Abstract
Vaccine production and initiation of mass vaccination is a key factor in rapid response to new influenza pandemic. During the 2009–2010 H1N1 pandemic, several bottlenecks were identified, including the delayed availability of vaccine potency reagents. Currently, antisera for the single-radial immunodiffusion (SRID) potency assay are generated in sheep immunized repeatedly with HA released and purified after bromelain-treatment of influenza virus grown in eggs. This approach was a major bottleneck for pandemic H1N1 (H1N1pdm09) potency reagent development in 2009. Alternative approaches are needed to make HA immunogens for generation of SRID reagents in the shortest possible time. In this study, we found that properly folded recombinant HA1 globular domain (rHA1) from several type A viruses including H1N1pdm09 and two H5N1 viruses could be produced efficiently by using a bacterial expression system and subsequent purification. The rHA1 proteins were shown to form functional oligomers of trimers, similar to virus derived HA, and elicited high titer of neutralizing antibodies in rabbits and sheep. Importantly, the immune sera formed precipitation rings with reference antigens in the SRID assay in a dose-dependent manner. The HA contents in multiple H1N1 vaccine products from different manufacturers (and in several lots) as determined with the rHA1-generated sheep sera were similar to the values obtained with a traditionally generated sheep serum from NIBSC. We conclude that bacterially-expressed recombinant HA1 proteins can be produced rapidly and used to generate SRID potency reagents shortly after new influenza strains with pandemic potential are identified.
Keywords: Pandemic influenza, Vaccine potency, Single-radial immunodiffusion assay, H5N1, H1N1, Vaccine
1. INTRODUCTION
The production, clinical testing and licensure of pandemic 2009-H1N1 (H1N1pdm09) vaccines in a limited amount of time was a remarkable achievement in the year 2009, which was fostered in part by close cooperation between WHO, national health authorities and the vaccine industry. It nevertheless took several months before H1N1 vaccine was available, and therefore vaccination was initiated only after the first wave of infections in both the northern and southern hemispheres.
The hemagglutinin (HA) of the influenza virus is the major surface protein that induces protective immune responses and is a major component of the currently licensed inactivated influenza vaccines. It is therefore used as the “potency marker” for each lot of inactivated influenza vaccine being released. The approved method used to measure the potency of inactivated-virus vaccine is the single-radial immunodiffusion (SRID) assay [1–3]. This assay utilizes influenza strain-specific antisera to measure the content of virus hemagglutinin (HA) in vaccine lots in comparison with the corresponding reference antigen. This assay relies on the availability of reference materials for standardization of vaccine potency, i.e., HA reference antigen and the corresponding anti-serum generated in sheep. Annually, it takes 2–4 months from the time of designation of influenza strains for vaccine production to prepare these reference reagents [4]. The production of strain-specific antibody involves preparation of purified HA, which is done by release of HA (without the transmembrane domain) from either wild type or vaccine virus strain by bromelain treatment followed by its purification [5, 6]. To generate sufficient quantities of sera, several sheep are immunized multiple times with purified HA until sufficiently high titers are achieved. During vaccine preparation for the 2009-H1N1 pandemic, it became evident that the virion-associated HA molecules of the H1N1pdm09 strain were extremely sensitive to bromelain digestion, making the purification of released HA for sheep immunization very difficult and resulting in a delay in the availability of these potency reagents [7]..
When faced with impending pandemics caused by the emergence of new zoonotic influenza strains from wild birds or pigs into the human populations, time is of the essence. Any alternative approach to generating vaccine seed and/or potency reagents for use in formulation and vaccine release that can shorten the time to vaccine availability for mass use is a high priority for health authorities around the world. To that end, several groups suggested alternative methods for preparing of either strain specific [7] or universal antibodies for quantitative determinations of HA in Influenza A and Influenza B vaccines [8].
Recombinant technology can save time in generating HA as it does not require the availability of virus or the isolation of vaccine virus through reassortment or reverse genetic processes. However, it is important to ensure that the recombinant HA proteins are properly folded and resemble structurally and functionally, the virion-associated oligomeric spikes. The antibodies generated against such functional recombinant HA should demonstrate virus neutralization and interact with the inactivated split virion or subunit vaccines in SRID quantitatively and qualitatively similar to the sheep sera generated by immunization with bromelain-released HA.
Our laboratory has recently described the expression and purification of recombinant HA1 globular domains from pandemic H1N1 (A/California/07/2009) as well as avian influenza H5N1 virus (A/Vietnam/1203/2004) in E. coli. Importantly the bacterially expressed HA1 proteins were shown to be properly folded, contained oligomers, bound sialic acid receptor, caused hemagglutination, generated neutralizing antibodies, and protected ferrets from pandemic influenza virus challenges [9] [10].
In the current study we demonstrate that the bacterially expressed proteins from group 1 type A influenza viruses elicited high titer antibodies in sheep that can be used in SRID assays to quantitate HA in pandemic influenza vaccine lots. The HA content determined with the new sheep sera were very similar to the values obtained with the currently available sheep serum generated at NIBSC against bromelain-released HA.
2. MATERIAL AND METHODS
Expression vector and cloning of rHA1 derivatives
cDNA corresponding to the HA gene segment of H1N1pdm09-A/California/07/2009, H5N1-A/Vietnam/1203/2004 and H5N1-A/Indonesia/5/2005 was generated from RNA isolated from egg-grown wild type viruses, and were used for cloning. pSK is a T7 promoter based expression vector where the desired polypeptide can be expressed as a fusion protein with His6 tag at the C-terminus[11]. DNA encoding HA1 of the influenza-A viruses were cloned as NotI-PacI inserts in the pSK expression vector.
