Abstract
Off-target gene silencing is a major concern when using RNA interference. Imperfect pairing of the antisense strand with unintended mRNA targets is one of the main causes of small interfering RNA (siRNA) off-target silencing. To overcome this, we have developed “bulge-siRNA,” a modified siRNA backbone structure with a single nucleotide (nt) bulge placed in the antisense strand. We found that siRNAs with a bulge at position 2 of the antisense strand were able to discriminate better between perfectly matched and mismatched targets, with no loss in silencing of the intended target. Genome-wide analysis also revealed that the bulge-siRNAs significantly reduced off-target silencing of transcripts with complementarity to the seed region of the siRNA antisense strand. When compared to 2′-methoxy ribosyl (2′-OMe) modified siRNAs previously developed to alleviate antisense off-target silencing; the bulge modification could better discriminate between on- versus off-targets. Our results suggest that the bulge-siRNA structure is a simple, yet superior alternative to chemical modifications for minimizing off-target silencing triggered by conventional siRNA structures.
Introduction
Gene silencing triggered by small interfering RNAs (siRNAs) has become the method of choice for gene function studies as well as therapeutic interventions due to the potency and specificity of the target gene silencing.1,2 However, since the implementation of siRNAs for gene silencing in mammalian cells, a number of studies have demonstrated unexpected nonspecific effects3 triggered by conventional siRNA structure, which harbors a 19 base pair duplex region with 2 nucleotide (nt) 3′-overhangs at both ends.4 These include off-target gene silencing (i.e., silencing of unintended target genes),5,6,7 activation of innate immune responses,8,9,10 and saturation of endogenous RNAi machinery.11,12,13,14,15
siRNA off-target gene silencing can be due to unintended incorporation of sense strand into RNA-induced silencing complex (RISC) or due to the imperfect pairing of mRNA with the antisense stand, both leading to the cleavage of unintended targets.6,16 To circumvent sense off-target silencing, various chemical as well as backbone modifications have been introduced to the siRNA structure, most of which were successful.17,18,19,20,21 Evidence for the existence of antisense off-target silencing was first given by Jackson et al.,5,22 who demonstrated that limited complementarity between the siRNA duplex and the mRNA transcripts is sufficient to initiate mRNA cleavage by RISC. Studies involving sequence analysis of off-target transcripts reveal that they have high complementarity with the 5′-end of the guide strand of transfected siRNAs, more specifically the first 8 nt region.5,23 This part of the guide strand resembles the seed region of microRNAs, which play key role in target recognition.24,25 Any modification that interferes with seed-target pairing has a more dramatic reduction in off-target silencing than on-target silencing, where such interference could be easily compensated with perfect pairing for the rest of the duplex.22 The seed region-dependent off-target silencing also extends to short hairpin RNAs and is independent of the type of regulatory RNA and the method of delivery.22 These off-targets can induce a measurable amount of phenotypic changes that can account for up to 30% of the false positive hits in RNAi-based phenotypic screens, complicating data interpretation.23,26 In view of this widespread antisense off-target silencing mediated by conventional siRNAs, development of a chemical or backbone structure modification, which can maintain the on-target silencing efficiency but minimize the off-target silencing, is of great interest. Several modified siRNAs have been made to reduce antisense off-target silencing. Among them, the most widely used siRNA modification was the introduction of a 2′-methoxy (2′-OMe) group in the ribosyl ring of the second nt of the antisense strand.27 This chemical modification significantly reduced antisense off-target silencing of several siRNAs. Besides 2′-OMe, other chemical modifications were also tested, among which locked nucleic acid28 and unlocked nucleic acid29 were found to be quite encouraging. Another study showed that siRNAs with the seed arm replaced with a cognate DNA sequence, can reduce off-target silencing.30 Although these chemical modifications successfully reduce antisense off-target silencing of siRNAs, some of these, such as 2′-OMe and DNA, can reduce on-target silencing efficiency as well. Moreover, these chemical modification strategies cannot be extended to intracellularly expressed siRNAs or short hairpin RNAs.
In this study, we designed a novel siRNA backbone structure that can minimize antisense off-target silencing without introducing any chemical modifications. We hypothesized that a minimal structural perturbation in the antisense strand and its target pair that weakens the duplex formation could destabilize the RISC, and that this destabilization would be more pronounced in the case of imperfectly paired mismatched targets, which would lead to inefficient silencing of off-target mRNAs. To achieve this, we introduced an additional nt in the antisense strand of siRNA duplexes, generating a 20 nt antisense that forms a bulge with the 19 nt sense at the position of insertion. We termed this modified siRNA structure as “bulge-siRNA.” Using a luciferase reporter system containing antisense on-target or wild-type (WT), and off-target or mismatch/mutant (Mut) sequences, we found that the introduction of a single nt bulge at position 2 of the antisense strand significantly reduces off-target silencing while maintaining or even enhancing on-target gene silencing. Importantly, the bulge-siRNAs were more efficient in off-target discrimination when compared with 2′-OMe modified siRNAs. A genome-wide microarray with the modified siRNAs further confirmed that bulge-siRNAs reduces conventional siRNA-mediated off-target silencing.
Results
siRNAs carrying a single nt bulge at positions 2, 18, and 19 of the antisense strand trigger on-target gene silencing comparable to unmodified siRNA
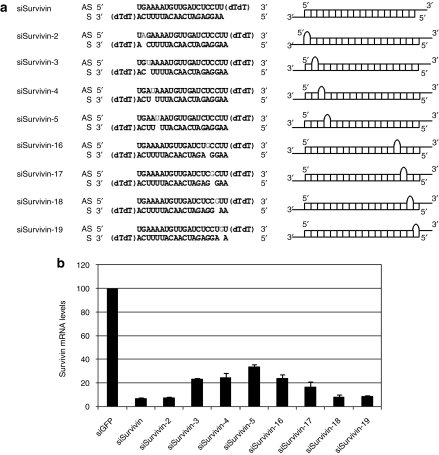
To design siRNAs that maintain the on-target silencing activity but have reduced antisense off-target silencing, we first tested various bulge-siSurvivin duplexes with a single nt bulge at either of the positions 2, 3, 4, 5, 16, 17, 18, 19 of the antisense strand, for antisense on-target silencing. We created group I single nt bulges in the siRNA, where the identity of the bulge was not identical to the neighboring nucleotides. This was necessary to avoid formation of ambiguous pairs with the adjacent nucleotides of the sense strand. In addition, the bulge nt was chosen such that the desired asymmetry in the siRNA structure is maintained. Hence, bases A and U were preferred toward the 5′-end and G and C toward 3′-end of the antisense strand. Bulges at or near the center of the siRNA construct were not studied as this region is considered a low-tolerance region and any modification at these positions has been shown to abolish most of the gene silencing activity.31,32,33 HeLa cells were transfected with 10 nmol/l of siRNA targeting GFP expression (siGFP), bulge-modified or unmodified siSurvivin (Figure 1a) and the on-target silencing activity of the siRNAs is given in Figure 1b. Unmodified siSurvivin showed 92% silencing of the target mRNA. Bulge-siRNAs show a biphasic profile with increasing loss in silencing as the bulge is positioned toward the center of the construct. siSurvivin with a single nt bulge placed at position 2, 18, and 19 showed no loss in silencing. For the other bulge-siRNAs, the silencing activity continued to decrease as the bulge became closer to the center of the siRNA (positions 3, 4, 5, 16, and 17). A comparison of Tm and free energy change (ΔG) of these bulge-siSurvivin constructs to unmodified siSurvivin shows that when bulges are placed at the extreme ends (position 2, 18, and 19), the thermodynamic stability of the duplex is not affected significantly (Supplementary Table S2). However, at all other positions the ΔG is more, indicating thermodynamic instability, which could contribute to the loss in gene silencing. Positions that maintained silencing activity up to 80%, i.e., siSurvivin-2, -18, and -19, were chosen to further study their antisense off-target silencing.
