Abstract
Despite the promise of RNA interference (RNAi) therapeutics, progress toward the clinic has been slowed by the difficulty of delivering short interfering RNA (siRNA) into cellular targets within the body. Nearly all siRNA delivery vehicles developed to date employ a single cationic or ionizable material. In order to increase the material space available for development of siRNA delivery therapeutics, this study examined the possibility of using binary combinations of ionizable lipid-like materials to synergistically achieve gene silencing. Interestingly, it was found that ineffective single lipid-like materials could be formulated together in a single delivery vehicle to induce near-complete knockdown of firefly luciferase and factor VII in HeLa cells and in mice, respectively. Microscopy experiments suggested that synergistic action resulted when combining materials that respectively mediated cellular uptake and endosomal escape, two important steps in the delivery process. Together, the data indicate that formulating lipid-like materials in combination can significantly improve siRNA delivery outcomes while increasing the material space available for therapeutic development. It is anticipated that this binary formulation strategy could be applicable to any siRNA delivery material in any target cell population that utilizes the two-step endosomal delivery pathway.
Introduction
The discovery of RNA interference (RNAi) in mammalian cells1 has enabled the development of short interfering RNA (siRNA) therapeutics,2 which have the potential to treat a wide variety of human diseases, including viral infections3,4,5,6,7 and cancer,8,9,10 through genetic modulation. Theoretically, siRNA can be used to alter the expression of nearly any gene in the body through the silencing of complementary messenger RNA. Such precise genetic control offers a broad therapeutic potential that is typically not attainable using conventional small molecule drugs. It has already been reported that synthetic siRNAs are capable of treating a variety of disease targets in vivo, including hypercholesterolemia,11,12 liver cirrhosis,13 intestinal colitis,14 human immunodeficiency virus,4,15 and ovarian cancer.9,16
Advances in siRNA delivery vehicle technology must be made before RNAi can attain its broadest potential in the clinic. Efficacious delivery materials facilitate several steps in the delivery process. In addition to escorting therapeutic cargo through the bloodstream and extracellular matrix, delivery vehicles must mediate siRNA transport across the cellular membrane of the target cell and facilitate endosomal escape before lysosomal digestion.17,18 It is only once all of these barriers have been breached that siRNA can interact with the RNAi machinery in the cytoplasm and trigger the gene silencing process.19
To date, siRNA delivery has been achieved primarily using vehicles that contain a single cationic or ionizable delivery material.3,10,20,21,22 On the cellular level, these efficacious materials must mediate several distinct steps in the siRNA delivery process, including cellular entry and intracellular compartmental escape. As such, most siRNA delivery efforts have focused on materials able to mediate all steps in the delivery pathway, particularly in vitro. In order to increase the material space available for therapeutic development, we started with a one-dimensional space of materials and studied them in binary combination to explore a two-dimensional space of materials. Using this technique, we sought to identify synergistic combinations of siRNA delivery materials capable of facilitating complete delivery. It was hypothesized that gene silencing could be achieved by formulating a material capable of cellular entry with a material capable of compartmental escape together into one delivery nanoparticle.
This hypothesis was interrogated using a library of lipid-like materials, termed lipidoids.20 Lipidoids, which contain tertiary amines, are among the ionizable lipids and polymers identified as promising nonviral siRNA delivery vehicles.20,23,24,25,26 Previously, over 1,000 lipidoids have been analyzed for delivery efficacy, and several leading candidates have been employed for siRNA delivery applications.9,16,20 However, the library also included hundreds of members unable to induce gene silencing. Using these compounds as model delivery materials, we sought out synergistic delivery behavior through the analysis of nanoparticles formulated with binary combinations of ineffective lipidoids. Herein, the search for and identification of synergistic lipidoid combinations is described.
Results
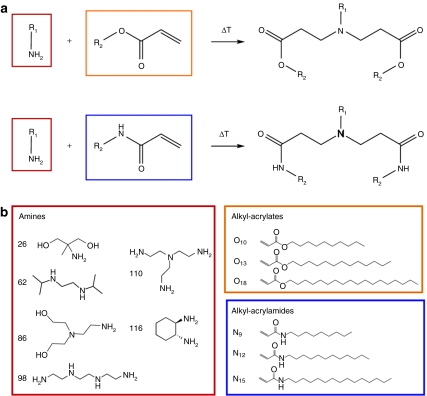
In order to explore the potential of binary lipidoid combinations for enhanced siRNA delivery, 36 lipidoids from the original library20 were selected for further examination. These lipidoids were synthesized through the Michael addition of an amine to an alkyl-acrylate or alkyl-acrylamide tail (Figure 1a). The lipidoid building blocks employed in this study (Figure 1b) were chosen to incorporate structural diversity (e.g., number and length of tails) into the 36 resulting lipidoids, many of which had been previously determined to lack transfection capability in vitro.20
Figure 1.