Protein expression, refolding and purification
E. coli Rosetta Gami cells (Novagen) were used for expression of influenza rHA1. Following expression, inclusion bodies were isolated by cell lysis and multiple washing steps with 1% Triton X-100. The final Inclusion Bodies (IBs) pellet was resuspended in denaturation buffer containing 6 M Guanidine Hydrochloride and dithioerythreitol (DTE) at final protein concentration of 10 mg/ml and centrifuged to remove residual debris. For refolding, supernatant was slowly diluted 100-fold in redox folding buffer. The renatured protein solution was dialyzed against 20 mM Tris HCl pH 8.0 to remove the denaturing agents. The dialysate was filtered through a 0.45 µM filter and was subjected to purification by HisTrap Fast flow chromatography.
Gel filtration Chromatography
Protein at a concentration of 5 mg/ml was analyzed on a Superdex S200 XK 16/60 column (GE-Healthcare) pre-equilibrated with PBS, and the protein elution was monitored at 280 nm. Protein molecular weight marker standards (GE healthcare) were used for column calibration and generation of standard curves, to identify the molecular weights of the test protein samples.
Transmission Electron Microscopy
Carbon-coated formvar films mounted on copper grids (EMS) were floated on a 10-µl droplet of sample for 2 min and then negatively stained with 1% uranyl acetate. Grids were studied in a CM120 transmission electron microscope (FEI, Hillsboro, OR) operating at 120 kV. Micrographs were recorded on SO-163 film (Kodak) with defocus values between 1.5 and 2.5 µm, at a nominal magnification of 45,000 and digitized with a Super COOLSCAN 9000 ED (Nikon). The images used for averaging were binned 4 times giving a final pixel size of 6.2 Å/pixel. Image analyses were then carried out using Bsoft [12], EMAN [13], and SPIDER [14]. Particles with rosette-like shapes that we take to represent HA oligomers were selected (n=1196) and extracted for further analysis, using a box size of 64 pixels. Initial classification was done using the refine2d python macro in EMAN, and iterative refinement was performed using Principal Component Analysis as implemented in SPIDER.
Hemagglutination Assay
Human erythrocytes were separated from whole blood (Lampire Biologicals). After isolation and washing, 30 µl of 1% human RBC suspension (vol/vol in 1% BSA-PBS) were added to 30 µl serial dilutions of purified HA1 proteins or influenza virus or vaccine in 1% BSA-PBS in a U-bottom 96-well plate (total volume, 60 µl). Agglutination was read after incubation for 60 min at room temperature
Microneutralization assay
Viral-neutralizing activity was analyzed in a microneutralization assay based on the methods of the pandemic influenza reference laboratories of the Center for Disease Control and Prevention (CDC). The H1N1 vaccine strain, generated by classical reassortment, was obtained from CDC (X-179A). Low pathogenicity H5N1 viruses, generated by reverse genetics, were obtained from CDC: A/Vietnam/1203/2004 (SJCRH, clade 1), A/Indonesia/5/2005 (PR8-IBCDC-RG2; clade 2.1), A/Turkey/1/05 (NIBRG-23; clade 2.2), A/Anhui/1/05 (IBCDC-RG5, clade 2.3.4). The experiments were conducted with three replicates for each serum sample and performed at least twice.
Animal Immunization
New Zealand rabbits were immunized three times intra-muscularly at 21-days interval with 100 µg of purified HA1 proteins with Titermax adjuvant (TiterMax Inc, Norcross, Georgia, US).
Sheep were immunized initially with 50 µg of purified rHA1 proteins intra-muscularly with Complete Freund’s adjuvant (Sigma Aldrich, US). After 28-days, two 50 µg boosts of purified rHA1 proteins were administered intra-muscularly at 21-day intervals with Incomplete Freund’s adjuvant (Sigma Aldrich, US). Plasma was collected from immunized sheep, defibrinated and serum separated.
All procedures were in accordance with the National Research Council (NRC) Guidelines for the Care and Use of Laboratory Animals, the Animal Welfare Act, and the Centers for Disease Control (CDC)/National Institutes of Health (NIH) Bio-Safety Guidelines in Microbiological and Biomedical Laboratories and approved by the Institutional Animal Care and Use Committee (IACUC).
SRID Assay
The SRID assay was performed essentially as previously described [1, 3]. Briefly, 1% agarose (Lonza) in PBS (KD Medical) gels were cast on GelBond film (Lonza). These gels also contained an optimal amount of HA antibodies. Four millimeter wells were punched in the solidified gel. Appropriately diluted solutions of antigens were prepared, treated with Zwittergent 3–14 (Calbiochem; final concentration 1%) and incubated for 30 minutes at room temperature. Such detergent treated samples were further diluted (1.5, 2 and 4 fold) and loaded in the previously punched wells. Gels were placed in a humidified chamber for 18–24 hours to allow diffusion of the antigens. Following incubation, the gels were washed in saline solution, followed by a water rinse, dried and stained with Coomassie Brilliant Blue. Dried gels were scanned and two perpendicular diameters of the precipitin rings were measured using an Immulab system (GT Vision). The concentration of HA in the vaccine lots was calculated by generation of dose response curves with the reference antigen (using the assigned HA content of the reference) and the test vaccine lots in the same assay. HA content in µg/ml was calculated from the linear region of the parallel dose response curves [1].
Adsorption of Reference antiserum (NIBSC) with rHA1 protein
The H1N1pdm09-NIBSC reference antiserum (100 µl) was added to 0.2 mg of purified rHA1-His6 proteins or to control GST-His6 protein, and incubated for 1 hr at RT. Ni-NTA magnetic beads (200 µl; Qiagen, Valencia, CA) were added for 20 min at RT on end-to-end shaker to capture the His6-tagged proteins and the antibodies bound to them, followed by magnetic separation. Supernatants containing the unbound antibodies were collected. The sera before and after adsorption with rHA1 were subjected to SRID assay.