Figure 1.
On-target silencing activity of bulge-small interfering RNA (siRNA) depends upon position of bulge. (a) The sequence and structure of siRNAs targeting survivin mRNA and the corresponding hypothetical structures. The number in the siRNA nomenclature represents the position of the single nucleotide bulge (gray) from the 5′-end of the antisense sequence. (b) Bulge-modified siSurvivin show position-dependent silencing activity. HeLa cells were transfected with 10 nmol/l of siRNA targeting GFP expression (siGFP) or siSurvivin siRNAs. Results from real-time quantitative PCR (qPCR) are shown as mean ± SE from three independent experiments.
siRNAs with a single nt bulge at position 2 shows significant discrimination between perfectly matched and mismatched antisense targets
To analyze antisense off-target activity of siSurvivin structural variants, eight different luciferase reporters were prepared, each with a single nt mismatch mutation in the antisense target sequence of siSurvivin (Figure 2a). We used mismatched targets that differ from the on-target sequence by a single nt to provide a highly stringent condition for studying antisense off-target discrimination. siRNAs that could discriminate even a single nt mismatch with the target in the assay format would provide much better discrimination de novo, where widespread off-target silencing is observed with various degrees of mismatches.
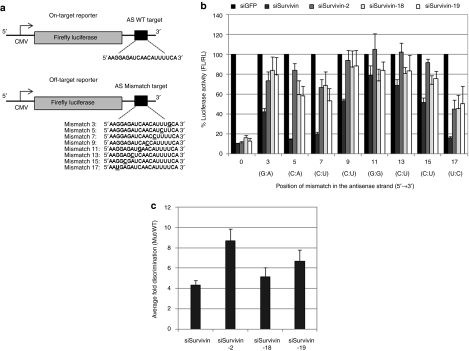
Figure 2.
Bulge modification at position 2 shows significant discrimination between perfectly matched and mismatched antisense targets. (a) Schematic view of pMIR lucifersase reporter used to monitor small interfering RNA (siRNA) on-target and off-target silencing. A single copy of the antisense target site, wild-type (WT) or mismatch/Mut was cloned in the 3′-untranslated region (UTR) of the Firefly luciferase reporter to generate an on-target or off-target reporter, respectively. Drawings are not to scale. (b) On-target and off-target silencing of unmodified and bulge-modified siSurvivin. HeLa cells were co-transfected with on-target or off-target pMIR reporter plasmid and 10 nmol/l of siSurvivin constructs for 24 hours. Renilla luciferase vector was transfected as an endogenous control to normalize differences in cell number. After 24 hours of transfection, Firefly luciferase signals were normalized with Renilla luciferase signals. The normalized luciferase intensity relative to siRNA targeting GFP expression (siGFP) (shown as 100%) is given here. The position and type of mismatch formed are indicated on the x axis. (c) Fold discrimination between on- and off-target silencing. Ratio of luciferase activities for the mismatched target versus wild-type target was calculated as fold discrimination and average of discrimination for all the targets is shown for each siRNA. Results are represented as mean± SE of three independent experiments.
HeLa cells were co-transfected with an on-target or off-target luciferase reporter and 10 nmol/l of siSurvivin or siSurvivin-2, -18, and -19 for 24 hours. As seen earlier with the bulges, the mismatched target gives a similar position-dependent loss in silencing; the central regions are more intolerant to mismatches than the terminal regions (Figure 2b). Remarkably, a mismatch at position 3 was highly intolerable, resulting in a greater loss in silencing compared to the neighboring positions. This exceptional intolerance at position 3 has not been previously observed.32,34 Even replacing the G:A mismatch with a more tolerable A:A mismatch, did not reduce the suppression in silencing at this position (data not shown). This effect could be siRNA sequence-dependent, which still needs to be confirmed. The bulge siSurvivin-2 sequence showed significantly better reduction in antisense off-target silencing compared to unmodified siSurvivin and siSurvivn-18 and 19 for seven out of eight mismatches tested. The results have been further compiled as fold discrimination where a ratio of luciferase activities for the mismatched versus WT target was calculated for each siRNA and the average of discrimination for all the eight targets is shown in Figure 2c. The higher the fold discrimination value is, the more specific the siRNA. It is noteworthy that in most previous studies, chemical modifications in siRNA led to some loss in on-target silencing activity, which is undesirable. An ideal modification would not reduce antisense off-target silencing at the cost of on-target silencing. Therefore, we believe that fold discrimination values give a clear picture of siRNA specificity and is the most reliable way to evaluate modified siRNAs. As seen in Figure 2c, siSurvivin-2 gave better discrimination of mismatched and WT targets than unmodified siRNA and the other bulge-siRNAs tested.
Bulge-siSurvivin is superior to 2′-OMe modified siSurvivin in off-target discrimination
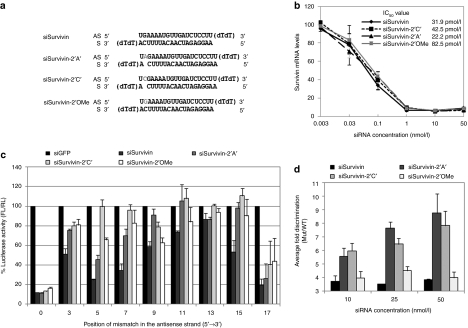
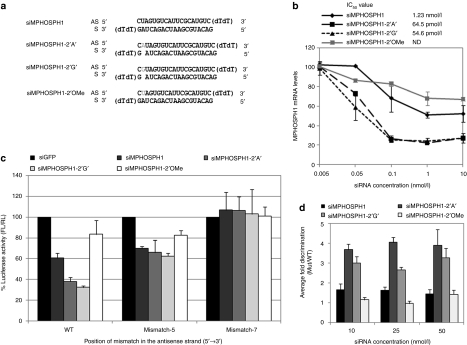
Among the chemical modifications that reduce off-target silencing, the 2′-OMe modification at position 2 in the antisense strand is the most widely used. We therefore compared the bulge-siRNA with 2′-OMe modified siRNA. In addition, we tested whether the nt identity of the siRNA bulge affects antisense off-target discrimination. Therefore, along with the previously tested siSurvivin-2′A′, we constructed an additional bulge structure siSurvivin-2′C′ (Figure 3a). Furthermore, to see whether the discrimination of on- versus off-target is maintained even at higher concentrations the siRNAs were tested at 10, 25, and 50 nmol/l concentrations.
