Abstract
Connective tissue growth factor (CTGF) is a cysteine-rich protein the synthesis and secretion of which are hypothesized to be selectively regulated by activins and other members of the TGF-β superfamily. To investigate the in vivo roles of CTGF in female reproduction, we generated Ctgf ovarian and uterine conditional knockout (cKO) mice. Ctgf cKO mice exhibit severe subfertility and multiple reproductive defects including disrupted follicle development, decreased ovulation rates, increased numbers of corpus luteum, and smaller but functionally normal uterine horns. Steroidogenesis is disrupted in the Ctgf cKO mice, leading to increased levels of serum progesterone. We show that disrupted follicle development is accompanied by a significant increase in granulosa cell apoptosis. Moreover, despite normal cumulus expansion, Ctgf cKO mice exhibit a significant decrease in oocytes ovulated, likely due to impaired ovulatory process. During analyses of mRNA expression, we discovered that Ctgf cKO granulosa cells show gene expression changes similar to our previously reported granulosa cell-specific knockouts of activin and Smad4, the common TGF-β family intracellular signaling protein. We also discovered a significant down-regulation of Adamts1, a progesterone-regulated gene that is critical for the remodeling of extracellular matrix surrounding granulosa cells of preovulatory follicles. These findings demonstrate that CTGF is a downstream mediator in TGF-β and progesterone signaling cascades and is necessary for normal follicle development and ovulation.
Ovarian follicle development requires tightly controlled endocrine, paracrine, and autocrine interplay among oocytes, granulosa cells, and thecal cells. This process is coordinated by gonadotropins and local factors such as activin, inhibin, and steroids, which are involved in the continuous remodeling of extracellular matrix (1–4). A paracrine factor related to this process in the ovary is connective tissue growth factor (CTGF) (5). CTGF belongs to a protein family of six distinct members: CTGF, CYR61, NOV, WISP1, WISP2, and WISP3 (6–13). Although all the members were initially classified as immediate early gene products or growth factors, additional activities have been revealed recently. Because CTGF, CYR61, and NOV were the first identified proteins in this family, this group has also been called the “CCN family” (14). CTGF is expressed and acts on a wide variety of cell types including chondrocytes (10, 15–18), fibroblasts (19, 20), (vascular) endothelial cells (6, 21–23), vascular smooth muscle cells (6, 21–23), epithelial cells (24), tumor cells (12, 25, 26), renal mesangial cells (27), and the central nervous system (28–30). CTGF was originally identified as a mouse fibroblast mitogen encoded by a growth factor-inducible immediate-early gene (6, 31). However, at present, CTGF is thought to take part in diverse cellular functions such as proliferation, differentiation, attachment, migration, apoptosis, and extracellular matrix remodeling (7, 32, 33). These wide-ranging biological activities, together with mRNA and protein-expression studies, have proposed that CTGF may be involved in many important processes such as embryo development, cell proliferation, tissue differentiation, angiogenesis, osteogenesis, and wound healing, in addition to various pathologies such as fibrosis, inflammation, and tumor growth (5, 18, 20–23, 32, 34). However, the precise biological roles, mechanisms of action, and physiological functions are largely unknown. In addition, its relation to the female fertility and reproductive functions remain elusive.
In the female reproductive tract, a previous report has shown that Ctgf mRNA is abundantly expressed in granulosa cells of preantral and early antral follicles (35) Additionally, Ctgf mRNA levels increase in granulosa and thecal cells of porcine ovaries during early antral follicle development (36). In antral follicles, Ctgf mRNA expression becomes maximal but is down-regulated in preovulatory follicles (35). FSH reduces Ctgf mRNA expression in granulosa cells both in vivo and in vitro, yet induces follicle maturation (37). In addition, CTGF treatment of ovaries induces the expression of many genes related to cell-cycle progression and cell differentiation, thereby enhancing the growth of immature follicles (38). Moreover, CTGF was found to be present in human corpus luteum and to have a developmental role in luteal vasculature and tissue remodeling associated with luteolysis (39). CTGF is inhibited by human chorionic gonadotropin (hCG); during early pregnancy, this inhibition appears to be mediated by paracrine factors (39). Taken together, these reports indicate that CTGF may function in not only follicle development but also corpus luteum formation and early embryo development.
Because Ctgf-null newborn mice demonstrate the neonatal lethality resulting from respiratory skeletal defects (40), it has not been possible to establish the function of CTGF in the female reproductive system. In this study, we overcame this difficulty by using conditional knockout (cKO) strategies directed by a knockin of the Cre recombinase gene into the anti-Müllerian hormone receptor type 2 (Amhr2) locus or progesterone receptor (Pgr) locus to generate female mice lacking functional CTGF in different cell types of the female reproductive tract. Both types of Ctgf conditional-null female mice are subfertile and demonstrate decreased ovulation rates, with a greater effect seen with Pgr-cre deletion. In addition, Ctgf cKO female mice with Amhr2-cre deletion show a marked decrease in numbers of preantral and antral follicles and an increase in atresia. Meanwhile, both types of cKO mice retain normal uterine function, as shown by normal decidual response, indicating that the decreased fertility observed in Ctgf conditional-null mice arises solely from impairments in ovarian function. This study thus revealed novel and important roles for CTGF-dependent pathways in follicle development and ovulation.
Results
Generation of Ctgf cKO mice
Because of embryonic lethality between embryonic d 11.5 (E11.5) and E14.5 in mice null for Ctgf (41, 42), we generated Ctgf cKO mouse models using Cre-mediated recombination under the control of Amhr2 or Pgr promoters. Amhr2-cre is expressed during development in the mesenchyme of the Müllerian ducts, in the adult ovarian granulosa cells, and in the smooth muscle and stromal cells of the oviduct and uteri (43–46). Previous reports indicated that the activity of the Amhr2-cre recombinase was detected as early as E12.5 as shown by ROSA reporter mice (45). Meanwhile, Pgr-cre is expressed postnatally in anterior lobe of pituitary glands, in epithelial, stromal, and myometrial cellular compartments of uteri, and in granulosa cells of the preovulatory follicles in the ovaries (47, 48). Previous reports indicated that the activity of the Pgr-cre recombinase in the ovary was not detected before 21 d of age without hormone treatment using pregnant mare serum gonadotropin (PMSG), followed by hCG using LacZ reporter mice (49). Ctgf cKO (Amhr2cre/+) female mice (Ctgfflox/− Amhr2cre/+) were generated by crossing Ctgf+/− Amhr2cre/+ male mice to Ctgfflox/flox female mice, whereas Ctgf cKO (Pgrcre/+) female mice (Ctgfflox/− Pgrcre/+) were generated by crossing Ctgf+/− Pgrcre/+ male mice to Ctgfflox/flox female mice. Representative genotype analyses of both Ctgf cKO female mice are depicted in Fig. 1, A and B.
Fig. 1.