3. RESULTS
H1N1 rHA1 (1–320) produced from E. coli forms functional oligomers and adsorb antibodies responsible for precipitin rings in the SRID assay
DNA fragment encoding amino acid sequence 1–320 of HA from A/California/07/2009 (H1N1pdm09) with His6 tag at the C-terminus was cloned as NotI-PacI insert in the T7 promoter based expression vector. The H1N1 HA1 protein expressed in E. coli Rosetta Gami cells (Novagen) partitioned into the insoluble fraction (inclusion bodies, IB). IBs were solublized, refolded under controlled redox conditions and purified by HisTrap Fast flow chromatography. This process was previously shown to generate highly purified properly folded HA1 fragments from H1N1 and H5N1 [9] [10] [11]. We next determined if the bacterially expressed protein oligomerized into higher molecular forms using gel filtration chromatography on Superdex S200 XK 16/60 column (GE-Healthcare) (Fig 1). As shown in Fig. 1A, the majority (≥75%) of the HA1 (1–320) protein eluted at a position consistent with oligomeric forms. This elution profile was compared with that of an H1N1 2009 inactivated vaccine, which runs as a single sharper high MW oligomer peak in gel filtration (Fig. 1B). Negative stain EM images showed that the oligomers mainly presented a globular shape with an average diameter of ~ 25 nm (Fig. 1C, top); this is bigger than the size of monomeric (~14 nm long and ~3 nm wide) and trimeric (~14 nm long and ~5 nm wide) HA1 molecules [15]. Oligomers presenting rosette-like structures were then analyzed further (white circles, Fig. 1C top; ~20% of all the oligomers were selected). As these images were not homogeneous, we classified them into like sets and separately averaged the particles from each class to enhance the images. This approach revealed rosettes consisting of 4–6 spikes, ~ 10 nm long and ~ 5 nm wide (Fig 1C, bottom). The difference in length between a spike and the complete HA trimer can be explained by the flexibility of the N-terminal region of the HA1 domain without the presence of HA2. The EM images are very similar to the rosettes formed by intact HA oligomers released from influenza viruses [16].
FIGURE 1. Characterization of purified H1N1pdm09 rHA1 protein from E. coli and H1N1pdm09 vaccine by gel filtration chromatography, transmission electron microscopy, hemagglutination, and SRID assay following rHA1 adsorption.
Superdex S-200 gel filtration chromatography of bacterial H1N1pdm09 rHA1 and H1N1pdm09 vaccine. Purified H1N1pdm09 HA1 with intact N-terminus (1–320) (A) and H1N1pdm09 vaccine (B) from the reassorted virus strain were subjected to gel filtration. The panels present superimposed elution profiles of purified HA proteins (red line) overlaid with calibration standards (grey line). The elution volumes of protein species are shown. (C) Negative stain electron micrograph (top) and class average images (bottom) of bacterially purified H1N1pdm09 HA1 protein oligomeric forms. Oligomers showing a clear rosette-like structure are encircled. Scale bars, 20 nm. (D) Agglutination of human RBC by properly folded bacterial H1N1pdm09 rHA1 (1–320) protein along with H1N1pdm09 vaccine. Serial dilutions of purified rHA1 were mixed with washed RBC and hemagglutination was read after 60 min at RT. Reassorted H1N1pdm09 virus was used as a positive control. H1N1pdm09 vaccine was used at a starting concentration of 15µg/ml. Dilutions of H1N1pdm09 potency reference antigen were analyzed by SRID assay using NIBSC (anti-H1N1pdm09) reference antiserum before (E) and after (F) adsorption with bacterially produced H1N1pdm09 rHA1.
Hemagglutination of red blood cells (RBC) is a surrogate assay to measure the functionality of the influenza hemagglutinin. To that end, the functional activity of the purified bacterially expressed HA1 protein was assessed in a hemagglutination assay using human red blood cells (RBC). Efficient HA1-mediated hemagglutination was observed, at concentrations of ≥390 ng/ml. Hemagglutination with a licensed H1N1pdm09 vaccine and H1N1pdm09 virus, used as positive controls, are shown in the bottom two rows (Fig. 1D). The RBC agglutination confirmed that the recombinant HA1 domain refolded properly into native conformations required for stable binding to its sialic-acid receptor substrate, and the oligomeric forms bound to multiple RBC to create the lattices structures that are measured in the HA.
Since most anti-HA neutralizing antibodies are conformation-dependent and the SRID potency assay is dependent on HA conformation and higher order structures, we tested whether the bacterially produced rHA1 can remove the antibodies responsible for SRID precipitin rings in the NIBSC anti-H1N1pdm09 sheep serum, currently used in the H1N1pdm09 SRID assay. To that end, The H1N1pdm09-NIBSC reference antiserum (100 µl) was added to 0.2 mg of purified rHA1-His6 proteins or to control GST-His6 protein, and incubated for 1 hr at RT, followed by addition of Ni-NTA magnetic beads for 20 min at RT to capture the His6-tagged proteins and the antibodies bound to them. As can be seen in Fig. 1E and 1F, the rHA1 protein completely absorbed the SRID activity of the NIBSC reagent resulting in no precipitation rings with reference antigen. Based on these findings, it was decided to investigate if the rHA1 protein could elicit H1N1pdm09-specific antibodies functional in the SRID assay.