Figure 3.
Bulge-siSurvivin is superior to 2′-methoxy ribosyl (2′-OMe) modified siSurvivin in off-target discrimination and reduction in off-target silencing is independent of the identity of the bulge nucleotide. (a) Sequence of unmodified and modified siSurvivin constructs with bulge and 2′-OMe modifications at position 2. Bulge and 2′-OMe modified nucleotides are marked in gray, respectively. (b) IC50 values of unmodified and modified siSurvivin were determined by real-time quantitative PCR (qPCR). (c) On-target and off-target silencing of siSurvivin constructs at 10 nM concentrations. HeLa cells were co-transfected with on-target or off-target pMIR reporter plasmid and 10 nM of siSurvivin constructs for 24 hr. The Firefly to Renilla normalized luciferase intensity relative to the siRNA targeting GFP expression (siGFP) is given here. (d) Fold discrimination between on- and off-target silencing. Ratio of luciferase activities for the mismatched versus wild-type target was calculated as fold discrimination and the average discrimination of both the mismatched targets is shown for each siRNA at three different concentrations. Results are shown as mean ± SE of three independent experiments.
The dose–response curve of siSurvivin variants against endogenous survivin mRNA shows that both the bulge-siRNAs had IC50 values similar to that of unmodified siSurvivin whereas the 2′-OMe modified siRNA showed some loss in silencing resulting in higher IC50 value (Figure 3b). The bulge and 2′-OMe modified siRNAs could reduce off-target silencing at all the three concentrations tested as shown in Figure 3c (10 nmol/l), Supplementary Figure S2a (25 nmol/l) and S2b (50 nmol/l). Upon further calculations of fold discrimination, the 2′-OMe modified structure gave a marginal and insignificant discrimination in comparison to unmodified siRNA, at all concentrations tested, which is mainly due to some loss in on-target silencing activity (Figure 3c and d). In contrast, both the siSurvivin-2 bulge structures showed significant discrimination in comparison to both unmodified and 2′-OMe siRNA, suggesting that bulge-siRNAs can efficiently discriminate perfect versus imperfect targets and this discrimination is independent of the identity of bulge nt.
Bulge modifications can reduce antisense off-target silencing of other siRNAs
We further tested the bulge modifications in other siRNAs to confirm that bulge-mediated reduction in off-target silencing is not limited to specific siRNA sequences. To analyze this, we referred to some of the siRNAs used by Jackson et al. for testing 2′-OMe modifications.27 We tested two bulge modifications for each siRNA that differed by the identity of the bulge nt and compared them with 2′-OMe modified siRNAs. For the antisense off-target sequence, we used position 5- and position 7- mismatched antisense targets for each siRNA. The siRNAs were tested at 10, 25, and 50 nmol/l concentrations.
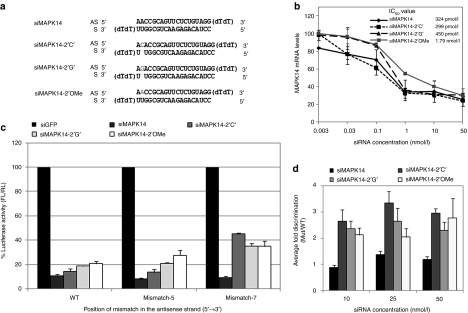
First, we tested bulge-siMAPK14 (Figure 4a). On-target gene silencing activity analysis showed that, compared with unmodified siMAPK14 (IC50 324 pM), 2′-OMe modified siMAPK14 results in greatly reduced IC50 values (1.79 nmol/l). In contrast, siMAPK14-2′C′ showed IC50 values similar to unmodified siRNA (Figure 4b). Although siMAPK14-2′G′ showed some loss in silencing activity with reduced potency (IC50 450 pmol/l), it is noteworthy that the bulge nt ′G′ placed in this construct can also form a wobble pair with the adjacent nucleotides in the sense strand, creating a bulge at position 3 or a overhang at position 1 (see Figure 4a), both capable of reducing the siRNA potency. For mismatch targets, both bulge and 2′-OMe modified siMAPK14 showed significant loss in antisense off-target silencing, especially at mismatch position 7 (Figure 4c,d and Supplementary Figure S2c,d). Next, siRNA targeting PIK3CB was modified with a bulge. The bulge-PIK3CB siRNA used here was found to be very potent with 93% silencing efficiency. Both the bulge modifications siPIK3CB-′C′ and siPIK3CB-′G′ maintained silencing efficiency (Supplementary Figure S1a–e). In this particular case, even a 2′-OMe modification did not affect the silencing efficiency of the siRNA. A significant reduction in off-target silencing of position 5- and 7- mismatched targets was seen with both the bulge and 2′OMe modification. However, for this siRNA alone, the 2′-OMe modification showed better discrimination than the bulge-modified siRNA (Supplementary Figure S1f).
Figure 4.
Bulge modification reduces off-target silencing of siMAPK. (a) Sequences of unmodified and modified siMAPK-14 with bulge and 2′-OMe modifications at position 2 of antisense strand. (b) IC50 values of MAPK14 small interfering RNAs (siRNAs). (c) On-target and off-target silencing of siMAPK-14 constructs at 10 nmol/l concentrations. (d) Average of fold discrimination between on- and off- target silencing of both the mismatched targets is shown for each siRNA at three different concentrations. For details see descriptions of Figure 3.
We then tested another siRNA targeting MPHOSPH1 mRNA (Figure 5a). Similar to a previous report,27 the unmodified siRNA was not very effective, resulting in only 45% on-target gene silencing at 10 nmol/l, with no further increase in silencing at higher concentrations (Figure 5b and Supplementary Figure 2e,f). Also, as previously observed, the 2′-OMe modification further reduced the gene silencing activity of siMPHOSPH1 (Figure 5b). However, to our surprise, both bulge modifications increased the silencing activity of siMPHOSPH1 up to 70% at 10 nmol/l (Figure 5b). Notably, in spite of a substantial increase in on-target silencing, the antisense off-target activity of bulge-siMPHOSPH1 did not increase (Figure 5c and Supplementary Figure S2e,f), resulting in enhanced fold discrimination (Figure 5d). We speculate that a bulge modification at position 2 could impart some structural asymmetry toward the 5′-end, causing preferential loading of the antisense strand into the RISC complex and improved silencing.