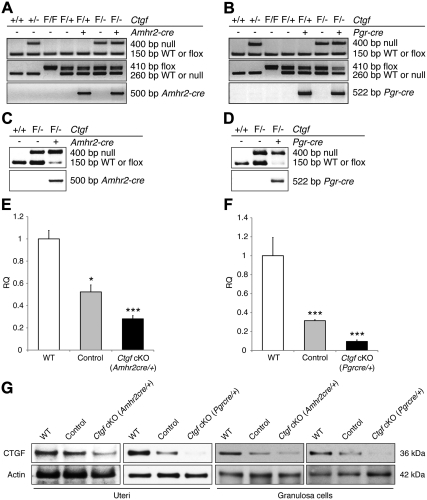
Recombination and deletion of the Ctgf conditional allele in the uterus and ovary. A and B, PCR analyses for Ctgf cKO genotyping. Representative PCR images are shown. Flox/− and Flox/+ are abbreviated as F/− and F/+, respectively. C and D, Recombination of Ctgf-flox alleles in the genomic DNA from granulosa cells. E and F, Ctgf transcript levels measured in granulosa cells by real-time qPCR in WT (n = 3), control (n = 3), and both Ctgf cKO female mice (n = 3 for each genotype). Levels of Ctgf relative quantity (RQ) are shown relative to the WT. Ctgf expression is efficiently ablated in the granulosa cells of Ctgf cKO female mice. The data are shown as means ± sem. WT values were set to equal 1. *, P < 0.05; ***, P < 0.001 (vs. WT female mice). G, Representative Western blots of total CTGF protein levels in uterine tissues and granulosa cells from control and both Ctgf cKO female mice. Compared with WT and control female mice, CTGF was markedly lower or undetectable in both Ctgf cKO female mice.
To determine whether the conditional alleles of Ctgf can undergo recombination in granulosa cells from both Ctgf cKO, genomic PCR analyses were performed using primers that can detect and distinguish the null, flox, and recombined alleles. As expected, the Ctgf-null or recombined alleles were detected in granulosa cells from control and both Ctgf cKO ovaries (Fig. 1, C and D). On the contrary, the Ctgf wild-type (WT) or flox alleles were barely detectable in granulosa cells from both Ctgf cKO female mice expressing Amhr2-cre or Pgr-cre, but not in granulosa cells from control female mice lacking the Amhr2-cre and Pgr-cre knockin alleles (Fig. 1, C and D). In addition, we also analyzed the mRNA abundance of Ctgf in the granulosa cells from exogenous hormone-primed mice using real-time quantitative PCR (qPCR) with predesigned TaqMan Assays-On-Demand PCR primer and probe sets. Ctgf mRNA expression was significantly reduced in both Ctgf cKO female mice, compared with WT and control female mice (Fig. 1, E and F). However, as shown in Fig. 1, C–F, granulosa cells from both Ctgf cKO female mice still contained a low level of Ctgf mRNA expression even though Ctgf-flox alleles and mRNA transcripts were markedly decreased. Accordingly, tissue-specific recombination of CTGF was further assessed by Western blot analysis using the uterine tissues and ovarian granulosa cells collected from WT, control, and both Ctgf cKO female mice. Compared with those of WT cells, CTGF protein levels in cells from control female mice were decreased, and a further decrease was observed in the cells from both Ctgf cKO female mice as expected (Fig. 1G). Complete absence of CTGF protein synthesis in uteri was not expected because Amhr2-cre is known to direct recombination only in mesenchyme-derived structures (i.e. smooth muscle and stromal cells) of the uterus (43–45, 50), whereas Pgr-cre recombination occurred in epithelial cells, stromal cells, and smooth muscle cells of uteri (48).
Subfertility of Ctgf cKO female mice
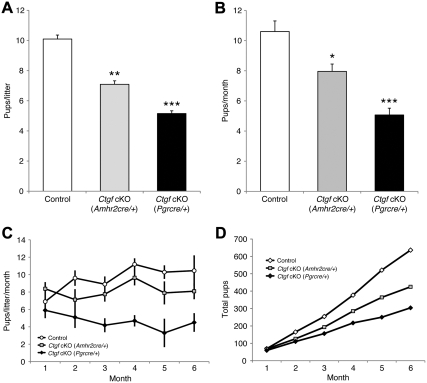
To evaluate the fertility of Ctgf cKO female mice, we conducted a continuous breeding study in which sexually mature female controls and mice of both Ctgf cKO strains (n = 10 for each genotype) at 6 wk of age were mated with known fertile WT male mice for 6 months. The control mice exhibited normal breeding activity and demonstrated a continuous accumulation of pups during the test period. The pups/ litter (10.1 ± 0.3; Fig. 2A) and pups/month (10.6 ± 0.7; Fig. 2B) of the control mice were not statistically different from the average of litter size and pups/month for our genetic mixed hybrid C57BL/6J;129S5 mouse colony (51). Meanwhile, Ctgf cKO (Amhr2cre/+) female mice exhibited reduction of fertility with a decrease in pups per litter (7.1 ± 0.2, P < 0.01) (Fig. 2A) and pups/month (8.0 ± 0.5, P < 0.05) (Fig. 2B) compared with control female mice after 1 month of mating (Fig. 2C). Additionally, Ctgf cKO (Pgrcre/+) female mice exhibited a more significant reduction of fertility by showing a decrease in pups per litter (5.2 ± 0.2, P < 0.001) (Fig. 2A) and pups/month (5.1 ± 0.5, P < 0.001) (Fig. 2B) with control female mice after 1 month of mating (Fig. 2C). The control mice produced the same level of pups per litter over the 6-month period, and an approximately equal numbers of pups were generated every 3–4 wk for the period of breeding trial (Fig. 2D). In contrast, Ctgf cKO (Amhr2cre/+) female mice produced fewer pups per litter than control female mice over the same period. Furthermore, the pups per litter of Ctgf cKO (Pgrcre/+) female mice continued to decline over the same period (Fig. 2C). Overall, the cumulative number of pups produced over the breeding period was reduced by 33% (425 pups) in Ctgf cKO (Amhr2cre/+) female mice and 52% (325 pups) in Ctgf cKO (Pgrcre/+) mice compared with control female mice (636 pups) (Fig. 2D). There were no differences in the frequency of litters per month among the three genotypes (data not shown). Thus, both Ctgf cKO female mice display severely disrupted fertility and fecundity.
Fig. 2.
Fertility reduction in Ctgf cKO female mice. Known fertile males were bred with 6-wk-old female littermates for 6 months. A, Litter sizes of Ctgf cKO (Amhr2cre/+) female mice (n = 10) and Ctgf cKO (Pgrcre/+) female mice (n = 10) vs. control female mice (n = 10). The litter sizes of both Ctgf cKO female mice were reduced compared with control female mice. B, Pups/month in Ctgf cKO (Amhr2cre/+) female mice (n = 10) and Ctgf cKO (Pgrcre/+) female mice (n = 10) vs. control female mice (n = 10). The pups/month of both Ctgf cKO female mice were also reduced compared with control female mice. C, A time course of the litter sizes in Ctgf cKO (Amhr2cre/+) female mice (n = 10) and Ctgf cKO (Pgrcre/+) female mice (n = 10) vs. control female mice (n = 10) during a 6-month testing period. Both Ctgf cKO female mice became subfertile after 1 month of breeding. Ctgf cKO (Pgrcre/+) female mice were more severe subfertile than Ctgf cKO (Amhr2cre/+) female mice. D, Total accumulated pups at each month of breeding vs. control female mice during a 6-month testing period. The control female mice (n = 10) have a uniform accumulation of pups at each month, whereas Ctgf cKO (Amhr2cre/+) female mice (n = 10) and Ctgf cKO (Pgrcre/+) female mice (n = 10) have fewer pups. The data are shown as means ± sem. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (vs. control female mice).