Lipidoid synthesis reaction and library. (a) A Michael addition reaction was used to synthesize lipidoids from the conjugate addition of amino molecules to alkyl-acrylate or alkyl-acrylamide molecules. (b) Six amines, three alkyl-acrylates, and three alkyl-acrylamides were used to create a library of 36 lipidoids. Lipidoids are named for the two molecules from which they were synthesized (e.g., 26O10 or 98N9).
It was hypothesized that certain binary combinations of lipidoids may form better siRNA delivery vehicles than either lipidoid alone. To test this hypothesis, the 36 synthetic lipidoids were combined with one another during the formulation process before the addition of siRNA. These combinations yielded 630 binary pairs, which were each generated at six weight fractions varying from 0 to 1 with a step size of 0.2. This resulted in a total of 3,780 unique formulations, which were tested for their ability to deliver siRNA to a HeLa cell line that stably expressed both Renilla and firefly luciferase proteins. All in vitro experiments were performed at a lipioid: siRNA weight ratio of 5:1 and a siRNA concentration of 20 nmol/l. Efficacy was determined by delivering an anti-firefly luciferase siRNA and measuring the ratio of firefly to Renilla luciferase activity. Renilla activity served as a built-in control, as lipidoids with cytotoxic or nonspecific silencing properties resulted in reduced activity of both types of luciferase. Results are presented as relative firefly luciferase activity, which is the ratio of firefly luciferase activity relative to the untreated control after normalization to Renilla luciferase activity.
The primary goal of the in vitro screen was to determine whether binary combinations of lipidoids offered improved transfection over their individual lipidoid counterparts. In order to quantify any improvements in gene silencing enabled by a lipidoid formulation comprised of two distinct lipidoids (A and B), we defined a synergy value (S value) as follows:
S value = XA · FA + (1−XA) · FB−FAB (1)
where XA is the weight fraction of lipidoid A, and FA, FB, and FAB are the relative firefly luciferase activity levels achieved with lipidoids A, B, and the A-B combination at weight fraction XA, respectively. In other words, the S value represents the difference between the expected value and the actual value of luciferase expression for lipidoid combinations. S values can range from 1 to −1, with 1 representing a combination that, while capable of maximal gene silencing in vitro, is comprised of two lipidoids that have no transfection ability on their own. An S value of 0 indicates that the combination offers no advantage over its single counterparts, whereas a negative S value signifies that a combination is less effective than its corresponding individual components.
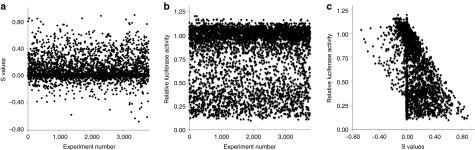
By plotting S values, the wide variety of transfection results obtained when lipidoid combinations were screened in HeLa cells can be observed (Figure 2a). Many binary formulations offered no significant improvement over their individual lipidoids (S values near 0), as is evidenced by the clustering of data points near the middle of the graph. Interestingly, however, 5% of the specific binary combinations screened in vitro exhibited S values ≥0.5 without inducing cytotoxicity, providing proof-of-principle that ineffective individual lipidoids can synergistically enable significant levels of gene silencing when formulated in combination.
Figure 2.
The in vitro screen identified numerous efficacious and synergistic lipidoid delivery formulations. (a) Although the transfection properties of most binary lipidoid combinations were not improved over their single lipidoid counterparts (S values close to zero), a small fraction of lipidoid combinations enabled markedly improved gene silencing (S values close to one). (b) Library members exhibited a broad range of transfection abilities. While the majority of materials lacked efficacy (relative luciferase values near 1), one quarter of the library was capable of inducing >50% firefly luciferase silencing. (c) High S values correlated well with higher levels of gene silencing, and low S values indicated poor gene silencing. Intermediate S values (−0.2 < S < 0.2) had little to no correlation with in vitro luciferase knockdown ability.
The in vitro gene silencing efficacy of the nearly 4,000 binary lipidoid combinations tested is shown in Figure 2b. A range of transfection behavior is seen across the library, with the majority of materials lacking efficacy (relative luciferase values near 1). A subset of the library, however, exhibited significant firefly luciferase knockdown efficiency. 26% and 8% of the materials tested induced >50% and >75% gene silencing, respectively. As a point of reference, the leading compound from the original lipidoid library, 98N12, mediates 61% silencing under the in vitro conditions used in this study.20 Figure 2c shows the relationship between S values and in vitro silencing efficiency for all members of the library. High S values correlated well with higher levels of gene silencing, and low S values indicated poor gene silencing. Intermediate S values (−0.2 < S < 0.2) had little to no correlation with in vitro luciferase knockdown ability.