Immunization of sheep with rHA1 protein generated high titers of H1N1pdm09-neutralizing antibodies and strain specific SRID reagents
Two sheep (#7870 and #270) were immunized three times with rHA1 in accordance with an approved CBER protocol. H1N1 neutralization was measured using the vaccine strain (X-179A) in MDCK cells according to the CDC protocol. As shown in Table 1, sheep-7870 and sheep-270 responded to the first immunization with titers of 320 and 80, respectively. The second and third immunizations resulted in very high virus-neutralizing titers of 20, 480 and 81,920, respectively. In comparison, the virus neutralization titer for the NIBSC traditional SRID reagent was 16,000 in multiple assays.
Table 1.
Mean reciprocal neutralizing titers of Sheep anti-HA sera
| SHEEP | SERA | END-POINT TITERS* |
|---|---|---|
| Sheep - 7870 | Pre Vaccine | <20 |
| Post- 1 | 320 | |
| Post- 2 | 20,480 | |
| Post- 3 | 10,240 | |
| Sheep - 270 | Pre Vaccine | <20 |
| Post- 1 | 80 | |
| Post- 2 | 81,920 | |
| Post- 3 | 81,920 | |
| NIBSC Sheep | Post-5th | 16,000 |
End-point titers (mean of three replicates) using polyclonal sheep sera in a microneutralization assay performed with A/California/07/2009 (X-179A)
The suitability of the rHA1-generated sheep sera for use in the SRID assay was assessed by comparing it to the traditionally-generated SRID reagent from NIBSC using the same reference antigen (Fig. 2). Similar to the NIBSC sheep serum, the rHA1-generated sheep sera generated precipitation rings with the H1N1pdm09 reference antigen in a dose-related manner. Importantly, no precipitation rings were observed with a panel of seasonal influenza strains of either H1N1 (n=7) or H3N2 subtypes (n=5) (Fig. 2 A & B, and data not shown).
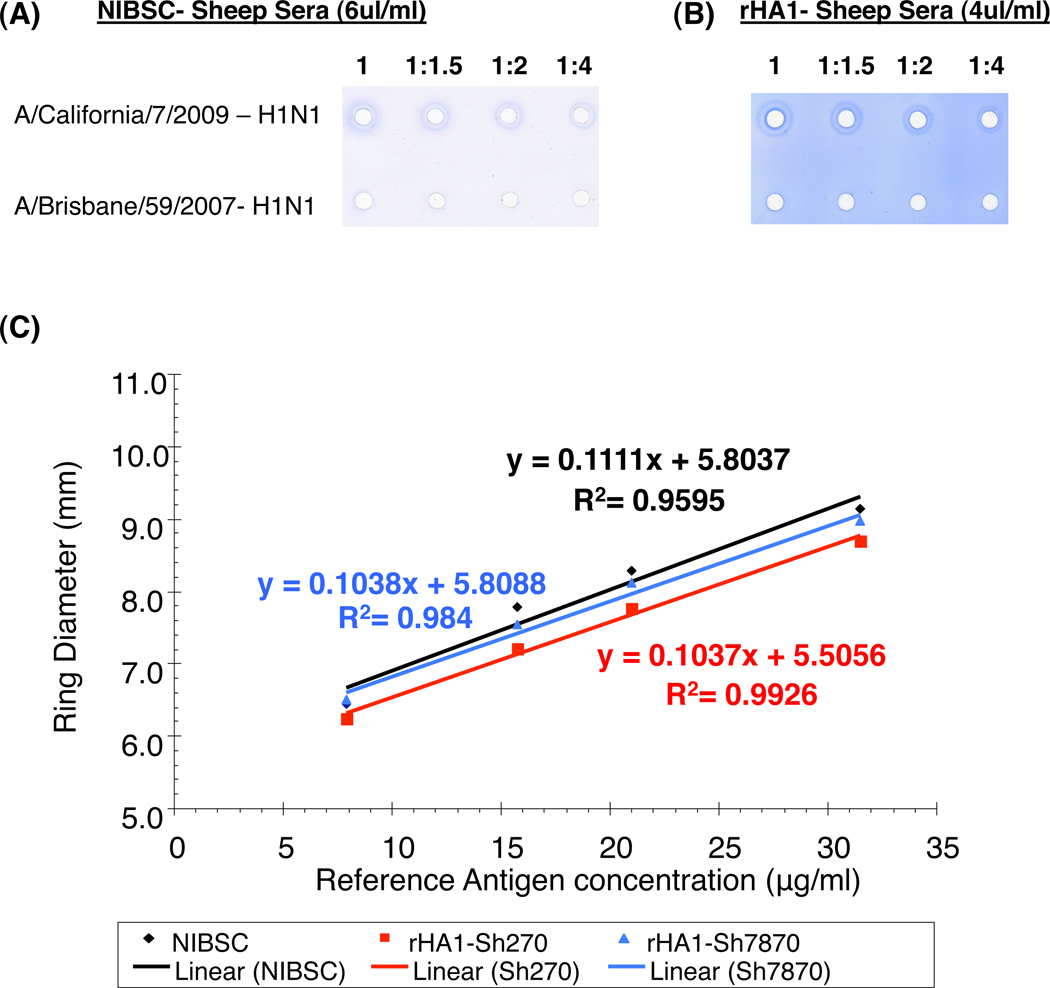
Figure 2. SRID analysis of H1N1pdm09 potency reference antigen using NIBSC (anti-H1N1pdm09) reference antiserum and antiserum prepared by the alternative method using the bacterially purified rHA1-H1N1pdm09.
Dilutions of A/California/7/2009 (H1N1pdm09) or A/Brisbane/59/2007 (H1N1) reference antigens were analyzed by SRID using either the homologous NIBSC reference antiserum (A) or the alternative CBER rHA1-sheep antiserum (B). (C) Comparison of NIBSC and CBER anti-rHA1 H1N1pdm09 sheep sera. Precipitin rings in the SRID assay were measured in two directions to the nearest 0.1 mm for determination of diameter with the serial dilutions of H1N1pdm09 reference antigens with the NIBSC sheep sera (in black) and the two sheep [Sh-270 (in red) & Sh-7870 (in blue)] immunized with bacterially purified rHA1. Slopes and correlation coefficient calculated for each sheep sera are represented in their respective colors.
