSM, 82% (unpublished data); heterosexual contact, 10%; and injection drug use, 6%. In the 2010 data set of the UKHIVRDB, of 17 334 individuals infected with subtype B, 83% are recorded as MSM (S. Hue, private communication, 28 April 2011), 12% as heterosexual contact, and 3% as injection drug use. From this we can reasonably assume that a little more than 80% of individuals analyzed in this data set were MSM.
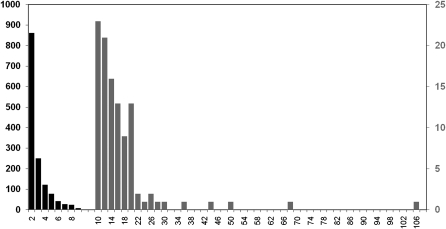
At a genetic distance threshold of 4.5%, 7617 individuals (52%) had a link to 1 or more other individuals (Figure 1). Sequences from 5809 of these individuals could be resolved in bootstrapped neighbor-joining trees. Of these, 1728 (29%) were linked only to 1 other, 2408 (41%) were linked to 2–10 others, and 1673 (29%) were linked to ≥10 others, in 97 clusters. As this indicates, the cluster size distribution (Figure 2) is highly right-skewed, as has been described previously for this risk group [12].
Figure 2.
Cluster size distribution of the human immunodeficiency virus transmission network among men who have sex with men in the United Kingdom. Horizontal scale unit, 2; vertical scale, number of clusters; left axis, cluster size of 2–9 (black); right axis, cluster size of 10–106 (gray).
Transmission Dynamics
For a complete analysis of the dynamics of HIV transmission in this population, we used BEAST to sample the 1524 clusters randomly in 30 pools containing all 5809 individuals and obtain dated phylogenies for all of them. The epidemic dynamics were reconstructed from these dated phylogenies. As discussed elsewhere [12], because each sequence represents a different individual, the estimated internode intervals represent the maximum intervals between transmissions. The median of the internode intervals for the entire population was 22 months, with 16% of intervals being ≤6 months. Equivalent estimates of 14 months and 25%, respectively, were obtained earlier for large clusters (≥10 individuals) identified in a single large London clinic [12]. Restricting the comparison to large clusters from the current study gives a median of 17 months and 20% of intervals being ≤6 months. These figures confirm the very important role for early infection in onward transmission in this risk group. The full distribution of internode intervals is given in Supplementary Figure 1.
Network Structure
The network structure was reconstructed using the time to the most recent common ancestors estimated in BEAST for all individuals at a given time depth. Time depths of 1–16 years were examined and compared with the maximal depth of 31 years to test whether cluster size was affected by time depth. Even within the maximum depth network, 688 separate clusters could be identified, indicating that the process of creating networks from pooled data did not artificially create large clusters. The node degree statistics for networks of time depths 1–16 and 31 are displayed in Supplementary Table 2; Figure 3 shows the structure of a large 42-node cluster as revealed at time depths of 3, 5, and 7 years.
Figure 3.
Estimation of network structure. The reconstruction of the largest network cluster is shown at 3 time depths: 3 years, 5 years and 7 years. Networks were initially reconstructed from the dated trees generated by BEAST at all time depths of 1–16 years. The networks for time depths of 3–7 years were used in the distribution fitting (Table 1; Figure 2).