Figure 5.
Bulge modification enhances on-target silencing activity of siMPHOSPH1 but reduces off-target silencing. (a) Sequences of unmodified and modified siMPHOSPH1 with bulge and 2′-methoxy ribosyl (2′-OMe) modifications at position 2 of the antisense strand. (b) IC50 values of MPHOSPH1 small interfering RNAs (siRNAs). (c) On-target and off-target silencing of siMPHOSPH1 constructs at 10 nmol/l concentrations. (d) Average of fold discrimination between on- and off-target silencing of both the mismatched targets is shown for each siRNA at three different concentrations. For details see descriptions of Figure 3.
Similar enhancement in siRNA activity has been observed earlier with “fork-siRNAs” that have single nt mismatches in the sense strand toward the 3′ end.35 These forked or frayed siRNAs are known to favor antisense incorporation into RISC and reduce sense strand-mediated RNAi. One possibility is that the bulge induces the generation of a structure similar to “fork-siRNA.” According to asymmetry rules, the 5′ end of the guide strand is AU rich and the nt pair at position 1 is generally A/U. The presence of a bulge at position 2 in the guide strand could interfere or inhibit the already weak Watson–Crick pairing at position 1, leading to the formation of a fork.35,36 To confirm this, fork-siRNAs for all the four mRNA targets were made based on the earlier published designs35 and tested for their on- and off- target silencing. No reduction in off-target silencing was seen with fork modification (Supplementary Figure S3). Moreover, against our expectations, the fork modification could not improve the silencing efficiency of siMPHOSPH1 (Supplementary Figure S3d). This result clearly demonstrates that the two modifications are different both structurally and functionally.
Taken together, bulge modifications could reduce off-target silencing of all the four siRNAs tested. However, the extent of off-target reduction differs with the siRNA sequence. Such variations have been commonly seen with the earlier reported modifications as well and are likely due to the siRNA sequence features or the mRNA target. The reduction in the off-target silencing by bulge modification does not seem to be influenced by the identity of bulge nt and is independent of siRNA concentration. Furthermore, unlike 2′-OMe modified siRNAs (three out of four), the on-target silencing activity was not compromised upon introduction of a bulge at position 2 of the antisense strand, which is also one of the main causes of better target discrimination observed with bulge-siRNAs than 2′-OMe modified siRNAs. Lastly, bulge modification could provide a new method of increasing the silencing activity of an otherwise inefficient siRNA and thus, enhanced silencing with off-target discrimination could be achieved.
Bulge-siRNAs reduce siRNA-mediated toxicity
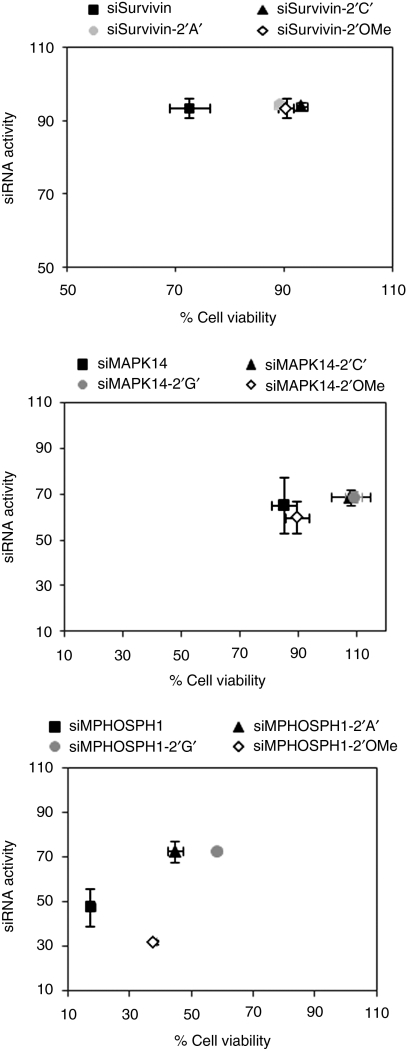
Antisense strand-driven silencing of undesired, imperfectly matched mRNAs is a siRNA-mediated side effect. Some siRNAs have been shown to inhibit cell viability without significantly silencing their target mRNAs, indicating that these are false positive phenotypes.37 Modifications that reduce antisense off-target silencing should alleviate such phenotypes. Therefore, we assessed the effect of modifications on siRNA-mediated loss in cell viability using MTT assay. Furthermore, to delineate target-dependent and -independent changes in cell viability, silencing activity was correlated with cell viability. As shown in Figure 6, HeLa cells transfected with 10 nmol/l of siSurvivin, siMPHOSPH1, and siPIK3CB-induced significant loss in cell viability while no cell death was seen in case of siMAPK-14 (Figure 6 and Supplementary Figure S1g). Unmodified siSurvivin showed 72% cell viability in comparison to Lipofectamine 2000-treated cells (considered to be 100% viable). Upon bulge modification, the cell viability increased to around 92% with no loss in silencing activity, suggesting considerable rescue of antisense mediated off-target phenotypes. siSurvivin 2′-OMe modification could also reduce siRNA-mediated cell death. siPIK3CB resulted in a ~30% loss in cell viability and siRNA-mediated cytotoxicity was completely abolished with both bulge and 2-′OMe modifications (Supplementary Figure S1g). In contrast to siSurvivin and siPIK3CB, siMPHOSPH1 was found to be highly toxic. siMPHOSPH1 resulted in a high level of cell death (85%), which was independent of the antisense silencing activity. With the bulge modification, in addition to the increase in silencing activity, we could also reduce the cell death considerably. The cytotoxicity data again confirms that introduction of a bulge at position 2 of the antisense strand imparts on-target silencing efficiency and specificity to an otherwise very poor siRNA. Although, presence of a bulge in the siRNA duplex, especially a central uridine bulge, has been shown to impart immunostimulatory properties,38 we did not find any upregulation in the expression of innate immune response associated genes with bulge modification at 2nd position in the siRNAs (Supplementary Figure S4a–d).
Figure 6.
Bulge modification reduces small interfering RNA (siRNA)-mediated cellular toxicity. Cells were transfected with 10 nmol/l siRNAs. After 4 days, cell viability was determined by MTT assay, considering lipofectamine transfection control as 100% viable. The silencing efficiency of the siRNAs at 10 nmol/l concentration was calculated by real-time quantitative PCR and a correlation of siRNA activity to cell viability is shown. Results are represented as mean ± SE of three independent experiments.