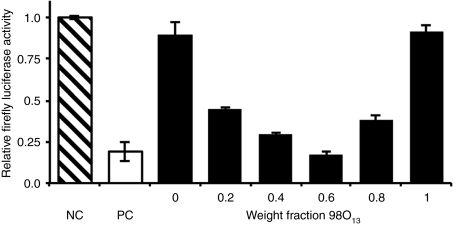
Although many binary combinations yielded high S values, we will use one particular lipidoid pair, 86N15–98O13, to illustrate the synergistic potential of a binary delivery material approach. It is representative of other synergistic combinations. Figure 3 demonstrates the in vitro ability of various weight fractions of the 86N15–98O13 combination to silence the production of firefly luciferase in HeLa cells. Firefly luciferase levels are reported relative to normal expression in HeLa cells (negative control). Although neither 86N15 (X98O13 = 0) nor 98O13 (X98O13 = 1) demonstrated any detectable transfection capability on their own in vitro, combinations of the two lipidoids at all weight fractions exhibited marked improvement in firefly luciferase silencing, with the best performing combination of 86N15–98O13 (X98O13 = 0.6) yielding 83% knockdown and an S value of over 0.7.
Figure 3.
The binary lipidoid combination 86N15–98O13 mediated synergistic gene silencing in vitro. The negative control (NC), nontreated HeLa cells, maintained full firefly luciferase activity (hatched bar), while the positive control (PC), HeLa cells transfected with Lipofectamine RNAiMax, exhibited ~80% knockdown (white bar). Binary lipidoid combinations significantly enhanced short interfering RNA (siRNA) delivery over single lipidoid counterparts (black bars). All compounds were tested at an siRNA concentration of 20 nmol/l. Error bars represent s.d. (n = 4).
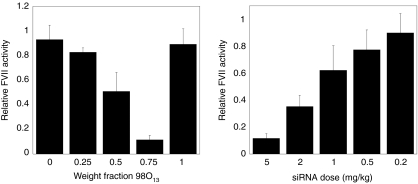
In order to further assess the potential of lipidoid siRNA delivery vehicles, in vivo testing was performed on the leading hits from the in vitro screen. Binary lipidoid combinations were formulated with cholesterol, polyethylene glycol, and siRNA into nanoparticles, which were then injected into mice via the tail vein. The siRNA used in in vivo formulations was specific against the factor VII gene, which encodes a model protein produced in hepatocytes. Silencing was assessed at 48 hours postinjection through the measurement of factor VII levels in the serum. Several binary lipidoid combinations were capable of inducing gene silencing in mouse hepatocytes, with the combination 86N15–98O13 demonstrating the most significant synergistic siRNA delivery capability (Figure 4).
Figure 4.
The binary lipidoid combination 86N15−98O13 synergistically improved gene silencing in mice. (a) Factor VII silencing of >85% is achieved with weight fraction 98O13 = 0.75, despite the ineffectiveness of the single lipidoid constituents (weight fractions = 0, 1). Formulations were delivered systemically at a total short interfering RNA (siRNA) dose of 5 mg/kg. (b) The factor VII silencing capability of the lead combination, 86N15−98O13 (weight fraction 98O13 = 0.75), is dose-dependent. Error bars in both figures represent s.d. (n = 3–6).
The combination 86N15–98O13 was first analyzed at a total siRNA dose of 5 mg/kg, shown in Figure 4a. The single lipidoid components, 86N15 (X98O13 = 0) and 98O13 (X98O13 = 1), did not facilitate factor VII silencing. However, each of the binary combinations of 86N15 and 98O13 offered some level of gene silencing improvement. The most efficacious delivery to mouse hepatocytes was achieved at a 98O13 weight fraction of 0.75, which resulted in ~85% factor VII knockdown at 5 mg/kg siRNA. A dose–response analysis was performed for this particular combination (Figure 4b). 86N15–98O13 (X98O13 = 0.75) mediated dose-dependent protein silencing, with an IC50 dose of ~1.5 mg/kg. The toxicity of lipidoid nanoparticles was assessed by monitoring mouse body weight at the time of injection and 48 hours postinjection. No changes in bodyweight for any of the combinations depicted in Figure 4 were detected compared to negative control mice.