Model Fitting to Degree Distribution
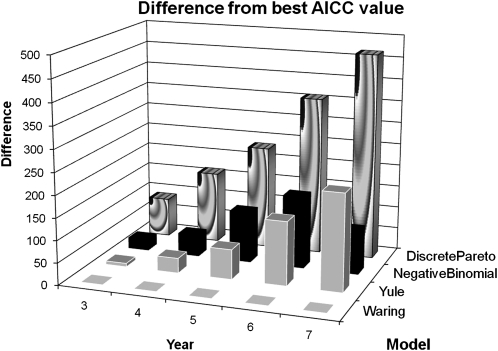
In our previous analysis of the UK non-B subtype HIV infection epidemic, we tested the degree distribution observed for fit to a power law [13]. A power law is an approximation to a scale-free network, which can arise from a preferential attachment process, whereby individuals are more likely to link to individuals who already have multiple links. There are explicit models that describe the distribution arising from a preferential attachment process, and in order to apply a more rigorous test, we have compared the HIV transmission network of the UK MSM population to negative binomial, discrete Pareto, Yule, and Waring distributions. These distributions reflect different processes; the simplest is the negative binomial distribution, which is generated if each individual acquires partners at a constant rate, drawn from a γ distribution. The discrete Pareto distribution is a power law distribution, whereas the Yule and Waring distributions arise from specific preferential attachment models [6]. Both the Yule and Waring distributions incorporate (1) the probability that a new link is made to a previously inactive individual and (2) the probability that a new link is made to a person with k partners. The Waring distribution additionally allows for a third class of random, nonpreferential attachments [23, 24]. We used maximum likelihood to estimate parameter values and AICC to compare goodness of fit and found that for all time depths the Waring distribution was the preferred distribution (Figure 4). The greatest difference, at all time depths, was with the discrete Pareto distribution, which clearly fitted less well than any other distribution. The Yule and negative binomial distributions provided the next best fits to the Waring distribution, with only slight differences in AICC for time depths of 3–4 years. An increasing difference for 5 and 6 years in the negative binomial distribution made the Yule distribution our second choice, but at a time depth of 7 years the negative binomial distribution was again preferred of these 2 distributions. The values of the parameter estimates for the Waring and negative binomial distributions are given in Table 1.
Figure 4.
Comparison of the fit of different distributions to the observed network structure. The fit of 4 different distributions, including 2 based on explicit preferential attachment models, to the observed data is shown as the difference between the Akaike information criterion for small sample sizes (AICC) value for each distribution at each time depth and the lowest AICC value at that time depth for time depths 3–7 years (see Table 1).
Table 1.
Parameters of the Waring and Negative Binomial Distributions Estimated by Maximum Likelihood From the HIV Transmission Network for Men Who Have Sex With Men in the United Kingdom
| Distribution, parameter |
||||
| Waring |
Negative binomial |
|||
| Year | ρ | α | k | μ |
| 3 | 3.8 | 0.4 | 0.33 | 0.7 |
| 4 | 3.2 | 0.5 | 0.33 | 1.3 |
| 5 | 2.8 | 0.7 | 0.32 | 1.8 |
| 6 | 2.7 | 1.0 | 0.34 | 2.4 |
| 7 | 2.7 | 1.2 | 0.35 | 2.9 |
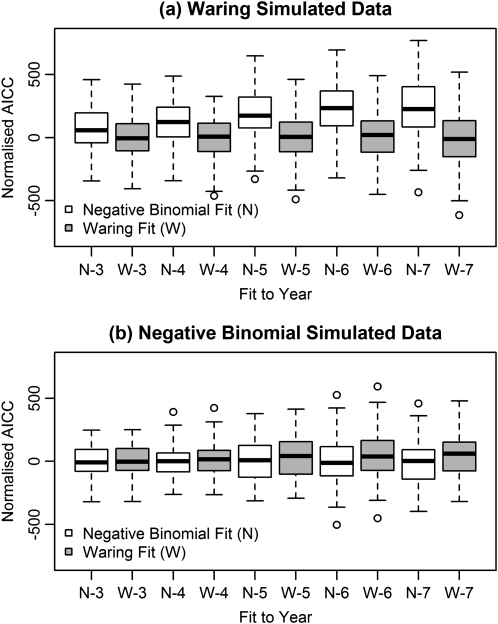
The difference in fit between the Waring and negative binomial distributions was examined by simulation, using parameter values from the observed data. The bootstrap confidence intervals from these simulations (Figure 5) indicate that the difference in fit was in many cases not significant. Although the negative binomial distribution never gave a lower AICC value than that given by a Waring distribution to a simulated Waring distribution (Figure 5), approximately 10% of bootstrap samples from a negative binomial distribution were fitted better by a Waring distribution. This indicates the level of uncertainty that remains over the existence of a preferential attachment effect in this population.
Figure 5.