An increased trend of serum progesterone levels in Ctgf cKO female mice
To evaluate the hormone profiles of both Ctgf cKO mice, serum data were collected from mice of each genotype. At 4 to 5 months of age, no statistically significant difference was seen for estradiol. In contrast, serum progesterone levels appeared to be increased in Ctgf cKO (Amhr2cre/+) female mice compared with the age-matched control female mice; however, this increase was not statistically significant (P = 0.069; Table 1).
Table 1.
Serum hormone data in 4- to 5-month-old control and Ctgf cKO female mice
| Hormone | Control mice (n) | Ctgf cKO (Amhr2cre/+) mice (n) | Ctgf cKO (Pgrcre/+) mice (n) |
|---|---|---|---|
| Estradiol (pg/ml) | 13.09 ± 1.24 (9) | 12.78 ± 3.99 (6) | 9.52 ± 0.97 (5) |
| Progesterone (ng/ml) | 16.03 ± 4.40 (9) | 29.25 ± 4.30 (6) | 12.77 ± 3.71 (5) |
Enhanced corpus luteum formation and disrupted follicle development in Ctgf cKO female mice
To study the causes of the subfertility in the Ctgf cKO female mice, ovaries were examined histologically in control and both Ctgf cKO mice at 6 months of age. No gross changes in ovarian size were noticeable between control and both Ctgf cKO female mice. However, both Ctgf cKO ovaries contained several histological abnormalities. The control ovaries contained follicles of all developmental stages as well as corpora lutea (Fig. 3A). In contrast, both Ctgf cKO ovaries contained more corpora lutea than the control ovaries (Fig. 3, B and C).
Fig. 3.
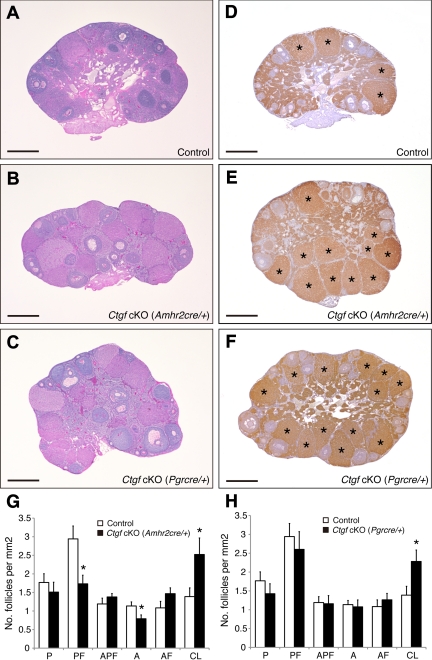
Morphological comparison of ovaries from control and both Ctgf cKO female mice. A–C, Comparison of ovarian histology from sexually mature (age 4–5 months) control and both Ctgf cKO female mice shows that Ctgf-mutant ovaries contain follicles at all stages of folliculogenesis as well as corpora lutea. Despite these observations, it is noted that there are reduced numbers of preantral and antral follicles, and the trend toward increased numbers of atretic preantral and atretic antral follicles in Ctgf cKO (Amhr2cre/+) ovaries. In addition, high numbers of functional corpora lutea are shown in both Ctgf cKO female mice ovaries. D–F, Immunohistochemistry for 3ß-HSD showing functional corpora lutea in both Ctgf cKO female mice ovaries. An asterisk marks the corpus luteum. G, Statistical analysis of the numbers of the follicle compartments in control and Ctgf cKO (Amhr2cre/+) female mice (n = 6 for each genotype). H, Statistical analysis of the numbers of the follicle populations in control and Ctgf cKO (Pgrcre/+) female mice (n = 6 for each genotype). Statistical significance was determined by using Student's unpaired and two-tailed t test. Representative sections are shown. P, Primordial and primary follicle; PF, preantral follicle; APF, atretic primordial, primary, and preantral follicle; A, antral follicle; AF, atretic antral follicle; CL, corpus luteum. The data are shown as means ± sem. *, P < 0.05 (vs. control female mice). All bars correspond to 500 μm.
To examine whether corpora lutea in Ctgf cKO ovaries could undergo functional regression, immunohistochemical analysis of 3ß-hydroxysteroid dehydrogenase (3ß-HSD), which catalyzes the synthesis of progesterone from pregnenolone, was examined in the ovaries at 6 months of age. Immunostaining for 3ß-HSD identified functional corpora lutea in all of the ovaries (Fig. 3, D–F). More regressing corpora lutea were found in both Ctgf cKO ovaries than in the control ovaries (Fig. 3, E–H).
We also compared follicle numbers in control and Ctgf cKO ovaries (n = 6 for each genotype). As shown in Fig. 3G, the follicle count showed reduced numbers of preantral (1.7 ± 0.2; P < 0.05) and antral follicles (0.8 ± 0.1; P < 0.05), and a trend toward increased numbers of atretic preantral (1.5 ± 0.1; P = 0.066) and atretic antral follicles (1.6 ± 0.2; P = 0.054) in Ctgf cKO (Amhr2cre/+) ovaries compared with control ovaries (preantral follicle, 2.9 ± 0.3; antral follicle, 1.1 ± 0.2; atretic preantral follicle, 1.2 ± 0.2; atretic antral follicle, 1.0 ± 0.2). In contrast, there were no differences between control and Ctgf cKO (Pgrcre/+) ovaries, with the exception of the corpus luteum numbers (Fig. 3H). Because Pgr-cre is expressed in preovulatory and later stage follicles (48, 49), it is unlikely to affect CTGF expression in granulosa cells of early-stage follicles in Ctgf cKO (Pgrcre/+).
Significant increase of granulosa cell apoptosis in Ctgf cKO female mice
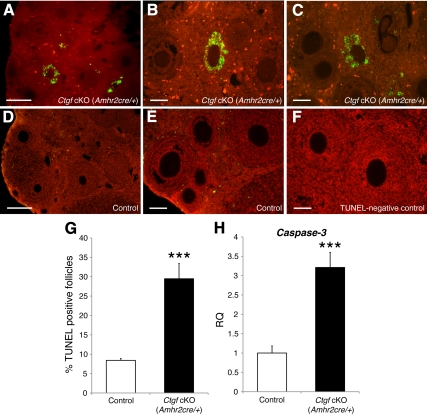
Quantitative analyses of follicle development in both Ctgf cKO female mice detected similar numbers of primordial and primary follicles in control and both Ctgf cKO female mice (Fig. 3, G and H). Therefore, if the defect in Ctgf cKO (Amhr2cre/+) granulosa cells is limited to proliferation rates, reduced numbers of preantral and antral follicles would be expected in Ctgf cKO (Amhr2cre/+) ovaries; however, we also consistently observed a trend toward increased numbers of atretic preantral (P = 0.066) and atretic antral follicles (P = 0.054) in Ctgf cKO (Amhr2cre/+) ovaries (Fig. 3G) compared with control ovaries. These results indicate that the observed decline of preantral and antral follicle numbers is due not only to impairment of granulosa cell proliferation, but also to excessive follicle atresia. Accordingly, to directly analyze follicle atresia in Ctgf cKO (Amhr2cre/+) ovaries, we performed a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay for the assessment of apoptosis in the follicles of control and Ctgf cKO (Amhr2cre/+) female mice at 4–5 months of age. Consequently, we detected a significant increase in the proportion of TUNEL-positive granulosa cells in Ctgf cKO (Amhr2cre/+) ovaries (Fig. 4, A–C and G; 29.5 ± 4.0%; P < 0.001) relative to the control ovaries (Fig. 4,D, E, and G; 8.4 ± 0.5%). To further elucidate the significant increase of apoptotic granulosa cells in Ctgf cKO (Amhr2cre/+) ovaries at a molecular level, we assessed Caspase-3 activation, which is a reliable marker of apoptosis, in granulosa cells by real-time qPCR using gene-specific primers. Consequently, we detected a significant increase of Caspase-3 mRNA in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice relative to control mice (Fig. 4H). Taken together, these data indicate that the reduction of survival of preantral and antral follicles in Ctgf cKO (Amhr2cre/+) ovaries is likely due to an increase in apoptotic granulosa cells, which is consistent with the trend toward increased numbers of atretic preantral and atretic antral follicles in Ctgf cKO (Amhr2cre/+) ovaries.