Bulge-siSurvivin reduces siSurvivin-mediated genome-wide off-target silencing
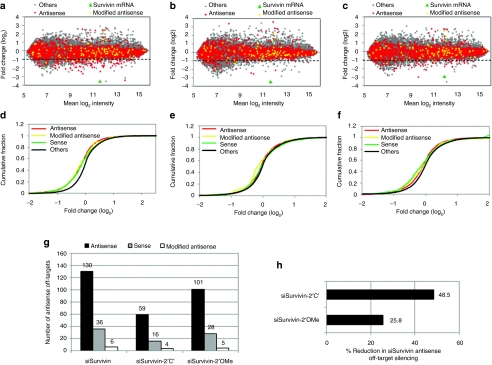
To confirm the findings of the luciferase reporter assays and to analyze the effect of bulge modification on overall cellular off-target silencing, genome-wide expression profiling was carried out using complementary DNA (cDNA) prepared from HeLa cells treated with unmodified and modified siSurvivin constructs. A total of 18,978 transcripts that represented RefSeg human mRNA sequences were examined from the microarray data. Homology between the seed region of the guide strand (positions 1–8, 2–8, and 1–7 from the 5′-end) and the 3′-untranslated region (UTR) of the mRNA is considered to be important for off-target silencing.22 Hence, 3′-UTR of transcripts with antisense seed matches were identified, which comprised 10.7% (n = 2,031) of the total transcripts analyzed. MA plots show overall changes in mRNA level upon transfection of unmodified and modified siSurvivin (Figure 7a). The expression levels of transcripts with siSurvivin antisense seed homology in their 3′-UTR were reduced more than the transcripts with no canonical matches (represented as others). However, reduction in the mRNA level of siSurvivin seed-matched transcripts was observed to be much less in the case of siSurvivin-2′C′, showing that bulge modifications can alleviate seed-matched off-target silencing (Figure 7b). Some reduction in off-target silencing of seed-matched transcripts was also observed with 2′-OMe modifications, but it was less evident than as seen with bulge modifications (Figure 7c). Furthermore, the 2′-OMe modification showed reduced Survivin mRNA silencing compared to the unmodified and bulge-siRNA (Figure 7c). Results from the cumulative fraction plots (Figure 7d–f) also confirm that transcripts with antisense seed homology in their 3′-UTRs have significantly higher expression in the case of bulge-modified siSurvivin when compared with unmodified or 2′-OMe modified siSurvivin. In general, transcripts downregulated by 50% or more are believed to have significant effects on cellular changes. Therefore, we also calculated the number of antisense seed-matched transcripts that were downregulated by 50% or more. When compared with unmodified siSurvivin (n = 130), the number of these downregulated transcripts were reduced significantly in the case of siSurvivin-2′C′ (n = 59) (Figure 7g). Not only the number, but also the magnitude of antisense off-target silencing observed with siSurvivin was reduced up to 50% by the bulge modification (Figure 7h). In contrast, siSurvivin-2′OMe was not found to reduce the number and magnitude of antisense off-target silencing to the same extent.
Figure 7.
Microarray-based genome-wide off-target profiling of siSurvivin. MA plots showing the changes in the expression of transcripts with antisense seed match (red dots) or no canonical matches in their 3′-UTR (represented as others, gray dots) upon siRNA transfection. (a) siSurvivin, (b) siSurvivin-2′C′, (c) siSurvivin-2′OMe. Transcripts with homology to new seed generated by the bulge are also shown (modified antisense, yellow dots). Cumulative distribution of antisense, sense, or modified seed-matched transcripts in comparison to transcripts with no seed matches are shown as log2 of fold change in expression upon treatment with (d) siSurvivin, (e) siSurvivin-2′C′, and (f) siSurvivin-2′OMe. (g) Number of transcripts with respective seed homology downregulated by 50% or more. (h) Reduction in siSurvivin-mediated off-target silencing by bulge or 2′-OMe modifications. Antisense seed-matched transcripts that were downregulated by more than twofold upon siSurvivin treatment (siSurvivin off-targets) were collected and average inhibition in their expression was calculated in siSurvivin, bulge, and 2′-OMe-modified constructs. The reduction in the silencing of these off-targets upon bulge or 2′-OMe modifications is shown.
Jackson et al. have shown that base substitutions in the siRNA seed region reduce silencing of off-target transcripts complementary to the WT siRNA sequence. However, they found that such base substitutions trigger new off-target transcripts complementary to the new seed, generated by the mismatch sequence.22 Because the introduction of a single nt bulge in the seed can have similar consequences, in our dataset we also identified siSurvivin-2′C′ seed matches (represented as modified antisense). These comprised 0.5% (n = 95) of the overall mRNA. Interestingly, transcripts with new seed matches did not give any differential expression pattern in the siSurvivin-′2′C transfected cells (Figure 7b,d–f). The number of these transcripts with expression reduced by 50% or more also did not increase with bulge-siRNA (Figure 7g). To confirm that siSurvivin-2′C′ do not cause silencing of its seed-matched targets we randomly picked some of the modified seed-matched target mRNAs and validated them using luciferase assay (Supplementary Figure S5). In addition, studies with seed-only target pMIR reporter clearly showed that seed-based off-target silencing caused by siSurvivin was completely abolished by bulge modification whereas no modified seed-based silencing was observed (Supplementary Figure S6).
The siSurvivin construct used here was also shown to have sense off-target silencing activity despite having a thermodynamically asymmetric sequence feature.17 Therefore, along with antisense off-target silencing, we also checked for sense off-target silencing. siSurvivin sense seed-matched transcripts comprised of 2.7% (n = 521) of the total transcripts. Like antisense transcripts, sense seed-matched transcripts also showed reduced expression in comparison to the remaining transcripts with no seed matches (Figure 7d). To our surprise, this sense seed-matched off-target silencing was greatly reduced with siSurvivin-2′C′, suggesting that the bulge modification at position 2 of the antisense strand could also control sense off-target silencing (Figure 7e). In contrast, such a change in the sense mediated off-target silencing was observed with the 2′OMe modified construct (Figure 7f). Although it is not clear how sense off-target effects are controlled by bulge modification, we speculate that the presence of a bulge at position 2 could impart some additional asymmetry to 5′-end of the antisense strand that enhances its preferential loading into RISC, thereby reducing sense strand driven RNAi. Taken together, these microarray results clearly demonstrate the advantage of using bulge-siRNAs in reducing both sense and antisense off-target gene silencing.
Bulge-siRNAs duplexes show increased Ago-2 incorporation and produce target mRNA cleavage product
In vitro Ago-2 incorporation assay carried to compare RISC loading of unmodified and bulge modified survivin and MAPK14 siRNAs demonstrates that the bulge modification significantly enhances the incorporation of siRNA duplexes into Ago-2 (Supplementary Figure S7). Although this could be a result of increased interaction of Ago-2 with the siRNA itself or due to increased binding with other RISC-associated proteins, it seems to be mostly due to the thermodynamic instable 5′-end of siRNA. siMAPK-2′G′ which could form a wobble instead of a bulge, shows much less incorporation into Ago-2 than siMAPK-2′C′, supporting the above speculation. However, the increased siRNA loading did not lead to increased silencing efficiency.