Test of the difference in fit between the Waring and negative binomial distributions. Populations were simulated using the parameter values estimated from the data under a Waring distribution (A) and a negative binomial distribution (B), and both distributions were tested for goodness of fit using the Akaike information criterion for small sample sizes (AICC). The simulations show that the negative binomial never fits as well to a Waring distribution as the Waring distribution, although the difference is not great. However, the converse does not hold: the Waring distribution can provide a fit that is not perceptively poorer to a negative binomial distribution.
DISCUSSION
The 14 560 HIV subtype B sequences analyzed here represented two-thirds (68%) of the 21 700 MSM in the United Kingdom who accessed health care for HIV infection in 2006 [25]. Among these sequences, just over one-half (52%) had a recent connection with another sequence in the same data set, inferred as a genetic distance of <4.5%. We established the phylogenetic relationships between the sequences of the virus infecting nearly 6000 individuals, from which we estimated the degree distribution of the entire transmission network. With nearly 30% of these sequences found in 97 groups of 10 or more individuals, this distribution was highly right-skewed (Figure 2), as seen in an earlier study based on a single London clinic [12]. Exhaustive comparisons of cluster numbers at different time depths showed that cluster size was not affected by the time depth chosen (Supplementary Table 2), but the retrospective nature of this study imposes a limitation to our conclusions due to censoring of the database at 2009 and the difficulty of locating recently entered patients correctly in the cluster pattern.
The analysis we have performed reveals the overall rate and distribution of HIV transmission within this population group. In the population of nearly 6000 MSM that was analyzed, a median interval between transmissions of at most 22 months was observed, with 16% of transmissions occurring within 6 months of infection. In large clusters (≥10 individuals), 20% of transmissions occurred within 6 months; our previous study based on a population at a central London clinic showed this proportion could reach 25% [12]. Overall, these findings suggest that the higher infectiousness associated with acute infection [4] plays a general role in accelerating transmission in the MSM population as a whole but that this could differ between populations.
There have been at least 2 large studies of sexual partnerships among MSM in the United Kingdom that can be compared with the results presented here, the representative (probability sampling) National Survey of Sexual Attitudes and Lifestyles (NATSAL) [26] and a community-based survey [27, 28]. The distribution of partner number in the past year reported by the NATSAL survey was as follows: 0 partners, 22%; 1 partner, 32%; 2 partners, 8%; 3–9 partners, 23%; and ≥10 partners, 15%. Substantially higher proportions with higher numbers of partners were reported in the community survey [29], with 53% reporting >5 partners in the past year as opposed to 19% for all MSM in the NATSAL survey. The distribution reported here from the HIV transmission network lies between those of the NATSAL and community-based surveys, with 29% linked to only 1 and 41% linked to 2–10 other individuals with a time depth of 5 years, but a much higher proportion with ≥10 links (29%) than that seen in the NATSAL survey. It could be expected that the transmission network would have a more right-skewed distribution than that of the total contact network because it is conditioned on HIV infection. For that reason, it may not be surprising that the distribution does not differ greatly from that reported by the community-based survey because that would be expected to be biased toward inclusion of a higher proportion of the more highly connected individuals.
Right-skewing of networks has been noted before as a prominent feature of degree distributions derived from surveys of sexual contacts and other networks, and in several cases it has been shown that the degree distribution fits a power law [5, 28, 30]. This has led to the concept that a “small world” model might underlie their dynamics. Because there are distributions that represent more precisely the preferential attachment process [23, 31, 32], we wished to test the distributions predicted for these models against the observed transmission network reconstructed for the HIV infection epidemic among MSM in the United Kingdom. We observed that both of the distributions specifically associated with a preferential attachment process, the Yule and Waring distributions [33, 34], could be fitted to the data, but at all time depths the Waring distribution fitted best (Figure 4). However, the data were also reasonably well fitted by a negative binomial distribution [35], which would not imply preferential attachment. After investigating this result in more detail, we found that a Waring distribution with parameters estimated from the data fitted a binomial distribution simulated with those parameters on approximately 10% of occasions. We cannot completely rule out the possibility that partner choice in this population may be random, but it is substantially more likely to be preferential.