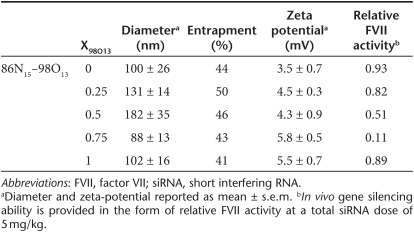
Nanoparticle characterization data for the 86N15–98O13 combination lipidoids can be found in Table 1. Sizes ranged from 78 to 182 nm in diameter, while zeta potentials were all slightly positive, reflecting the slightly protonated state of the lipidoid amines at a neutral pH. siRNA entrapment values were all ~50%. Neither the size, siRNA entrapment ability, nor surface charge appear to lend any obvious advantage to the most efficacious combination, 86N15–98O13 (X98O13 = 0.75).
Table 1. Characterization of selected lipidoid binary combinations.
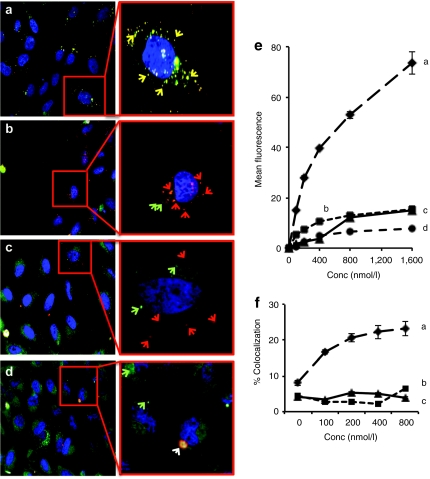
To elucidate the in vitro mechanism of action of the leading lipidoid combination, 86N15–98O13, uptake and the final fate of the delivery material was monitored using automated high throughput confocal microscopy. The lipidoids 86N15 and 98O13, and the synergistic combinations 86N15–98O13 (X98O13 = 0.5) and 86N15–98O13 (X98O13 = 0.75), were analyzed in this study. HeLa cells were transfected with lipidoid nanoparticles containing Alexa 594-labeled siRNA at doses ranging from 50 to 1,600 nmol/l. Cells were imaged at 60 minutes post-transfection, and fluorescence intensity was quantified (Figure 5). Both the images (Figure 5a–d) and the quantification (Figure 5e) indicate that 86N15 was internalized in greater amounts than the other lipidoids. In contrast, cells were not able to internalize 98O13 at any concentration (Figure 5d,e). Combinations of 86N15 and 98O13, however, showed intermediate uptake, with higher weight fractions of 86N15 corresponding with higher levels of internalization (Figure 5b,c,e). A similar internalization pattern of lipidoid nanoparticles was observed with alternative dyes (Cy3 and Cy5) and with longer incubation times (24 hours) at low nanoparticle concentrations (50 nmol/l) (data not shown).
Figure 5.
Synergistic 86N15–98O13 lipidoid combinations were uptaken into cells and avoided accumulation in the lysosomes. Confocal images were taken of the lipidoids (a) 86N15, (b) 86N15–98013 (weight fraction 98O13 = 0.5), (c) 86N15–98O13 (weight fraction 98O13 = 0.75), and (d) 98O13 formulated into nanoparticles with Alexa-594-short interfering RNA (siRNA) and exposed to HeLa cells in the presence of Lysotracker Green. Representative images of the 400 nmol/l siRNA dose are shown. Inset-magnified images have been labeled with red and green arrows indicative of lipidoid and lysosomes, respectively. Yellow arrows show colocalization between the lipidoid nanoparticles and the endosomes, while the white arrow in d shows aggregated nonspecific binding of lipidoid to plate. (e) Quantification of lipidoid internalization. (f) Quantification of internalized lipidoid nanoparticle colocalization with lysosomes. All quantification data presented was averaged from 20 different fields per well using Acapella software. Experiments were performed in triplicate, and error bars represent s.e.
Next, to assess the intracellular trafficking fate of lipidoid nanoparticles, we quantified the localization of the nanoparticles with LysoTracker Green, a marker for lysosomes, at 1 hour post-transfection. 98O13 was not included in this portion of the study, as it did not internalize sufficiently to provide useful trafficking data. 86N15 nanoparticles experienced a dose-dependent increase in the % colocalization with cell lysosomes (Figure 5f), indicating that these nanoparticles are being trafficked into degradative pathways. The percent colocalization for 86N15 is quantitatively lower than what appears in Figure 5a because quantitation has been performed using Acapella software (see Materials and Methods section), which has a strict definition of colocalization. In contrast, combinations of 86N15–98O13 did not show appreciable localization with lysosomes (~3%), even at higher doses (Figure 5f).