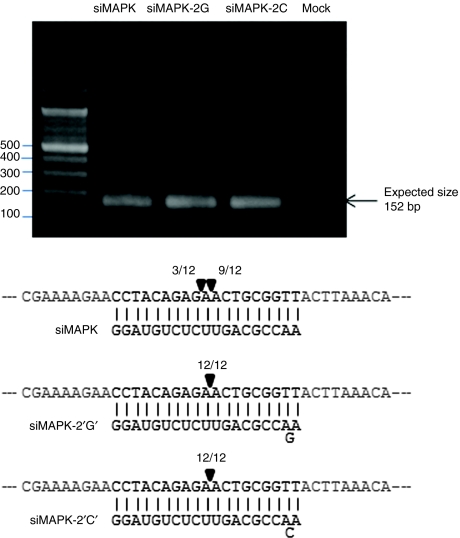
5′-RACE assay with unmodified and bulge-modified MAPK14 siRNAs suggest that both the siRNA types cleave the mRNA at nearly identical positions. In case of siMAPK14, cleavage between position 10 and 11 of the target was mainly observed while some RACE products from cleavage between position 9 and 10 were also obtained. However, both the bulge-siMAPK14 resulted in mRNA cleavage between position 10 and 11 (Figure 8).
Figure 8.
5′-RACE assay shows similar cleavage products for unmodified and bulge-modified MAPK14 small interfering RNA (siRNAs). PCR products of 5′-RACE assay from siMAPK14 transfected cells. The mRNA cleavage sites were analyzed by 5′-RACE assay and sequencing. The MAPK14 target mRNA (upper strand) sequence with the antisense strand (lower) are given. The cleavage sites are marked with arrowheads and the numbers show the frequency of clones sequenced.
Discussion
In this study, we systematically evaluated single nt bulge modifications in the antisense strand of a conventional siRNA duplex structure. Among all the positions tested, position 2 of the guide strand (from the 5′-end) was found to be the ideal position for bulge modification. This position-specific single nt bulge modification reduces silencing of off-targets without compromising silencing of perfectly matched intended targets by all the siRNAs tested. In most cases, bulge-siRNAs were found to be superior to 2′-OMe modified constructs in discriminating between perfect versus imperfect targets. The bulge modification also reduced siRNA-mediated false positive phenotypes in the cell death assay and was shown to dramatically increase the silencing efficiency of an otherwise poor siRNA. We believe that bulge-siRNA could be an appropriate siRNA backbone structural variant to reduce antisense mediated off-target silencing and will aid accurate functional analysis and microarray data profiling.
The single nt bulge-siRNA structure we developed, does not affect the silencing of perfectly matched targets, especially when the bulge is located at position 2, suggesting that it does not interfere with siRNA loading into RISC, target recognition or RISC catalytic activity. However, it does affect silencing of an imperfectly paired target, and the effect is more pronounced when the mismatches are in the seed region. This particular observation hints upon the mechanism of discrimination between on- and off-target silencing in the case of bulge modifications; a single nt bulge creates a structural perturbation in the seed region, which may also affect the thermodynamic stability of the seed. This is supported by the fact that group I bulges generally destabilize double-stranded regions of RNA.39,40 Free energy increments for the introduction of such bulges vary between 1.3 and 5.2 kcal/mol at physiological conditions. Moreover, destabilization caused by group I bulges is neither affected by the identity of the bulge nor by the neighboring sequence of the RNA duplex.40 However, the stem length does seem to correlate with the destabilization incurred by the bulge. A position 2 bulge, being at the terminus, is not expected to cause dramatic destabilization, as would be expected in the case of other bulges that lie closer toward the center of the siRNA duplex. Therefore, more strongly hybridizing targets may be resistant to seed instability caused by position 2 bulge modifications; however, in the case of weakly paired targets, an additional disruptive effect of the bulge modification would increase the thermodynamic and duplex structure instability, resulting in the formation of inefficient RISC and loss in silencing. When bulge and mismatches are both placed in seed, the instability would be larger, causing greater loss in silencing than when both are placed far apart (Figure 3b). Current evidence also supports the fact that siRNA seed and target duplex stability is an important determinant in off-target silencing. The melting temperature of siRNA seed-target pairs correlates with the extent of off-target silencing.41 Recently, based upon this finding, an updated siRNA design algorithm has been proposed whereby siRNAs with lower seed-target duplex stabilities and a minimal of two mismatches anywhere throughout the guide strand and target duplex, are capable of eliminating off-target effects to a large extent.42 In our case, bulge-mediated siRNA seed-target instability along with a single mismatch in the siRNA target pair were seen to be sufficient to reduce off-target silencing.
It can be argued that an antisense bulge in the siRNA duplex might not remain as a bulge in the antisense-mRNA pair in the RISC complex. Alternatively, the bulge nt insertion could increase the length of the guide strand, shifting the seed sequence by a positional increment of one toward the 3′-end, thus generating a new seed in the RISC. However, we disagree with this assumption for two reasons. Firstly, recent experiments have shown that both strands of siRNA get loaded onto Ago-2 protein and Ago-2 likely cleaves the sense strand after siRNA binding.43,44 This means that the guide strand interactions with the Piwi-Mid domains of Ago-2 would have already occurred before the separation of the sense strand, allowing the bulge in the guide strand to be recognized as a bulge. This is also the case for Ago1-mediated microRNA recognition, where mismatches and central bulges in the pre-Ago1-RISC are retained in the mature Ago1-RISC.45 Secondly, in the genome-wide expression analysis with siSurvivin-2′C′, we did not find any reduction in expression levels of transcripts with matches to the new seed that would have been generated due to the bulge nt, suggesting that the bulge nt may not form a part of the seed.
Jackson et al. proposed that 2′-OMe modifications at position 2 of the guide strand affect the ternary complex with Argonaute and target RNA. Conformational alteration of RISC is required to accommodate a methoxy residue at this particular position, which preferentially reduces the silencing of partially complementary transcripts without affecting cleavage of the fully-complementary target.27 It is possible that similar conformational changes are required to occur in RISC to accommodate the bulge nt, which could lead to differential silencing of on- and off-targets. Although at the present it is difficult to clarify the key mechanism by which a bulge modification at position 2 reduces off-target silencing, it is likely an additive effect and further structural and thermodynamic studies would be needed to provide a clear understanding of this mechanism.
Materials and Methods
siRNAs. Chemically synthesized, high-performance liquid chromatography-purified RNAs were purchased from Samchully Pharma (Seoul, South Korea) and annealed according to the manufacturer's instructions. The quality of annealed siRNAs was verified on 15% polyacrylamide gel before use. The sequence of the siGFP sense strand is 5′-GGCUACGUCCAG GAGCGCA-3′. Sequences of other siRNAs used are given in the figures.