A critical property of truly scale-free networks is that for values of the parameter ρ between 2 and 3 they exhibit infinite variance [23]. Under these conditions, an infection will spread regardless of its transmissibility and no random intervention is sufficient to halt the epidemic. The estimates of ρ obtained here for a time depth of 5–7 years are within that range, implying that any intervention must be targeted to high-degree individuals to be effective [11]. The significance of this conclusion is heightened by the increasing awareness of the potential offered by high-efficacy therapeutic interventions for preexposure prophylaxis. One study performed among women has reported a reduction in the infection rate of 65% [36], whereas the multinational Preexposure Prophylaxis Initiative (iPrEx) trial among MSM reported a reduction of 45% [37]. In the latter study, inclusion criteria specified ≥4 episodes of unprotected anal sex in the past 6 months; however, partner number was not specified. The impact of a treatment program targeted in that way on the most connected individuals in the population could be estimated from the approach described here by incorporation of the quantitative description of the transmission network for the whole population derived from viral sequence data into structured epidemiological models [11]. This would then permit enhanced accuracy in estimates of the cost per infection saved over the population as a whole [38].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support.
This work was supported by the Medical Research Council and the Biotechnology and Biological Science Research Council. The UK HIV Drug Resistance Database is partly funded by the Department of Health; the views expressed in the publication are those of the authors and not necessarily those of the Department of Health. Additional support is provided by Boehringer Ingelheim, Bristol-Myers Squibb, Gilead, Tibotec (a division of Janssen-Cilag), and Roche.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Klovdahl AS. Social networks and the spread of infectious diseases: the AIDS example. Soc Sci Med. 1985;21:1203–16. doi: 10.1016/0277-9536(85)90269-2. [DOI] [PubMed] [Google Scholar]
- 2.Yirrell DL, Pickering H, Palmerini G, et al. Molecular epidemiological analysis of HIV in sexual networks in Uganda. AIDS. 1998;12:285–90. doi: 10.1097/00002030-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Resik S, Lemey P, Ping LH, et al. Limitations to contact tracing and phylogenetic analysis in establishing HIV type 1 transmission networks in Cuba. AIDS Res Hum Retroviruses. 2007;23:347–56. doi: 10.1089/aid.2006.0158. [DOI] [PubMed] [Google Scholar]
- 4.Wawer MJ, Gray RH, Sewankambo NK, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–9. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 5.Liljeros F, Edling CR, Amaral LA, Stanley HE, Aberg Y. The web of human sexual contacts. Nature. 2001;411:907–8. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton DT, Handcock MS, Morris M. Degree distributions in sexual networks: a framework for evaluating evidence. Sex Transm Dis. 2008;35:30–40. doi: 10.1097/olq.0b013e3181453a84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leigh Brown AJ, Lobidel D, Wade CM, et al. The molecular epidemiology of human immunodeficiency virus type 1 in six cities in Britain and Ireland. Virology. 1997;235:166–77. doi: 10.1006/viro.1997.8656. [DOI] [PubMed] [Google Scholar]
- 8.Brenner BG, Roger M, Moisi DD, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22:2509–15. doi: 10.1097/QAD.0b013e3283121c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yerly S, Junier T, Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23:1415–23. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 10.Kouyos RD, von Wyl V, Yerly S, et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis. 2010;201:1488–97. doi: 10.1086/651951. [DOI] [PubMed] [Google Scholar]
- 11.Keeling MJ, Eames KT. Networks and epidemic models. J R Soc Interface. 2005;2:295–307. doi: 10.1098/rsif.2005.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog. 2009;5:e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BHIVA Writing Committee on behalf of the BHIVA Executive Committee. British HIV Association (BHIVA) guidelines for the treatment of HIV-infected adults with antiretroviral therapy. HIV Med. 2003;4(suppl 1):1–41. [PubMed] [Google Scholar]