For comparison with the traditional SRID reagent from NIBSC, linear calibration curves were generated (reference antigen concentration vs. precipitin ring diameter) using the parallel line bioassay method (log antigen dilution vs. log zone diameter) for the NIBSC reagent and the rHA1-generated sera from sheep 270 and sheep 7870 (Fig. 2C). All three linear curves had high correlation coefficient values (R2) and equality of slopes (t) suggesting that the reference HA antigen interacts similarly with each of the three different polyclonal sera. This is important since parallel lines with high correlation coefficient and equality of slopes are required for alternative polyclonal sera to be used as a vaccine potency reagent, and only in such cases is it possible to replace the traditional sheep serum for SRID assay.
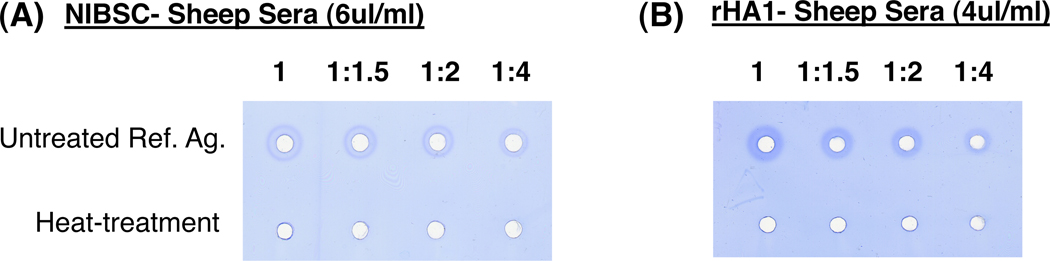
Since pandemic influenza vaccines may need to be stored for several years and seasonal influenza vaccines can have a shelf life for up to 1–2 years, it is important that the sheep sera used in the SRID assay should be indicative of vaccine lot HA stability. Loss of antigenic integrity tends to result in poorer precipitation rings or smaller diameter rings, suggesting lower amounts of intact HA. Different treatment can be used to induce loss of HA integrity in the vaccine lots. Accelerated stability testing often involves subjecting the vaccine lots to high temperature conditions (23–37°C) for a given time period (1–2 weeks). In the current study, the reference antigen was subjected to heat-treatment at 56°C for 3 hours to evaluate the antibody specificity in the NIBSC or rHA1-generated sheep sera responsible for antigen-antibody immune complexes in the SRID assay. As can be seen in Fig 3, no precipitation rings were observed in the SRID assay with either NIBSC or rHA1-generated sheep sera (Fig. 3A–B). These findings confirmed that both sera reacted with conformationally intact HA but not with denatured HA (which still contain linear epitopes).
Figure 3. Heat treatment of H1N1pdm09 reference antigen to evaluate antibody specificity by SRID using NIBSC (anti-H1N1pdm09) reference antiserum and rHA1-H1N1pdm09 sheep antiserum.
Dilutions of H1N1pdm09 (A/California/7/2009) reference antigen were analyzed by SRID assay before or after heat treatment at 56°C for 3 hours, using either the homologous NIBSC reference antiserum (A) or the alternative CBER rHA1-sheep antiserum (B).
HA content measured in different H1N1pdm09 vaccine products and multiple vaccine lots from the same manufacturer (pandemic and trivalent) using rHA1-generated sheep sera is comparable to traditional NIBSC sheep sera in SRID assay
The rHA1-generated sheep sera were used side by side with the NIBSC antibody reference reagent to determine the amount of HA in several H1N1pdm09 vaccine products derived from either X-179A or X-181 reassortant virus and produced by different influenza vaccine manufacturers. As shown in Table 2A, the HA concentrations measured in SRID using the rHA-1 sera (sheep-7870) ranged from 95% to 118% of the HA concentrations measured using the NIBSC reagent. Next we compared the HA concentration in multiple pandemic vaccine lots from the same manufacturer (Table 2B). Again the HA concentrations obtained with the rHA1-generated sheep sera ranged from 94% to 105% of the HA concentration determined with the NIBSC sheep serum.
Table 2.
| A- Determination of HA content (µg/ml) in H1N1pdm09 vaccine from different vaccine manufacturers determined by SRID using traditional and rHA1 potency antibody. | ||
|---|---|---|
| H1N1 Vaccine | NIBSC | rHA1 SERA |
| Product 1 (X-179A) | 29.0 | 30.4 |
| Product 2 (X-179A) | 35.0 | 33.2 |
| Product 3 (X-181) | 47.6 | 56.2 |
| Product 4 (X-181) | 44.6 | 56.6 |
| B- HA content (µg/ml) in vaccine lots determined by SRID using traditional and rHA1 potency antibody. | ||
|---|---|---|
| H1N1 Vaccine | NIBSC | rHA1 SERA |
| Lot 1 | 38.1 | 38.8 |
| Lot 2 | 37.4 | 35.9 |
| Lot 3 | 32 | 30.1 |
| Lot 4 | 29.1 | 30.6 |
| Lot 5 | 32.8 | 32.1 |
| C- Precipitin ring size (in mm) in SOIV-H1N1 monovalent & trivalent vaccine lots containing 15µg SOIV-H1N1 HA measured by SRID using traditional and rHA1 potency sera. | ||||
|---|---|---|---|---|
| Vaccine | H1N1 MONOVALENT POOL | H1N1, H3N2 & B TRIVALENT POOL | ||
| NIBSC | rHA1 SERA | NIBSC | rHA1 SERA | |
| Lot 1 | 6.3 | 6.4 | 5.6 | 5.6 |
| Lot 2 | 6.9 | 6.5 | 5.5 | 5.7 |
| Lot 3 | 6.8 | 6.7 | 5.7 | 5.5 |
| Average | 6.7 | 6.5 | 5.6 | 5.6 |
Finally, it was important to confirm that the rHA1- generated antibodies can be used to measure H1N1pdm09-HA as a component of trivalent vaccines with the same degree of sensitivity and specificity found for the monovalent H1N1pdm09 vaccine. To that end, three lots of monovalent H1N1pdm09 vaccine (2009) and three lots of Trivalent Influenza Vaccine (TIV, 2010) were used in SRID. The H1N1pdm09 HA content as determined either by NIBSC serum or the rHA1-generated sheep serum were virtually identical, confirming that these sera were equally suitable to measure HA content in both monovalent and trivalent vaccine lots (Table 2C).