Fig. 4.
Increase of apoptotic granulosa cells in Ctgf cKO (Amhr2cre/+) ovaries. A TUNEL staining of 4- to 5-month-old ovaries from control (D) and Ctgf cKO (Amhr2cre/+) (A) female mice reveal a significant increase with apoptotic granulosa cells (green/yellow). Nuclei are counterstained with propidium iodide (red). B and C, High magnification of TUNEL-positive follicles in the Ctgf cKO (Amhr2cre/+) ovary. E, High magnification of TUNEL-negative follicles in the control ovary. F, TUNEL-negative control. Note that granulosa cells are TUNEL positive, whereas oocytes are negative. G, The proportion of TUNEL-positive follicles was quantified and found to be significantly higher in Ctgf cKO (Amhr2cre/+) ovaries than control ovaries. H, Caspase-3 transcript levels measured in granulosa cells by real-time qPCR in control (n = 5) and Ctgf cKO female mice (n = 5). Caspase-3 mRNA expression is markedly higher in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice relative to control female mice. The data are shown as means ± sem. The control values were set to equal 1. ***, P < 0.05 (vs. control female mice). In panels A and D, bars correspond to 250 μm. The bars in panels B, C, E, and F correspond to 150 μm.
Impaired ovulation, but not cumulus expansion, in Ctgf cKO female mice
When performing follicle counts on control and both Ctgf cKO ovaries, we observed the presence of luteinizing follicles with trapped oocytes in both Ctgf cKO females (Fig. 5, A and B). This indicates that ovulatory function may be also impaired in Ctgf cKO female mice. We therefore performed pharmacological superovulation experiments, in which control and both Ctgf cKO females were injected with exogenous hormones (PMSG/hCG) to induce ovulation. The numbers of oocytes recovered from Ctgf cKO (Amhr2cre/+) female mice (45.0 ± 10.9) and Ctgf cKO (Pgrcre/+) female mice (40.9 ± 6.0) were not significantly different from the controls (45.0 ± 4.1) at 3 wk of age (Fig. 5C). However, at 4–5 months of age, the numbers of ovulated oocytes were dramatically reduced in Ctgf cKO (Amhr2cre/+) female mice (28.5 ± 1.6) and Ctgf cKO (Pgrcre/+) female mice (25.3 ± 2.4), which ovulated about half the number of oocytes of control female mice (43.6 ± 3.9) (Fig. 5C).
Fig. 5.
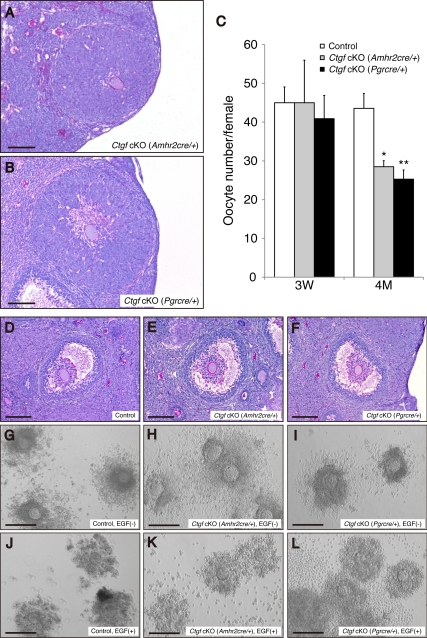
Impaired ovulation, but not cumulus expansion, in Ctgf cKO female mice. A and B, Ovaries from both Ctgf cKO female mice have large lutenized follicles with trapped oocytes. The bars in panels A and B correspond to 150 μm. C, Immature (3 wk) and mature (age 4 months) female mice of control (immature, n = 6; mature, n = 9), Ctgf cKO (Amhr2cre/+) (immature; n = 5; mature, n = 5), and Ctgf cKO (Pgrcre/+) (immature, n = 9; mature, n = 7) were subjected to a superovulation regimen. Oocyte/cumulus masses were then surgically extracted from their oviducts, and oocytes were counted after hyaluronidase digestion for 10 min. The numbers of ovulated oocytes was markedly lower in the independent samples from both Ctgf cKO female mice vs. control female mice. The data are shown as means ± sem. *, P < 0.05; **, P < 0.01 (vs. control female mice). D–F, Control (n = 3) and both Ctgf cKO female mice (n = 3 for each genotype) were treated with PMSG/hCG for in vivo cumulus expansion analysis. D, Note the outward radiation pattern of the expanded cumulus cells from the oocytes illustrated. E and F, The cumulus cells of both Ctgf cKO female mice undergo normal expansion in response to PMSG/hCG treatment. G–L, Control and both Ctgf cKO female mice were treated with PMSG alone, or followed by adding EGF, for in vitro cumulus expansion analyses. G–I, COC from control and both Ctgf cKO female mice cultured in the absence of EGF are illustrated. J, In vitro expansion of cumulus cells from control female mice stimulated by EGF (10 ng/ml). Note the expansion pattern of cumulus cells outward from the oocytes in the COC culture. K and L, Cultured COC from both Ctgf cKO female mice show normal expansion in response to EGF (10 ng/ml). The bars in panels D–F correspond to 150 μm. The bars in panels G–L correspond to 100 μm. 3W, 3-wk; 4M, 4-month.
To further assess the ovulatory function of both Ctgf cKO female mice, we performed in vivo and in vitro cumulus expansion analyses to define whether the Ctgf cKO mice have cumulus-cell defects during the preovulatory follicle stage. To study in vivo cumulus expansion, control and both Ctgf cKO female mice were given ip injections of PMSG, followed by ip injections of hCG 46 h later. The ovaries were collected after 6 h hCG injection and subjected to the histological analyses. Interestingly, cumulus cells within the preovulatory follicles underwent normal expansion in both control and Ctgf cKO females in vivo (Fig. 5, D–F). We also performed in vitro cumulus expansion analyses by culturing cumulus-oocyte complexes (COC) from control and both Ctgf cKO mice, and inducing the cumulus expansion by epidermal growth factor (EGF). The COC from control ovaries (Fig. 5G) and both Ctgf cKO ovaries (Fig. 5, H and I) remained compact and did not expand in the absence of EGF. On the contrary, adding EGF induced cumulus expansion in the COC from both control and Ctgf cKO ovaries (Fig. 5, J–L). The normal cumulus expansion in both Ctgf cKO ovaries thus indicates that the reduced ovulation is caused by an alternative defect in the ovulatory process.