Reporters and target modification. DNA oligonucleotides (Bioneer, Daejeon, Korea) corresponding to the antisense-strand target of siSurvivin, siMAPK14, siMPHOSPH1 were cloned into the 3′-UTR of the pMIR-Report luciferase vector (Ambion, Austin, TX) in the SpeI and HindIII cloning sites to make an on-target reporter. Eight different siSurvivin antisense targets with a single nt mutation tiling throughout the length of the 19 nt sequence at alternating positions that forms a mismatch upon pairing with the antisense strand were used to generate various off-target reporters. For siMAPK14, siMPHOSPH1, and siPIK3CB two antisense off-target reporters were made with mismatches at position 5 and 7. The sequences of the DNA oligos used to construct the targets are given in Supplementary Table S1.
Transfection and luciferase assay. HeLa cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were plated on 24-well plates and at 40–50% cell density and transfections were performed in complete medium without antibiotics. One hundred microgram of pMIR-Report Firefly luciferase vector, harboring the siRNA target sequence, was co-transfected with 1 µg of Renilla luciferase vector and siRNA. All transfections were carried out using the Lipofectamine-2000 reagent at the concentrations indicated by the manufacturer (Invitrogen). Twenty-four hours after transfection, cells were lysed using passive lysis buffer (Dual-Luciferase Reporter Assay System; Promega, Madison, WI). Firefly and Renilla luciferase activity was then measured in 20 µl of cell extract using a Victor3 plate reader (PerkinElmer, Boston, MA). The Firefly luciferase signal was normalized to the Renilla luciferase signal and the silencing efficacy of each siRNA construct was calculated relative to luciferase activity of siGFP transfected samples. All experiments were repeated three times. To calculate the efficiency of siRNAs to discriminate between perfectly matched (WT) and mismatched (Mut) targets, the ratio of silencing efficiency for the Mut to WT was calculated for each mismatched target and plotted as fold discrimination. Mean values of discrimination for all mismatches were also computed.
Quantitative real-time PCR. HeLa cells were transfected with siRNAs for 24 hours. Total RNA was extracted using TRI Reagent (Ambion) and 1 µg of RNA was used as a template for cDNA synthesis, using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) according to the manufacture's protocol. Aliquots of the cDNA reaction mixture were analyzed by quantitative real-time PCR on a Step-One real-time PCR machine (Applied Biosystems). All primer pairs (Supplementary Table S3) spanned an intron to avoid possible genomic DNA contamination. PCR was carried in MicroAmPTM Fast Optical 48-well Reaction Plate (Applied Biosystems). Quantitative Real-Time PCR data for each gene product was normalized with GAPDH transcript levels and are reported as mean ± SE relative changes compared to siGFP-treated samples.
Microarray and data analysis. Total RNA was extracted using the TRI Reagent and an RNeasy mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturers' protocols. Total RNA (10 µg) was used for each double-stranded cDNA synthesis, employing a commercial kit (Invitrogen). Reactions were stopped with EDTA and the double-stranded cDNA treated with RNase A. Samples were then ethanol-precipitated. One microgram of double-stranded cDNA was used for labeling by Klenow fragment (New England Biolabs, Beverly, MA) using Cy3-labeled random 9 mer (TriLink Biotechnologies, San Diego, CA) and labeled samples were precipitated using isopropanol. Cy3-labeled DNA (4 µg) containing sample tracking control and alignment oligo was hybridized to Nimblegen 385K 4-plex human microarray for 18 hours at 42 °C using the Nimblegen Hybridization system (Roche Nimbelgen, Madison, WI). Arrays were washed and array images were obtained using the InnoScan 900 scanner (Innopsys, Carbonne, France). Scanned images were imported into Mapix software (Innopsys). Expression data was normalized through quantile normalization46 and Robust Multichip Average algorithm.47 We removed 0.3% of transcripts from either end of the intensity distribution as outliers, leaving 23,856 transcripts to be used in this study. Of these, 18,978 sequences that represented the RefSeq human mRNA sequences (hg18 March 2006) were considered for further analysis. siSurvivin antisense and sense seed (nucleotides 2–8, 2–7, 1–7) were matched with all distinct human RefSeq 3′-UTRs using Target Rank48 (http://genes.mit.edu/targetrank/). This gave us a list of all RefSeq entries that had 3′-UTR seed matches. Using this sequence output, the seed-matched transcripts in the experimental dataset of 18,978 sequences were identified. Overall changes in the mRNA level of transcripts with and without siRNA seed matches were visualized with MA plots. Cumulative distribution of the log2 fold changes in gene expression of the mRNA with no seed matches or seed-matched transcripts was also calculated.
Cell viability assay. Cell viability was determined by MTT colorimetric assay (TaKaRa, Bio, Shiga, Japan) according to the manufacturer's instructions. Briefly, HeLa cells seeded in 96 well plates were transfected with 10 nmol/l of siRNAs and after 96 hours of transfection, 20 µl of MTT reagent was added to 100 µl of complete media. Cells were then incubated for 30 minutes at 37 °C in a humified atmosphere with 5% CO2. The growth medium was removed and 100 µl of complete dimethyl sulphoxide was added to the wells. The plates were kept on a shaker for 5 minutes and color development was measured on an ELISA plate reader at 540 nm with reference wavelength of 620 nm. All absorbance values were corrected against a blank that contained media alone. Percent cell viability was calculated considering lipofectamine-treated control as 100% viable
5′-RACE assay. RACE assay was carried out as described previously17 with some modifications. Briefly, Twenty-four hours after siRNA (10 nmol/l) transfection into HeLa cells, total RNA was extracted using Tri-reagent kit. Total RNA (5 µg) was then ligated to 0.25 µg of GeneRacer RNA oligo (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAG UAGAAA-3) without prior treatment. The GeneRacer RNA oligoligated total RNA was then reverse transcribed using a oligo dT and a SuperScript II RT kit (Invitrogen). After 25 cycles of PCR using the GeneRacer 5 primer and a gene-specific 3 primer, 20 cycles of nested PCR was performed using the GeneRacer 5′ nested primer and a gene-specific 3′ nested primer. The resulting PCR product was then cloned into a T&A cloning vector (RBC Bioscience, Tapei, Taiwan) and at least 12 independent clones from each PCR were sequenced. Sequences of primers used are provided in Supplementary Table S3.
SUPPLEMENTARY MATERIAL Figure S1. Reduction in antisense off-target silencing by bulge-modified PIK3CB siRNA. Figure S2. Antisense-mediated off-target silencing of unmodified and bulge-modified siRNAs at higher concentrations. Figure S3. Fork-siRNAs could not reduce antisense off-target silencing. Figure S4. Bulge-siRNA do not trigger any antiviral responses. Figure S5. Silencing efficiency of survivin siRNAs for a set of randomly selected mRNAs from the microarray dataset that have seed complementarity with siSurvivin-2′C′. Figure S6. Silencing efficiency of unmodified and bulge-modified survivin siRNAs for perfectly matched antisense target or the siRNA seed sequence. Figure S7. Bulge-siRNA show increased Ago-2 incorporation. Table S1. List of DNA oligos used to construct the on-target and off-target reporters. Table S2. Thermodynamic measurements for the unmodified and bulge-modified survivin siRNAs. Table S3. List of primers used for real-time PCR and 5′RACE assay. Materials and Methods.