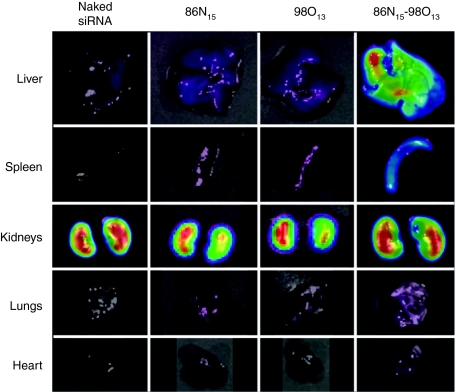
Lipidoid nanoparticles were also imaged in vivo in order to evaluate any differences between synergistic combinations and ineffective single lipidoids following intravenous delivery. Biodistribution profiles related to the synergistic 86N15–98O13 system obtained using an IVIS imaging system are shown in Figure 6. All nanoparticles were formulated with Cy5-labeled siRNA and were injected at a total siRNA dose of 1 mg/kg. One hour postinjection, organs were excised from sacrificed animals and imaged using an IVIS imaging system. The images in Figure 6 demonstrate that naked siRNA as well as siRNA introduced via 86N15 and 98O13 nanoparticles did not accumulate in the liver, spleen, lungs, or heart. On the other hand, the synergistic delivery combination 86N15–98O13 (X98O13 = 0.75) enabled siRNA localization to the target organ, the liver, as well as the spleen. All siRNA formulations were subject to some level of clearance at 1 hour postinjection, as was evidenced by siRNA accumulation in the kidneys of all injection groups.
Figure 6.
The synergistic binary combination 86N15–98O13 localizes to the liver. Mice were injected via the tail vein with lipidoid delivery formulations containing Cy5-labeled short interfering RNA (siRNA), and organs were harvested 60 minutes postinjection and imaged using an IVIS imaging system. Neither naked siRNA nor single lipidoid formulations accumulated in any organs besides the kidneys. However, the synergistic binary combination 86N15–98O13 mediated significant siRNA accumulation in the liver. Experiments were performed in triplicate, with representative images shown here.
In order to assess the importance of serum stability on in vivo gene silencing, all 86N15, 98O13, and the three combinations 86N15–98O13 (X98O13 = 0.25, 0.5, and 0.75) were exposed to serum for a period of 24 hours. Fluorescent siRNAs individually tagged with Alexa 594 and Alexa 647 were incorporated into each lipidoid nanoparticle. Fluorescence resonance energy transfer (FRET) was used to assess the proximity of siRNAs contained within the nanoparticle to one another. Decreases in FRET signal correspond to an increased distance between neighboring siRNAs, indicating that the nanoparticle is losing stability. Data obtained using this technique indicated that all of the nanoparticles tested were very stable in serum over a 24 hours period, with only 5–20% decreases in FRET signal (Supplementary Figure S1). At 24 hours, a 1% Triton-X solution was added to the nanoparticles to confirm extinguishment of FRET signal upon particle rupture.27 This suggests that serum stability was not specifically responsible for the synergistic gene silencing ability of 86N15–98O13 (X98O13 = 0.75).
Discussion
Since the discovery of RNAi, the identification of safe and efficacious siRNA delivery materials has proven to be a considerable challenge. Although progress has been made in identifying increasingly viable delivery candidates, either through rational design25 or high-throughput synthesis,12,20 no such material has yet been approved for use in the clinic. Most siRNA delivery vehicles developed to date employ a single cationic or ionizable material that negotiates the varied obstacles en route to the cytoplasm of target cells. In an attempt to rescue ineffective delivery materials while increasing the material space available for therapeutic development, this study examined the possibility of using combinations of ionizable materials to synergistically achieve gene silencing using lipidoid materials as a model delivery system. Interestingly, we have shown that it is possible to formulate two ineffective siRNA delivery materials together in a nanoparticle to facilitate near-complete protein silencing both in vitro and in vivo.
Our data suggest that, in vitro, synergistic gene silencing phenomena may involve two important steps in the siRNA delivery process. Although certain lipidoids are able to facilitate transport of siRNA across the cellular membrane, they may lack the ability to escape from the intracellular compartments once inside the cell. Similarly, other lipidoids may mediate intracellular delivery to the cytoplasm, but are not able to gain cellular entry. However, by formulating these materials together into one delivery vehicle, it may be possible to capitalize upon the unique capabilities of each lipidoid to achieve RNAi-mediated gene silencing.