Normal morphology and histology and normal hormonal and decidual response in Ctgf cKO uteri
We next investigated the effects of Ctgf loss in the uterus. First, to determine whether there are morphological changes in both Ctgf cKO uteri, we measured the size of uteri and compared the weight of uteri normalized to body weights in control and both Ctgf cKO female mice. Uterine size and weight were significantly reduced in Ctgf cKO (Pgrcre/+) female mice (3.1 ± 0.3 mg/g; P < 0.001) compared with control mice (4.7 ± 0.3 mg/g) (panels B and F of Supplemental Fig. 1 published on The Endocrine Society's Journals web site at http://mend.endojournals.org); however, this was not observed in Ctgf cKO (Amhr2cre/+) mice (3.6 ± 0.5 mg/g) (Supplemental Fig. 1, A and F). Hematoxylin and eosin staining revealed no histological changes between control and both Ctgf cKO uteri, and both Ctgf cKO uteri displayed normal myometrial and endometrial compartments (Supplemental Fig. 1, C–E). In addition, to determine the hormonal response of both Ctgf cKO uteri to endocrine stimulation, mice were given ip injections with PMSG, followed by ip injections with hCG, to induce an increase in ovarian steroid hormone production. The mice were killed after 18 h, and the uterine weights, which are an indication of the response to ovarian steroid hormones, were recorded and normalized to body weights. However, the relative increase in uterine weight after hormonal stimulation was similar between control female mice (5.1 ± 0.3 mg/g; 1.1-fold), Ctgf cKO (Amhr2cre/+) female mice (5.0 ± 0.0 mg/g; 1.4-fold), and Ctgf cKO (Pgrcre/+) female mice (4.9 ± 0.5 mg/g; 1.6-fold) (Supplemental Fig. 1F). Thus, although the sizes and weights of the Ctgf cKO uteri were smaller than those of the controls, the Ctgf cKO uteri retained the normal ability to respond to steroid hormones.
To initiate embryo implantation and maintenance of pregnancy, decidualization is the crucial transformation process, by which progestin-induced fibroblastoid stromal cells of estrogen-primed endometrium differentiate into decidual cells. Although the estradiol response of both Ctgf cKO uteri appears normal, progesterone responsiveness and stromal-cell differentiation of both Ctgf cKO uteri remained to be clarified. Therefore, we induced an artificial decidual reaction as previously described (52). Briefly, 10 d after ovariectomy, control and both Ctgf cKO female mice were subjected to a steroid hormone regimen. To induce artificial decidualization, one uterine horn was given a decidual stimulus, while the contralateral horn served as an unstimulated control. After daily treatment with estrogen and progesterone, the mice were killed on the fifth day after the decidual trauma (Supplemental Fig. 1G). As presented in Supplemental Fig. 1, H–J, decidual responses appeared to be normal in control and both Ctgf cKO female mice. All stimulated uterine horns showed a similar increase in size and weight compared with the unstimulated uterine horns (Supplemental Fig. 1, H–J and Q). In addition, histological analyses of the endometrial stroma of all the decidual horns showed the expected transformation of fibroblasts to epithelioid cells in control and both Ctgf cKO female mice (Supplemental Fig. 1, K–M). Moreover, alkaline phosphatase activity, a marker for stromal-cell differentiation in response to decidualization, was detected in the stimulated control and both Ctgf cKO uterine horns, but not in the unstimulated control and both Ctgf cKO uterine horns (Supplemental Fig. 1, N–P). Accordingly, absence of Ctgf in the uterus appears to affect uterine growth but not decidualization.
Gene-expression changes in granulosa cells from Ctgf cKO female mice
To investigate the molecular changes in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice, we performed real-time qPCR and verified the expression of genes that are important for female fertility during folliculogenesis and luteinization. Granulosa cells were collected from 46 h PMSG-primed adult (6–8 months of age) control and Ctgf cKO (Amhr2cre/+) female mice. A number of ovarian expressed genes showed significant changes (Fig. 6 and Table 2). Cyclin D1 (Ccnd1), a cell-cycle promoting gene, was significantly down-regulated in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice (2.4-fold; P < 0.01, Fig. 6A and Table 2); however, cyclin D2 (Ccnd2), another cell-cycle promoting gene, and cyclin-dependent kinase inhibitor 1b (Cdkn1b), a cell cycle-inhibitory gene, were unchanged in granulosa cells from control and Ctgf cKO (Amhr2cre/+) female mice (Table 2). In addition, cyclin-dependent kinase inhibitor 2b (Cdkn2b), a corpus luteum marker, was also unchanged in granulosa cells from all genotypes (Table 2).
Fig. 6.
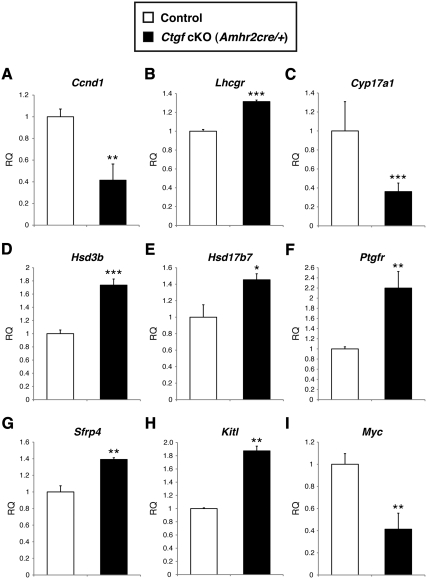
Gene-expression changes in granulosa cells from adult control and Ctgf cKO (Amhr2cre/+) ovaries after PMSG stimulation. Adult control and Ctgf cKO (Amhr2cre/+) female mice (age 6–8 months) were stimulated with PMSG for 46 h. Granulosa cells were then collected from five independent control and cKO ovaries. Total RNA was isolated and converted to cDNA. Real-time qPCR analysis of granulosa cell mRNA was performed using gene-specific primers. The data are shown as means ± sem. Significant changes in gene expression are seen for Ccnd1 (A), Lhcgr (B), Cyp17a1 (C), Hsd3b (D), Hsd17b7 (E), Ptgfr (F), Sfrp4 (G), Kitl (H), and Myc (I). Other genes examined are shown in Table 4. The control values were set to equal 1. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (vs. control female mice). RQ, Relative quantity.
Table 2.