Acknowledgments
This study was supported by a Global Research Laboratory grant (2008-00582) from the Korean Ministry of Education, Science, and Technology to D.-k. L.
Supplementary Material
Reduction in antisense off-target silencing by bulge-modified PIK3CB siRNA.
Antisense-mediated off-target silencing of unmodified and bulge-modified siRNAs at higher concentrations.
Fork-siRNAs could not reduce antisense off-target silencing.
Bulge-siRNA do not trigger any antiviral responses.
Silencing efficiency of survivin siRNAs for a set of randomly selected mRNAs from the microarray dataset that have seed complementarity with siSurvivin-2′C′.
Silencing efficiency of unmodified and bulge-modified survivin siRNAs for perfectly matched antisense target or the siRNA seed sequence.
Bulge-siRNA show increased Ago-2 incorporation.
List of DNA oligos used to construct the on-target and off-target reporters.
Thermodynamic measurements for the unmodified and bulge-modified survivin siRNAs.
List of primers used for real-time PCR and 5′RACE assay.
Ago-2 incorporation assay Melting temperature determination
REFERENCES
- Dorsett Y., and, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM., and, Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu Rev Biomed Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- Jackson AL., and, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W., and, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M.et al. (2003Expression profiling reveals off-target gene regulation by RNAi Nat Biotechnol 21635–637. [DOI] [PubMed] [Google Scholar]
- Jackson AL., and, Linsley PS. Noise amidst the silence: off-target effects of siRNAs. Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Clark PR, Pober JS., and, Kluger MS. Knockdown of TNFR1 by the sense strand of an ICAM-1 siRNA: dissection of an off-target effect. Nucleic Acids Res. 2008;36:1081–1097. doi: 10.1093/nar/gkm630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Yoo JW, Hong SW, Kim S., and, Lee DK. Inflammatory cytokine induction by siRNAs is cell type- and transfection reagent-specific. Biochem Biophys Res Commun. 2006;347:1053–1058. doi: 10.1016/j.bbrc.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S.et al. (2005Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7 Nat Med 11263–270. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR.et al. (2006Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways Nature 441537–541. [DOI] [PubMed] [Google Scholar]
- Vickers TA, Lima WF, Nichols JG., and, Crooke ST. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35:6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC., and, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JW, Kim S., and, Lee DK. Competition potency of siRNA is specified by the 5'-half sequence of the guide strand. Biochem Biophys Res Commun. 2008;367:78–83. doi: 10.1016/j.bbrc.2007.12.099. [DOI] [PubMed] [Google Scholar]
- Li X, Yoo JW, Lee JH, Hahn Y, Kim S., and, Lee DK. Identification of sequence features that predict competition potency of siRNAs. Biochem Biophys Res Commun. 2010;398:92–97. doi: 10.1016/j.bbrc.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Wei JX, Yang J, Sun JF, Jia LT, Zhang Y, Zhang HZ.et al. (2009Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells PLoS ONE 4e5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Yoo JW, Hong SW, Lee SE, Kang HS, Sun X.et al. (2009Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects Mol Ther 17725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y.et al. (2005Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality Nucleic Acids Res 33439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Rogoff HA., and, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- Sano M, Sierant M, Miyagishi M, Nakanishi M, Takagi Y., and, Sutou S. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T.et al. (2008Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity RNA 14263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L.et al. (2006Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity RNA 121179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW.et al. (2005siRNA-mediated off-target gene silencing triggered by a 7 nt complementation Nucleic Acids Res 334527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB., and, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K.et al. (2006Off-target effects by siRNA can induce toxic phenotype RNA 121188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J.et al. (2006Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing RNA 121197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Wang X, Varma R, Burnett C, Beauchamp L, Batten DM.et al. (2008LNA incorporated siRNAs exhibit lower off-target effects compared to 2'-OMethoxy in cell phenotypic assays and microarray analysis Nucleic Acids Symp Ser (Oxf) 25–26. [DOI] [PubMed]
- Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D.et al. (2010A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects Nucleic Acids Res 385761–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F.et al. (2008Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect Nucleic Acids Res 362136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T., and, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C., and, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K., and, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren C, Zhang HY, Du Q, Grahn M, Norstedt G, Wahlestedt C.et al. (2008Analysis of siRNA specificity on targets with double-nucleotide mismatches Nucleic Acids Res 36e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 2004;557:193–198. doi: 10.1016/s0014-5793(03)01492-3. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Tokunaga K., and, Hohjoh H. Influence of assembly of siRNA elements into RNA-induced silencing complex by fork-siRNA duplex carrying nucleotide mismatches at the 3'- or 5'-end of the sense-stranded siRNA element. Biochem Biophys Res Commun. 2005;329:516–521. doi: 10.1016/j.bbrc.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y.et al. (20063' UTR seed matches, but not overall identity, are associated with RNAi off-targets Nat Methods 3199–204. [DOI] [PubMed] [Google Scholar]
- Gantier MP, Tong S, Behlke MA, Irving AT, Lappas M, Nilsson UW.et al. (2010Rational design of immunostimulatory siRNAs Mol Ther 18785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znosko BM, Silvestri SB, Volkman H, Boswell B., and, Serra MJ. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges. Biochemistry. 2002;41:10406–10417. doi: 10.1021/bi025781q. [DOI] [PubMed] [Google Scholar]
- Blose JM, Manni ML, Klapec KA, Stranger-Jones Y, Zyra AC, Sim V.et al. (2007Non-nearest-neighbor dependence of the stability for RNA bulge loops based on the complete set of group I single-nucleotide bulge loops Biochemistry 4615123–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Nishi K, Juni A., and, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yoshimura J, Morishita S., and, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP., and, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F., and, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H., and, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M., and, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B., and, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J., and, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reduction in antisense off-target silencing by bulge-modified PIK3CB siRNA.
Antisense-mediated off-target silencing of unmodified and bulge-modified siRNAs at higher concentrations.
Fork-siRNAs could not reduce antisense off-target silencing.
Bulge-siRNA do not trigger any antiviral responses.
Silencing efficiency of survivin siRNAs for a set of randomly selected mRNAs from the microarray dataset that have seed complementarity with siSurvivin-2′C′.
Silencing efficiency of unmodified and bulge-modified survivin siRNAs for perfectly matched antisense target or the siRNA seed sequence.
Bulge-siRNA show increased Ago-2 incorporation.
List of DNA oligos used to construct the on-target and off-target reporters.
Thermodynamic measurements for the unmodified and bulge-modified survivin siRNAs.
List of primers used for real-time PCR and 5′RACE assay.
Ago-2 incorporation assay Melting temperature determination