Recombinant HA1 derived from H5N1 avian influenza strains generated strain specific SRID reagents
Recombinant HA1 (1–320) proteins derived from H5N1 A/Vietnam/1203/04 and A/Indonesia/5/05 were expressed in E. coli then purified under the same conditions as the H1N1pdm09 rHA1. Both proteins contained high fractions of oligomeric forms, bound sialic acid receptor and agglutinated human RBC ([10] and data not shown). Upon rabbit immunization, these proteins generated high-titer homologous as well as heterologous neutralizing antibodies (Fig. 4A). These rabbit sera were used in the SRID assay to evaluate the reactivity with different H5N1 reference antigens. Both sera gave precipitation rings with the homologous as well as with heterologous H5N1 virus, in agreement with the cross-clade neutralization observed with these sera (Fig. 4A). On the other hand, these anti-H5N1-rHA1 sera did not cross-react with seasonal influenza reference antigens (Fig. 4B–C). Importantly, the SRID results with the rHA1-generated polyclonal sera were very similar to those obtained with the CBER traditionally-generated SRID sheep reagents against the two H5N1 viruses as determined by the parallel lines bioassay method (Fig. 4D–E).
Figure 4. H5N1 rHA1 (1–320) from A/Vietnam/1203/04 and A/Indonesia/05/2005 elicits high titer cross-neutralizing antibodies and generate H5N1 specific SRID antiserum in rabbits.
(A) Animals were immunized with 100 µg proteins mixed with TiterMax adjuvant every three weeks. Sera were collected 8 days after each vaccination and analyzed in a microneutralization assay against various H5N1 virus strains. Representative of three experiments. Dilutions of A/Vietnam/1203/04 (H5N1; Clade-1), A/Indonesia/05/2005 (H5N1; Clade-2),, A/Victoria/210/2009 (H3N2) or A/California/7/2009 (H1N1pdm09) reference antigens were analyzed by SRID using either the CBER rHA1-A/Vietnam/1203/04 antiserum (B) or the CBER rHA1-A/Indonesia/05/2005 antiserum (C).
Parallel line bioassay method for comparison of traditional anti-H5N1 sheep sera with either CBER rHA1-A/Vietnam/1203/04 antiserum (D) or the CBER rHA1-A/Indonesia/05/2005 antiserum (E).
Precipitin rings in the SRID assay were measured in two directions to the nearest 0.1 mm for determination of diameter with the serial dilutions of homologous H5N1-reference antigens with the traditional anti-H5N1 sheep sera (in black) and the rabbits immunized with bacterially purified H5N1-rHA1. Correlation coefficient calculated for each animal serum is represented in their respective colors.
4. Discussion
In April 2009, the Centers for Disease Control and Prevention (CDC) announced the detection of a novel strain of H1N1 influenza virus in humans. The novel virus derived its genes from viruses circulating in the pig population [17, 18]. Due to sustained human-to-human transmission of this novel virus throughout the world, on June 11th the World Health Organization (WHO) raised the worldwide pandemic alert level to Phase 6 [19–21].
As H1N1pdm09 influenza enters the post-pandemic phase, health authorities around the world are reviewing the response to the pandemic to ensure this process enhances future vaccine preparations. These analyses identified several bottle-necks that require technical improvement including the need to more rapidly select optimal vaccine viruses (with high growth properties) and the need for alternative vaccine standardization technologies for potency assays and stability tests [22].
Recently, several approaches have been explored as alternatives for quantification of the HA content in vaccines [23–27]. To date, however, none of the newer techniques have been shown to measure the same HA antigenic form, and to be suitable for HA stability in vaccine lots. Therefore, while potentially useful, none of the newer techniques were implemented for potency determination of inactivated influenza vaccines. Furthermore, universal antibodies based on the fusion peptide in HA2 could be used in competitive ELISA, but required HA denaturation by low pH or 4–8M urea treatment [8] and could not be used for quantitation of conformationally intact HA as required from SRID and any alternative potency methods.
Various recombinant technologies have been used to generate HA in insect and mammalian cells [28–30]. Only one was approach was shown to function in SRID, but it required multiple vaccinations with alternative forms of the HA (plasmid, MVA vector, and VLP proteins) [7]. Therefore, it was important to compare our alternative approach to generate the HA antigen for sheep immunization with the activity of sheep sera generated by the traditional method (i.e. bromelain-released HA).