Summary of real-time qPCR: Ctgf cKO (Amhr2cre/+) granulosa cells
| Gene | Synonym | Fold change in Ctgf cKO granulosa cells (PMSG 46 h) |
|---|---|---|
| Cell cycle | ||
| Ccnd1 | bcl-1, cD1, Cyl-1, PRAD1 | −2.4b |
| Ccnd2 | 2600016F06Rik, cD2, Vin-1, Vin1 | No change |
| Cdkn1b | p27, p27Kip1 | No change |
| Cdkn2b | MTS2, p15, p15Ink4b | No change |
| Endocrine signaling | ||
| Fshr | FSH receptor | No change |
| Lhcgr | Gpcr19-rs1, Lhr, LH/CG receptor | +1.3c |
| Steroidgenesis | ||
| Cyp11a1 | cscc, Cyp11a, D9Ertd411e, Scc, p450scc | No change |
| Cyp17a1 | p450c17 | −2.8c |
| Cyp19a1 | Ar, ArKO, Cyp19, Int5, p450arom, Aromatase | No change |
| Hsd17b7 | 17β-HSD7 | +1.5a |
| Hsd3b | 3β-HSD | +1.7a |
| Star | D8Ertd419e | No change |
| Other | ||
| Grem1 | Cktsf1b1, Drm, Gremlin1 | No change |
| Ptgfr | FP, PGF | +2.2b |
| Sfrp4 | Secreted frizzled-related sequence protein 4 | +1.4b |
| Kitl | Gb, grizzle-belly, Mgf, SCF, SF, SI, SLF, Steel | +1.9b |
| Myc | bHLHe39, Niard, Nird | −2.4b |
| Ghr | GHBP, GHR/BP | No change |
| Taf4b | 105 kDa, 2610524B04Rik, 4932409F03Rik, Taf2c2, TAFII105 | No change |
Fold changes are expressed relative to control.
, P < 0.05;
, P < 0.01;
, P < 0.001 (Student's t test).
FSH receptor (Fshr) and LH receptor (Lhcgr) are necessary to mediate the actions of the pituitary gonadotropins during the antral stage of follicle development. Fshr levels were unchanged in granulosa cells from PMSG-stimulated Ctgf cKO (Amhr2cre/+) female mice (Table 2); however, Lhcgr transcripts were up-regulated approximately 1.3-fold (P < 0.001, Fig. 6B and Table 2).
A number of genes related to steroidogenesis and luteinization were also significantly changed in the granulosa cells from Ctgf cKO (Amhr2cre/+) female mice. Cyp17a1, which catalyzes the synthesis of 17α-hydroxypregnenolone from pregnenolone, was down-regulated (2.8-fold; P < 0.001, Fig. 6C and Table 2); however, a corpus luteum marker, Hsd3b, which catalyzes the synthesis of progesterone from pregnenolone, was up-regulated (1.7-fold; P < 0.001) in the granulosa cells from Ctgf cKO (Amhr2cre/+) female mice (Fig. 6D and Table 2). In addition, another three luteal markers, Hsd17b7, prostaglandin F receptor (Ptgfr) (53), and secreted frizzled-related sequence protein 4 (Sfrp4) (54), were also up-regulated (1.5-fold; P < 0.05, 2.2-fold; P < 0.01, and 1.4-fold; P < 0.01, respectively) (Fig. 6, E–G, and Table 2). These data indicate that the granulosa cells from Ctgf cKO (Amhr2cre/+) female mice inappropriately up-regulate a subset of luteal markers in response to gonadotropin stimulation. In addition, gene expression of Hsd17b7, Ptgfr, and Sfrp4 in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice is similar to the previously reported gene-expression patterns of Smad4 granulosa cell-specific knockout mice (55).
Expression levels of granulosa cell proliferation markers such as Cyp11a1, Cyp19a1, Ccnd2, and Fshr were unchanged in the granulosa cells from Ctgf cKO (Amhr2cre/+) female mice (Table 2). However, there was a significant increase of apoptotic granulosa cells in Ctgf cKO (Amhr2cre/+) ovaries, as shown in Fig. 4. Thus, these data suggest that one of the important roles of CTGF in granulosa cells is the regulation of preantral and antral follicle numbers by blocking granulosa cell apoptosis during proliferation period.
CTGF has been shown to be regulated by several TGF-β family members including activin (27, 37, 56). Similar to Ctgf cKO (Amhr2cre/+) female mice, activin-deficient mice have reduced fertility (57). In addition, there are similar morphological changes, such as an abundance of corpus lutea and large lutenized follicles with trapped oocytes, and similar ovulatory changes such as decreased ovulation rates (57). We thus examined whether there are similar gene-expression changes in granulosa cells in Ctgf cKO (Amhr2cre/+) and activin-deficient mice. The qPCR analyses verified that the gene-expression changes in Ctgf cKO (Amhr2cre/+) mice were similar to those reported in activin-deficient mice. Kit ligand (Kitl), a positive regulator of follicle growth (58) and an important growth-stimulatory effector on granulosa cells (59), was up-regulated (1.9-fold; P < 0.01), and myelocytomatosis oncogene (Myc) was down-regulated (1.7-fold; P < 0.01) in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice, as reported in activin-deficient mice (Fig. 6, panels H and I, and Table 2). However GH receptor (Ghr) and TATA-binding factor-associated factor (Taf4b) were not changed compared with activin-deficient mice (Table 2). These data suggest that CTGF is a potential downstream mediator of activin signaling during follicle development.
Adamts1 mRNA is decreased in preovulatory granulosa cells from Ctgf cKO female mice
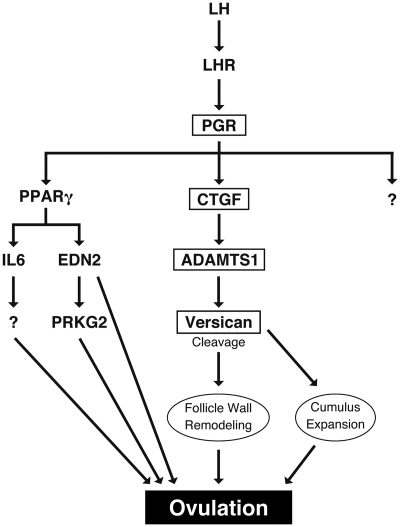
When antral follicles grow to reach the preovulatory stage by FSH (60), a timely surge of LH stimulates these preovulatory follicles to trigger ovulation. In preovulatory follicles, the LH surge initiates the orchestrated complex cascade of gene-expression events that culminate in the rupture of preovulatory follicles and the release of fertilizable oocytes (61). In addition, LH acts through its cellular receptor on mural granulosa cells of the preovulatory follicle to induce the expression of progesterone receptor (PGR) in these cells. Progesterone receptor and its steroid ligand progesterone play an important role in ovulation (62–64). In previous reports, Pgr-null mice fail to ovulate because of their inability to release oocytes from preovulatory follicles, even when ovulation is hormonally induced (63). Similarly, both Ctgf cKO mice have defects in ovulation in spite of the normal cumulus expansion (Fig. 5). Taken together, we hypothesized that the ovulatory dysfunction in both Ctgf cKO mice may be due to defects in expression of PGR-regulated genes, which in turn regulate the expression of downstream genes related to ovulation process.
To investigate this, we performed qPCR. Mice were given ip injections with 5 IU PMSG for 46 h, followed by ip injections with 5 IU hCG for an additional 11 h. Granulosa cells were collected from stimulated adult (6–8 months of age) control and both Ctgf cKO female mice for the examination of RNA levels.