This hypothesis is supported by the confocal microscopy experiments is shown in Figure 5. 86N15 entered cells readily in vitro but was not capable of inducing siRNA-mediated gene knockdown, indicating that it faltered as a delivery material at a step downstream of cellular entry. 98O13, on the other hand, was not adept at cellular entry. Yet, when combined together in one delivery vehicle, 86N15 and 98O13 work synergistically to deliver siRNA to the cytoplasm and enable gene silencing. The data, taken together, imply that 86N15 helps the combination enter cells while 98O13 facilities downstream delivery obstacles, allowing siRNA to reach the RNAi machinery in the cytosol. The asymmetry of the data presented in Figure 4a lends further insight into the in vitro delivery mechanisms of the synergistic combination 86N15–98O13. Only a small amount of 86N15 needed to be added to 98O13 to achieve effective factor VII silencing. As such, 86N15 is likely more efficient at gaining cellular entry than 98O13 is at mediating internal delivery.
Interestingly, the biodistribution data presented in Figure 6 suggest that the mechanism for synergistic delivery in vivo may be more complex than what had been observed in vitro. When siRNA was delivered using single lipidoids 86N15 and 98O13, it accumulated only in the kidneys of injected mice. This type of biodistribution is the same as that achieved with naked siRNA. Additionally, it was found that both the efficacious combination 86N15–98O13 (X98O13 = 0.75) as well as its individual nonpotent counterparts, 86N15 and 98O13, were very stable in serum over a 24-hour period (Supplementary Figure S1). This indicates that serum stability does not give the synergistic combination a delivery advantage over 86N15 and 98O13. Together, these data suggest that the combination 86N15–98O13 (X98O13 = 0.75) derives its synergistic in vivo siRNA delivery ability from two sources. It (i) facilities uptake into hepatocytes via 86N15, and (ii) mediates intracellular delivery of siRNA into the cytoplasm via 98O13, where the siRNA interacts with RNAi machinery to trigger gene silencing.
Results presented here establish the proof-of-principle that binary combinations of lipidoid materials can be formulated together in a single vehicle to synergistically achieve high levels of gene silencing both in vitro and in vivo. Because the multistep siRNA delivery process is not unique to lipidoid-mediated delivery, it is anticipated that a binary formulation strategy may be used achieve enhanced delivery in any target cell population with any delivery material that employs endosomal delivery, including lipids, polymers and siRNA conjugates. Such a discovery increases the material space available for therapeutic development of siRNA delivery materials and has the potential to hasten the advent of RNAi applications in the clinic.
Materials and Methods
Lipidoid synthesis. Lipidoids were synthesized through the addition of alkyl-acrylates or alkyl-acrylamides to amines. Amines were purchased from Sigma-Aldrich (St Louis, MO), acrylates were purchased from Scientific Polymer Products (Ontario, NY) and Hampford Research (Stratford, CT) and acrylamides were synthesized via the drop-wise addition of acryloyl chloride to the appropriate 1-aminoalkane in dry tetrahydrofuran at −15 °C with stirring for several hours. Two hundred milligram of amine was added to the maximal stoichiometric ratio of acrylate or acrylamide. The mixture was stirred in a glass scintillation vial at 90 °C for either for 1 or 7 days for acrylates or acrylamides, respectively.
In vitro testing of lipidoids. HeLa cells, which were modified to stably express both firefly and Renilla luciferase, were maintained in phenol-red free Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C and 5% carbon dioxide. Each lipidoid was mixed with 50 ng of anti-firefly luciferase siRNA (Dharmacon, Lafayette, CO) at a weight ratio of 5:1 in 25 mmol/l sodium acetate buffer. Then, 50 µl of siRNA-lipidoid complexes were applied to 1.5 × 10 HeLa cells in serum-containing medium. Transfections were performed at an siRNA concentration of 20 nmol/l, and the transfection medium (which included 10% serum) was kept on cells for the entire experiment. Luciferase silencing was assessed 24 hours post-transfection using a Dual-Glo Luciferase Assay kit (Promega, Madison, WI). Transfections were performed in quadruplicate. Lipofectamine RNAiMax (Invitrogen) was used as a positive control.
Formulation of lipidoid nanoparticles for in vivo applications. Lipidoid nanoparticles were formed by mixing lipidoids, cholesterol (Sigma-Aldrich), and mPEG2000-DMG (MW 2660, synthesized by Alnylam Pharmaceuticals20) at a molar ratio of 42:48:10 in ethanol and then diluting in 200 mmol/l sodium acetate buffer. Particles were extruded through a 100-nm filter membrane. Anti-factor VII siRNA was then mixed with the lipidoid nanoparticles at a total lipid:siRNA weight ratio of 10:1 and incubated for 30 minutes at 37 °C. Nanoparticles were dialyzed into phosphate-buffered saline (PBS) for 75 minutes in 3,500 MWCO cassettes (Pierce/Thermo Scientific, Rockford, IL). siRNA concentration and entrapment efficiency were determined using the Quant-iT RiboGreen RNA assay from Invitrogen.27 Nanoparticle size and zeta-potential were acquired in PBS using a ZETAPals analyzer (Brookhaven Instruments, Holtsville, NY). Each formulation was run in triplicate. Particle size and zeta potential are expressed as the average values of each run.