Our group evaluated the possibility of producing recombinant HA in bacterial systems for the purpose of rapidly generating hyperimmune sera for the SRID-based potency assays. Within two weeks of swine-like H1N1 virus isolation from acutely infected patient in California, two HA cDNA from the A/California/07/2009 virus, HA1 (1–330) and HA (1–480), were expressed and purified from E. coli under controlled redox refolding conditions that favoured proper protein folding. Surprisingly, only the recombinant HA1 (1–330) globular domain formed oligomers, showed specific binding to sialic acid receptor and agglutinated human red blood cells [9]. These proteins were used to vaccinate ferrets prior to challenge with the A/California/07/2009 virus. Both proteins induced neutralizing antibodies, and reduced viral loads in nasal washes. However, the HA1 (1–330) protein that had higher content of oligomeric forms provided better protection from fever and weight loss at a lower vaccine dose compared with HA (1–480) [9]
In a preliminary study, rabbit immune sera generated against the purified rHA1 proteins were tested in SRID assay against H1N1pdm09 reference antigen. Good precipitation rings were observed with the sera from rHA1(1–330)-vaccinated animals but not with rHA(1–480) immune sera after second vaccination (data not shown). Based on these findings it was concluded that the presence of functional oligomers in HA preparations may be crucial for rapid generation of antibodies suitable for the SRID assay. In agreement with our previous experience with the rHA1 (1–320) produced from H5N1-A/Vietnam/1203/2004 strain [10], we observed that the H1N1pdm09-rHA1 (1–320) protein produced in E. coli was more stable than the rHA1 (1–330), and also agglutinated RBC better than rHA1 (1–330). The deletion of 10 amino acid sequence at the carboxy-terminus of HA1 improved HA1-oligomer stability as judged by more efficient hemeagglutination in different influenza strains including group-1 (H1N1, H5N1) and group-2 (H7N7) viruses.
In the current study, we demonstrated that the rHA1 proteins preparations contained a large fraction of oligomeric forms (rosettes of trimers) as determined by gel filtration and electron microscopy, similar to native HA spikes isolated from influenza virus. These oligomeric rHA1 were functional as measured by receptor binding assay and RBC agglutination. Importantly, the rHA1 generated high titer neutralizing antibodies in rabbits and sheep. The sera from two sheep vaccinated with the rHA1 of H1N1pdm09 according to the CBER approved protocol were found to generate good precipitation rings with H1N1pdm09 reference antigen with parallel dose-response lines when compared with the traditionally-generated sheep serum from NIBSC. The HA content in several inactivated vaccine products and multiple lots as determined in SRID assays using the rHA1-generated sheep sera were very similar to the HA content measured using the NIBSC potency reagent (i.e. values within 94%–118%). The rHA1 immunized sheep sera showed similar capacity to measure H1N1pdm09 HA content in either monovalent or trivalent vaccine formulations and were stability sensitive. Lastly, rHA1 derived from the highly pathogenic avian influenza strains A/Vietnam/1203/04 and A/Indonesia/05/05 also contained oligomers and elicited strain-specific neutralizing antibodies that behaved in the SRID assays similar to the traditional sheep sera.
These results further demonstrate that the source of immunizing antigen for generation of SRID reagent is not limited to the traditional vaccine derived bromelain-released HA. Recombinant technology could help to overcome the severe bottleneck of producing HA from influenza virus for generation of vaccine potency reagents.
In emergency situation, the HA1 of an emerging influenza strain can be chemically synthesized and cloned within one week after the HA sequence is deduced without isolating, handling, growing a pathogenic novel influenza virus or developing a reassortant vaccine strain. The purified HA1 protein can be produced within two weeks and used for sheep vaccinations.
This methodology might also be helpful to quantify and differentiate among drifted seasonal influenza strains where most of the cross-reactivity is due to the antibodies that bind to highly conserved HA2 sequences. Moreover, with ongoing efforts to develop quadrivalent seasonal influenza vaccines that includes one strain each of closely matched HA sequence but antigenically diverse Victoria and Yamagata lineage influenza B strains, it will become even more challenging to generate strain specific non-cross reactive SRID potency reagents.
However, there are several critical quality criteria that could help to determine if a given rHA1 product could be used for this purpose. Glycosylation may play more important role in the proper folding of some subtypes compared with others. In our studies to date from several group-1 influenza-A strains, one group-2 strain and two influenza-B strains, the recombinant HA proteins produced in the E. coli system, folded properly into functional oligomers with conformation that bound receptor and caused hemagglutination.
Moreover, our studies to date demonstrated that the presence of stable functional oligomers that mimic vaccine preparations is a critical requirement for generation of potency reagents for the SRID assay. RBC hemagglutination could be used as a quality control method to confirm functional oligomers in recombinant HA preparations intended for generation of potency reagents. Our study clearly demonstrated that both in the case of H1N1pdm09 and the two H5N1 AIV strains, the rHA1 elicited strain specific SRID reagents. These findings suggest that the absence of HA2 in the immunogen used for sheep vaccination had no impact on the specificity and sensitivity of the antibodies generated. Importantly, the parallel lines generated with the reference antigens in the SRID assay, suggest that the rHA1 sera recognized very similar antigenic epitopes in the vaccine products.
CONCLUSIONS
In the face of impending influenza pandemic, alternative approaches are needed for generation of vaccine potency reagents to ensure timely release of influenza vaccines. The bromelain-released HA from vaccine virus was traditionally used as immunogen. During the 2009 pandemic response it was identified as one of the important bottlenecks in the generation of reference reagents for the SRID assay. Our studies demonstrated that rapid expression and purification of oligomeric rHA1 in a bacterial system could provide an alternative approach that will significantly reduce the time needed to generate immune sera by 1–2 months and could support early initiation of mass vaccination.