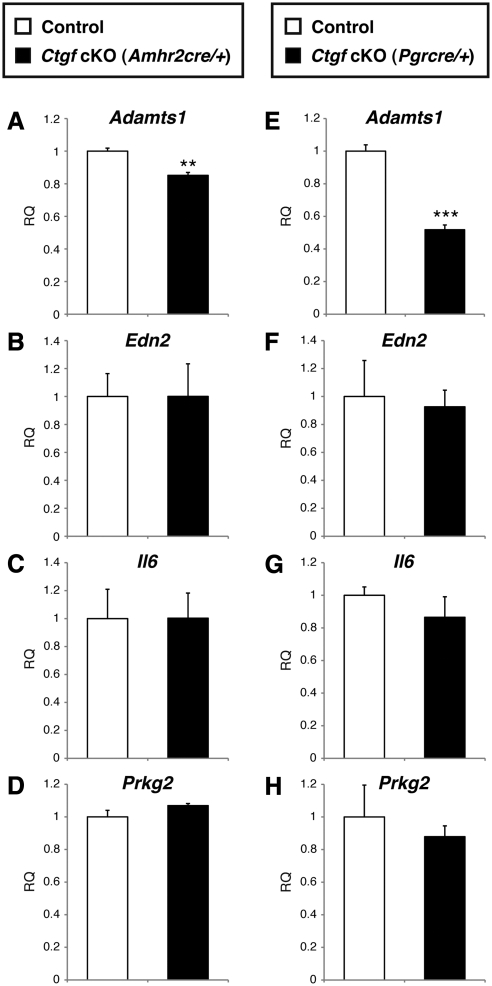
Endothelin-2 (Edn2), interleukin-6 (Il6), and cGMP-dependent protein kinase II (Prkg2), which are critical regulators of ovulation and induced in the preovulatory follicles in a PGR-dependent manner via the action of peroxisome proliferator activated receptor γ (PPARγ) (65–69), were unchanged between preovulatory granulosa cells from control and both Ctgf cKO female mice (Fig. 7, panels B–D and F–H). In contrast, a disintegrin and metalloprotease with thromboxane motif 1 (Adamts1), which is another PGR-regulated gene and independent from the PPARγ-regulated signaling pathway (67, 70), was significantly down-regulated in Ctgf cKO (Amhr2cre/+) female mice (1.2-fold; P < 0.01, Fig. 7A) and Ctgf cKO (Pgrcre/+) female mice (1.8-fold; P < 0.001, Fig. 7E). These findings indicate that CTGF acts as a downstream mediator of PGR-signaling pathway to control ovulation.
Fig. 7.
Ctgf Expression and regulation of Adamts1 during ovulatory period. Real-time qPCR analysis of granulosa cell mRNA from 6- to 8-month-old adult control and both Ctgf cKO female mice stimulated with hCG for 11 h after PMSG priming for 46 h. Granulosa cells were collected from five independent control and cKO ovaries. Total RNA was isolated and converted to cDNA. Adamts1 (A and E), Edn2 (B and F), Il6 (C and G), and Prkg2 (D and H) mRNA levels were analyzed. Adamts1 is lower in Ctgf cKO (Amhr2cre/+) and Ctgf cKO (Pgrcre/+) granulosa cells than control. There are no differences in other genes regulated by progesterone receptor. The control values were set to equal 1. **, P < 0.01; ***, P < 0.001 (vs. control female mice). RQ, Relative quantity.
The time-dependent changes of Ctgf and Adamts1 mRNA expression in granulosa cells from Ctgf cKO female mice during the preovulatory period
We performed additional qPCR studies to investigate time-dependent changes in mRNA expression of Ctgf, Adamts1, and other PGR-regulated genes in granulosa cells during the preovulatory period. Six- to eight-month-old adult control and Ctgf cKO (Pgrcre/+) female mice were given ip injections of 5 IU PMSG for 46 h, followed by ip injections with 5 IU hCG. Granulosa cells were collected at 0 h (46 h PMSG), 6 h, and 11 h after administration of hCG from every five independent control and Ctgf cKO (Pgrcre/+) ovaries and subjected to qPCR for the examination of mRNA levels.
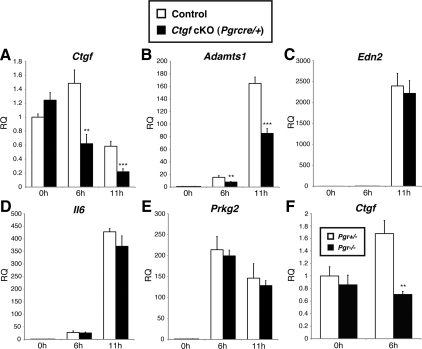
The expression of Ctgf was induced 6 h after hCG stimulation; this expression declined at 6 h and 11 h after hCG stimulation in preovulatory granulosa cells from control female mice (Fig. 8A). As expected, the expression of Ctgf was significantly down-regulated in Ctgf cKO (Pgrcre/+) female mice compared with control expression levels (Fig. 8A). Adamts1 was induced in a time-dependent manner after hCG stimulation in control mice (Fig. 8B). However, the expression of Adamts1 was significantly down-regulated in Ctgf cKO (Pgrcre/+) female mice, in accordance with the reduction of Ctgf expression (Fig. 8B). On the other hand, the expression of Edn2, Il6, and Prkg2 was induced after hCG stimulation and unchanged between control and Ctgf cKO (Pgrcre/+) female mice (Fig. 8, C–E). Taken together, CTGF can regulate the expression of Adamts1 in a time-dependent manner during preovulatory period, but does not appear to control expression of other PGR-regulated genes.
Fig. 8.
Time-dependent changes of Ctgf and Adamts1 mRNA expression in granulosa cells of Ctgf cKO and Pgr mutant mice during the preovulatory period. Real-time qPCR analysis of granulosa cell mRNA from 6- to 8-month-old adult control and Ctgf cKO (Pgrcre/+) female mice and from 3-wk-old Pgr+/− and Pgr−/− female mice treated with PMSG for 46 h before the stimulation of ovulation by hCG. Granulosa cells were collected at 0 h (only 46 h PMSG), 6 h, and 11 h after administration of hCG from ovaries of five independent mice. Total RNA was isolated and converted to cDNA. Ctgf, Adamts1, and other progesterone-regulated genes-mRNA levels were analyzed over time. A, Ctgf is induced at 6 h after hCG stimulation, followed by a continuing declines at 6 h and 11 h after hCG stimulation in preovulatory granulosa cells from control mice. In contrast, the expression of Ctgf is significantly down-regulated in Ctgf cKO (Pgrcre/+) female mice compared with the control expression. B, Adamts1 is induced in a time-dependent manner after hCG stimulation in preovulatory granulosa cells from control mice. However, the expression of Adamts1 is significantly down-regulated in Ctgf cKO (Pgrcre/+) female mice, according with the reduction of Ctgf expression. C–E, The expression of Edn2, Il6, and Prkg2 was induced after hCG stimulation and unchanged between control and Ctgf cKO (Pgrcre/+) female mice. The control values were set to equal 1. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (vs. control female mice). F, PGR regulates the induction of Ctgf mRNA during ovulation. Pgr+/− and Pgr−/ − female mice (age 3 wk) were treated with PMSG for 46 h before the stimulation of ovulation by hCG. Granulosa cells were collected at 0 h (only 46 h PMSG) and 6 h after administration of hCG from Pgr+/− and Pgr−/− ovaries (n = 3–5). Total RNA was isolated and real-time PCR was used to measure Ctgf mRNA levels. The expression of Ctgf mRNA in Pgr+/− granulosa cells at 0 h was set to equal 1. **, P < 0.01 (vs. Pgr+/− female mice). RQ, relative quantity.