siRNA delivery in mice using lipidoids. All animal experiments were conducted using institutionally approved protocols. Female C57BL/6 mice (Charles River Laboratories, Wilmington, MA) received tail vein injections of either PBS (negative control) or lipidoid nanoparticles containing anti-factor VII siRNA diluted in PBS at a volume of 0.01 ml/g. siFVII was synthesized by Alnylam Pharmaceuticals, and the sequence is as follows:
sense: 5′-GGAucAucucAAGucuuAcT*T-3′
antisense: 5′-GuAAGAcuuGAGAuGAuccT*T-3′,
where 2′-fluoro-modified nucleotides are in lower case and phosphorothioate linkages are represented by asterisks. Two days postinjection, mice were anesthetized via isoflurane inhalation and bled retro-orbitally into serum separator tubes. Serum levels of factor VII were analyzed using a Biophen FVII assay kit (Aniara, Mason, OH).
Whole-mouse fluorescent imaging. Female SKH1 hairless mice received tail vein injections of either PBS (negative control) or lipidoid nanoparticles containing Cy5-labeled siRNA (Integrated DNA Technologies, Coralville, IA) at a dose of 1 mg/kg and volume of 0.01 ml/g. For organ imaging experiments, mice were euthanized at various time points postdelivery, and the heart, lungs, liver, spleen, and kidneys were removed for imaging with an IVIS Spectrum system (Caliper Life Sciences, Hopkinton, MA). Excitation and emission wavelengths used for Cy5 imaging on the IVIS were 640 and 680 nm, respectively.
Automated confocal microscopy. HeLa cells were seeded at 1.5 × 104 cells/well in black 96-well plates (Greiner Bio-one, Longwood, FL). Cells were starved for 30 minutes in the presence of PBS/BSA and incubated with various concentrations (0–1,600 nmol/l) of lipidoid nanoparticles-containing BLOCK-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) for 60 minutes at 37 °C. Cells were washed with PBS and incubated with LysoTracker green (500 nmol/l) for 15 minutes. Cells were then counterstained in OPTIMEM-containing Hoescht (2 µg/ml) for nuclei identification. Triple-stained live cell imaging was performed with an automated spinning disk confocal microscope (OPERA; Perkin Elmer, Shelton, CT) with a ×40 objective. The same defined pattern of 20 fields from each well was acquired to eliminate bias and provide a statistically significant number of cells for analysis. After identification of cell location and perimeter, intracellular siRNA signal intensity and % colocalization over single field was calculated using Acapella software. Data presented are an average of intracellular intensity from 20 different fields.
FRET. FRET was used to assess the stability of nanoparticles in serum through the measurement between siRNA neighbors within the nanoparticle. Lipidoid nanoparticles were formulated for in vivo applications as described above, except that half of the siRNA that was used was labeled with Alexa 594, and the other half was labeled with Alexa 647 (Integrated DNA Technologies). Nanoparticles were incubated in 100% mouse serum (Invitrogen) or in PBS at 37°C, and their fluorescence intensity was measured once per hour (excitation = 540 nm, emission = 620 and 690 nm). The 690/620 nm signal ratio was normalized to that of naked, labeled siRNAs in serum or PBS. Data are presented normalized to FRET signal at t = 0. After 24 hours, a 1% Triton-X (Sigma-Aldrich, St Louis, MO) solution was added to nanoparticles27 to confirm the extinguishment of FRET signal upon particle destruction.
SUPPLEMENTARY MATERIAL Figure S1. The binary combination 86N15–98O13 is stable in serum. Lipidoid nanoparticles containing 50% Alexa 594-labeled siRNA and 50% Alexa 647-labeled siRNA were incubated in mouse serum over a 24-hour time period. FRET intensity, which corresponds to particle stability, was measured as a function of time. All particles retained >80% of their stability over a period of 24 hours. Data are presented normalized to FRET signal at t = 0. After 24 hours, a 1% Triton-X (Sigma) solution was added to nanoparticles to confirm the extinguishment of FRET signal upon particle destruction. Error bars represent standard deviation (n = 4).