- 15.Health Protection Agency. New HIV diagnoses in the UK. 2007. http://www.hpa.org.uk/web/HPAweb&HPAwebStandard/HPAweb_C/1203084373037. Accessed 28 April 2010. [Google Scholar]
- 16.De Oliveira T, Deforche K, Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 18.Swofford DL. PAUP*: phylogenetic inference using parsimony (and other methods) 1996. http://paup.csit.fsu.edu/. Accessed 15 March 2009. [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: a language and environment for statistical computing. http://www.R-project.org. Accessed 25 August 2011. [Google Scholar]
- 21.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 22.Handcock MS, Hunter DR, Butts CT, Goodreau SM, Krivitsky PN, Morris M. Statnet: software tools for the statistical modeling of network data. 2003. http://statnetproject.org. Accessed 26 August 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handcock MS, Jones JH. Likelihood-based inference for stochastic models of sexual network formation. Theor Popul Biol. 2004;65:413–22. doi: 10.1016/j.tpb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Irwin JO. The place of mathematics in medical and biological statistics. J Roy Stat Soc A. 1963;126:1–45. [Google Scholar]
- 25.Health Protection Agency. Testing times: HIV and other sexually transmitted infections in the United Kingdom. London: Health Protection Agency; 2007. [Google Scholar]
- 26.Mercer CH, Fenton KA, Copas AJ, et al. Increasing prevalence of male homosexual partnerships and practices in Britain 1990–2000: evidence from national probability surveys. AIDS. 2004;18:1453–8. doi: 10.1097/01.aids.0000131331.36386.de. [DOI] [PubMed] [Google Scholar]
- 27.Dodds JP, Nardone A, Mercey DE, Johnson AM. Increase in high risk sexual behaviour among homosexual men, London 1996–8: cross sectional, questionnaire study. BMJ. 2000;320:1510–1. doi: 10.1136/bmj.320.7248.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneeberger A, Mercer CH, Gregson SA, et al. Scale-free networks and sexually transmitted diseases: a description of observed patterns of sexual contacts in Britain and Zimbabwe. Sex Transm Dis. 2004;31:380–7. doi: 10.1097/00007435-200406000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Dodds JP, Mercer CH, Mercey DE, Copas AJ, Johnson AM. Men who have sex with men: a comparison of a probability sample survey and a community based study. Sex Transm Infect. 2006;82:86–7. doi: 10.1136/sti.2005.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colgate SA, Stanley EA, Hyman JM, Layne SP, Qualls C. Risk behavior-based model of the cubic growth of acquired immunodeficiency syndrome in the United States. Proc Natl Acad Sci U S A. 1989;86:4793–7. doi: 10.1073/pnas.86.12.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JH, Handcock MS. Social networks: sexual contacts and epidemic thresholds. Nature. 2003;423:605–6. doi: 10.1038/423605a. [DOI] [PubMed] [Google Scholar]
- 32.de Blasio BF, Svensson A, Liljeros F. Preferential attachment in sexual networks. Proc Natl Acad Sci U S A. 2007;104:10762–7. doi: 10.1073/pnas.0611337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones JH, Handcock MS. An assessment of preferential attachment as a mechanism for human sexual network formation. Proc R Soc Lond B. 2003;270:1123–8. doi: 10.1098/rspb.2003.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yule GU. A mathematical theory of evolution, based on the conclusions of Dr J.C. Willis F.R.S. Phil Trans R Soc Lond B. 1925;213:21–87. [Google Scholar]
- 35.Bliss CI, Fisher RA. Fitting the negative binomial distribution to biological data. Biometrics. 1953;9:176–200. [Google Scholar]
- 36.Peterson L, Taylor D, Roddy R, et al. Tenofovir disoproxil fumarate for prevention of HIV infection in women: a phase 2, double-blind, randomized, placebo-controlled trial. PLoS Clin Trials. 2007;2:e27. doi: 10.1371/journal.pctr.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant RM, Lama JR, Anderson PL, et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363:2587–99. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paltiel AD, Freedberg KA, Scott CA, et al. HIV preexposure prophylaxis in the United States: impact on lifetime infection risk, clinical outcomes, and cost-effectiveness. Clin Infect Dis. 2009;48:806–15. doi: 10.1086/597095. [DOI] [PMC free article] [PubMed] [Google Scholar]