Acknowledgements
We thank Vladimir Lugovtsev, and Falko Schmeisser for a thorough review of the manuscript. This work was partly supported funds from BARDA/HHS, by DMID IAA 224-10-1006, and by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wood JM, Schild GC, Newman RW, Seagroatt V. An improved single-radial-immunodiffusion technique for the assay of influenza haemagglutinin antigen: application for potency determinations of inactivated whole virus and subunit vaccines. J Biol Stand. 1977;5(3):237–247. doi: 10.1016/s0092-1157(77)80008-5. [DOI] [PubMed] [Google Scholar]
- 2.Wood JM, Dunleavy U, Newman RW, Riley AM, Robertson JS, Minor PD. The influence of the host cell on standardisation of influenza vaccine potency. Dev Biol Stand. 1999;98:183–188. [PubMed] [Google Scholar]
- 3.Williams MS, Mayner RE, Daniel NJ, Phelan MA, Rastogi SC, Bozeman FM, et al. New developments in the measurement of the hemagglutinin content of influenza virus vaccines by single-radial-immunodiffusion. J Biol Stand. 1980;8(4):289–296. doi: 10.1016/s0092-1157(80)80006-0. [DOI] [PubMed] [Google Scholar]
- 4.Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21(16):1776–1779. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 5.Compans RW, Klenk HD, Caliguiri LA, Choppin PW. Influenza virus proteins. I. Analysis of polypeptides of the virion and identification of spike glycoproteins. Virology. 1970;42(4):880–889. doi: 10.1016/0042-6822(70)90337-5. [DOI] [PubMed] [Google Scholar]
- 6.Brand CM, Skehel JJ. Crystalline antigen from the influenza virus envelope. Nat New Biol. 1972;238(83):145–147. doi: 10.1038/newbio238145a0. [DOI] [PubMed] [Google Scholar]
- 7.Schmeisser F, Vodeiko GM, Lugovtsev VY, Stout RR, Weir JP. An alternative method for preparation of pandemic influenza strain-specific antibody for vaccine potency determination. Vaccine. 2010;28(12):2442–2449. doi: 10.1016/j.vaccine.2009.12.079. [DOI] [PubMed] [Google Scholar]
- 8.Chun S, Li C, Van Domselaar G, Wang J, Farnsworth A, Cui X, et al. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine. 2008;26(48):6068–6076. doi: 10.1016/j.vaccine.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, et al. Properly folded bacterially expressed H1N1 hemagglutinin globular head and ectodomain vaccines protect ferrets against H1N1 pandemic influenza virus. PLoS One. 2010;5(7):e11548. doi: 10.1371/journal.pone.0011548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, et al. Bacterial HA1 Vaccine against Pandemic H5N1 Influenza Virus: Evidence of Oligomerization, Hemagglutination, and Cross-Protective Immunity in Ferrets. J Virol. 2011;85(3):1246–1256. doi: 10.1128/JVI.02107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khurana S, Suguitan AL, Jr, Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6(4):e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 14.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3(12):1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 16.Ruigrok RW, Wrigley NG, Calder LJ, Cusack S, Wharton SA, Brown EB, et al. Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J. 1986;5(1):41–49. doi: 10.1002/j.1460-2075.1986.tb04175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360(25):2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 18.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 19.Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol. 2009;45(3):174–8. doi: 10.1016/j.jcv.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Peiris JS, Poon LL, Guan Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol. 2009;45(3):169–173. doi: 10.1016/j.jcv.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Update: influenza activity --- United States, 2009–10 season. MMWR Morb Mortal Wkly Rep. 2010;59(29):901–908. [PubMed] [Google Scholar]
- 22.Abelin A, Colegate T, Gardner S, Hehme N, Palache A. Lessons from pandemic influenza A(H1N1): The research-based vaccine industry's perspective. Vaccine. 2010;29(6):1135–1138. doi: 10.1016/j.vaccine.2010.11.042. [DOI] [PubMed] [Google Scholar]
- 23.Kapteyn JC, Porre AM, de Rond EJ, Hessels WB, Tijms MA, Kessen H, et al. HPLC-based quantification of haemagglutinin in the production of egg- and MDCK cell-derived influenza virus seasonal and pandemic vaccines. Vaccine. 2009;27(9):1468–1477. doi: 10.1016/j.vaccine.2008.11.113. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Canas V, Lorbetskie B, Bertrand D, Cyr TD, Girard M. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Anal Chem. 2007;79(8):3164–3172. doi: 10.1021/ac0621120. [DOI] [PubMed] [Google Scholar]
- 25.Williams TL, Luna L, Guo Z, Cox NJ, Pirkle JL, Donis RO, et al. Quantification of influenza virus hemagglutinins in complex mixtures using isotope dilution tandem mass spectrometry. Vaccine. 2008;26(20):2510–2520. doi: 10.1016/j.vaccine.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Hashem AM, Van Domselaar G, Li C, Wang J, She YM, Cyr TD, et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Commun. 2010;403(2):247–251. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Shao M, Cui X, Song Y, Li J, Yuan L, et al. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals. 2010;38(2):284–289. doi: 10.1016/j.biologicals.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Treanor JJ, Wilkinson BE, Masseoud F, Hu-Primmer J, Battaglia R, O'Brien D, et al. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine. 2001;19(13–14):1732–1737. doi: 10.1016/s0264-410x(00)00395-9. [DOI] [PubMed] [Google Scholar]
- 29.Lakey DL, Treanor JJ, Betts RF, Smith GE, Thompson J, Sannella E, et al. Recombinant baculovirus influenza A hemagglutinin vaccines are well tolerated and immunogenic in healthy adults. J Infect Dis. 1996;174(4):838–841. doi: 10.1093/infdis/174.4.838. [DOI] [PubMed] [Google Scholar]
- 30.Wei CJ, Xu L, Kong WP, Shi W, Canis K, Stevens J, et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol. 2008;82(13):6200–6208. doi: 10.1128/JVI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]