Acknowledgments
K.A.W. would like to thank Kaitlin Bratlie (MIT), Christopher Levins (MIT), and Daniel Siegwart (MIT) for technical assistance, as well as Arturo Vegas (MIT) for helpful discussions and feedback on the manuscript. This work was supported by Alnylam Pharmaceuticals as well as NIH grants EB000244, R01CA115527, and R01CA132091. K.A.W. was also supported by an NIH fellowship (award number F32EB009623) from the National Institute of Biomedical Imaging and Bioengineering. C.Z. and W.Q. are current employees of Alnylam Pharmaceuticals. D.G.A and R.S.L. report receiving consulting fees from Alnylam Pharmaceuticals.
Supplementary Material
The binary combination 86N15–98O13 is stable in serum. Lipidoid nanoparticles containing 50% Alexa 594-labeled siRNA and 50% Alexa 647-labeled siRNA were incubated in mouse serum over a 24-hour time period. FRET intensity, which corresponds to particle stability, was measured as a function of time. All particles retained >80% of their stability over a period of 24 hours. Data are presented normalized to FRET signal at t = 0. After 24 hours, a 1% Triton-X (Sigma) solution was added to nanoparticles to confirm the extinguishment of FRET signal upon particle destruction. Error bars represent standard deviation (n = 4).
REFERENCES
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V.et al. (2010Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study Lancet 3751896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Ban H-S, Kim S-S, Wu H, Pearson T, Greiner, DL.et al. (2008T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice Cell 134577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W.et al. (2005Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs Nat Biotechnol 231002–1007. [DOI] [PubMed] [Google Scholar]
- Niu XY, Peng ZL, Duan WQ, Wang H., and, Wang P. Inhibition of HPV 16 E6 oncogene expression by RNA interference in vitro and in vivo. Int J Gynecol Cancer. 2006;16:743–751. doi: 10.1111/j.1525-1438.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- Okumura A, Pitha PM., and, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K.et al. (2005Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo PNAS 10212177–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Bao Y, Peng W, Goldberg M, Love K, Bumcrot DA.et al. (2009Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis Proc Natl Acad Sci USA 1063426–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A.et al. (2008Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates Proc Natl Acad Sci USA 10511915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR.et al. (2010Lipid-like materials for low-dose, in vivo gene silencing Proc Natl Acad Sci USA 1071864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y.et al. (2008Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone Nat Biotechnol 26431–442. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Dalmasso G, Wang L, Sitaraman SV, Merlin D., and, Murthy N. Orally delivered thioketal nanoparticles loaded with TNF-a-siRNA target inflammation and inhibit gene expression in the intestines. Nat Mater. 2010;9:923–928. doi: 10.1038/nmat2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya S, Armant M, Salkowitz JR, Nyakeriga AM, Haridas V, Hasan M.et al. (2010Enhanced induction of HIV-specific cytotoxic T lymphocytes by dendritic cell-targeted delivery of SOCS-1 siRNA Mol Ther 182028–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MS, Xing D, Ren Y, Orsulic S, Bhatia SN., and, Sharp PA. Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc Natl Acad Sci USA. 2011;108:745–750. doi: 10.1073/pnas.1016538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshayes S, Morris M, Heitz F., and, Divita G. Delivery of proteins and nucleic acids using a non-covalent peptide-based strategy. Adv Drug Deliv Rev. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Akinc A., and, Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: implications for lysosomal trafficking. Biotechnol Bioeng. 2002;78:503–508. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N.et al. (2008A combinatorial library of lipid-like materials for delivery of RNAi therapeutics Nat Biotechnol 26561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL.et al. (2007Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes Proc Natl Acad Sci USA 10412982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK.et al. (2007Mechanisms and optimization of in vivo delivery of lipophilic siRNAs Nat Biotechnol 251149–1157. [DOI] [PubMed] [Google Scholar]
- Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ., and, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhao B, Jiang H, Wang B., and, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK.et al. (2010Rational design of cationic lipids for siRNA delivery Nat Biotechnol 28172–176. [DOI] [PubMed] [Google Scholar]
- Han SE, Kang H, Shim GY, Suh MS, Kim SJ, Kim JS.et al. (2008Novel cationic cholesterol derivative-based liposomes for serum-enhanced delivery of siRNA Int J Pharm 353260–269. [DOI] [PubMed] [Google Scholar]
- Heyes J, Palmer L, Bremner K., and, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The binary combination 86N15–98O13 is stable in serum. Lipidoid nanoparticles containing 50% Alexa 594-labeled siRNA and 50% Alexa 647-labeled siRNA were incubated in mouse serum over a 24-hour time period. FRET intensity, which corresponds to particle stability, was measured as a function of time. All particles retained >80% of their stability over a period of 24 hours. Data are presented normalized to FRET signal at t = 0. After 24 hours, a 1% Triton-X (Sigma) solution was added to nanoparticles to confirm the extinguishment of FRET signal upon particle destruction. Error bars represent standard deviation (n = 4).