Because Adamts1 is regulated positively by progesterone (67, 70) and is suppressed in our Ctgf cKO (Pgrcre/+) mice (Fig. 8B), we examined whether CTGF is downstream of PGR in granulosa cells during the ovulatory process by performing hormonal stimulation experiments using Pgr+/− (control) and Pgr−/− female mice. We examined the expression of Ctgf in granulosa cells 46 h after PMSG or after both PMSG (46 h) and hCG (6 h). Whereas Ctgf levels were not altered after PMSG treatment alone, Ctgf levels were up-regulated in the control but not the Pgr-null females 6 h after hCG stimulation (Fig. 8F). These results suggest that CTGF is downstream of the PGR-signaling pathway (see Fig. 9 and Discussion).
Fig. 9.
A model of PGR-regulated CTGF signaling during ovulation. Ovulation is initiated by the surge of LH, which acts via its receptor on granulosa cells of preovulatory follicles to stimulate the expression of PGR in these cells. PGR induces expression of CTGF, PPARγ, and other PGR-regulated genes. Activated PPARγ induces the expression of a subset of PGR-target genes including Edn2, Il6, and Prkg2. CTGF mediates its effects, at least partially, by inducing the expression of ADAMTS1, which catalyzes the cleavage of versican to an active form. Activated versican participates in the remodeling of the preovulatory follicle wall, ultimately leading to ovulation. LHR, LH receptor.
Discussion
In this study, we have demonstrated, for the first time, the roles of CTGF in follicle development and ovulation in vivo. Our Ctgf cKO mice demonstrate decreased fertility, secondary to defects in follicle development and ovulation.
Our data suggest a direct role of CTGF in normal follicle development. If growth induction is one of the primary roles of CTGF in follicle development, loss of CTGF in granulosa cells would be predicted to result in arrested granulosa cell growth, early differentiation of granulosa cells, and/or retarded follicle development. Our data demonstrate that Ctgf cKO (Amhr2cre/+) ovaries have decreased numbers of preantral and antral follicles but increases in atretic preantral and antral follicles compared with control ovaries. Furthermore, we showed that a significant increase in granulosa cell apoptosis resulted in these follicle-number changes. Although no definitive function has been known for CTGF in follicle development, some previous studies suggest that CTGF plays an active role in follicle development. First, Ctgf mRNA levels in granulosa cells rise gradually through preantral stages to a maximum in antral follicles (71). Second, in vitro cultures of ovaries demonstrate that CTGF induces primary and primordial follicle growth alone and in combination with TGF-β1 (38). And third, CTGF expression is up-regulated by activin and growth differentiation factor 9, and down-regulated by FSH and cAMP in vitro (72). Taken together, it is indicated that there are active and inhibitory communications between thecal cells and granulosa cells in multilayered preantral follicles through CTGF expression, which may regulate preantral and antral follicle formation.
The increased number of functional corpora lutea is another striking phenotype of Ctgf cKO female mice. Recently, some studies suggest a relationship between CTGF and corpus luteum formation. First, Ctgf mRNA is highly expressed in corpora lutea (36). Second, CTGF has been proposed as a stimulating factor for endothelial-cell migration in corpora lutea. Ctgf mRNA is expressed in fibroblasts and endothelial cells of larger vessels in human corpora lutea (39). Therefore, CTGF may also play an important role in corpus luteum function. In our study, Ctgf cKO (Amhr2cre/+) female mice appeared to have increased serum progesterone levels, although this increase was not statistically significant (P < 0.069; Table 1). Follicles demonstrating marked luteinization of granulosa cells and increased number of corpora lutea were apparent in the ovaries of Ctgf cKO mice; however, there is a decrease in the number of preantral and antral follicles. This suggests that CTGF may play a role in corpus luteum regression. Conditionally deleting Smad4 in the ovary also leads to premature luteinization of the granulosa cells of antral follicles (55). In addition, a number of genes related to steroidogenesis and luteinization were also significantly changed in the granulosa cells from Ctgf cKO (Amhr2cre/+) female mice. The expression of Cyp17a1 was down-regulated, and corpus luteum markers, Hsd3b, Hsd17b7, Ptgfr, and Sfrp4, were up-regulated (Fig. 6, D–G, and Table 2). These results suggest that there is both an increase in the synthesis of progesterone and inappropriate up-regulation of a subset of luteal markers in Ctgf cKO (Amhr2cre/+) female mice in response to gonadotropin stimulation. Along with the up-regulation of Lhcgr, these gene-expression data are similar to the previously reported gene-expression patterns of Smad4 granulosa cell-specific knockout mice (55). Therefore, although follicles are slowly lost at early time points in the Ctgf cKO (Amhr2cre/+) female mice, the relative rate of disappearance of the corpora lutea is much slower, thereby leading to an increased production of progesterone. Also, similar to Ctgf cKO female mice, activin-deficient female mice show enhanced formation of corpora lutea (57). Several studies suggest that activin plays an active role in corpora lutea (73–76). We also found that several other genes are changed in Ctgf cKO (Amhr2cre/+) granulosa cells; Kitl was up-upregulated, and Myc was down-regulated (Fig. 6, H and I, and Table 2). These changes are similar to the expression patterns of activin-deficient female mice (57). Smad4 granulosa cell-specific knockout female mice and activin-deficient female mice also exhibit increased serum progesterone levels (55, 57). CTGF is thought to be regulated by activin, and studies of human and macaque luteal cells find that activin inhibits progesterone synthesis (77, 78). Therefore, it is possible that the enhanced formation of functional corpora lutea and the increase of serum progesterone levels in Ctgf cKO mice are due to the lack of CTGF activity in granulosa cells. However, the mechanisms leading to the increased numbers of corpora lutea in Ctgf cKO mice need to be further elucidated. In addition, CTGF may be a key mediator of activin and SMAD4 signaling that regulates normal corpus luteum formation and ovarian synthesis of progesterone.
Gene-expression analysis indicates that Kitl, a mitogen for granulosa cells (59), was up-regulated in Ctgf cKO (Amhr2cre/+) granulosa cells during follicle development. Interestingly, Kitl-2, an isoform of Kitl, is normally down-regulated during antral follicle formation (79). Blocking the interaction between c-KIT and Kitl disrupts granulosa cell proliferation in antral follicles in vivo (80). In our previous paper, it was shown that activin is capable of suppressing Kitl expression (57). Taken together with the relationship between activin and CTGF, CTGF may play an important role in the activin-Kitl signal cascade for down-regulation of Kitl expression, resulting in the transition of antral follicles from proliferation phase to differentiation phase, which is required for the preovulatory period.
We also found that Myc and Ccnd1, cell cycle and proliferation markers, are down-regulated in Ctgf cKO (Amhr2cre/+) granulosa cells. Myc is known to regulate G1 cyclin-dependent kinase (CDK) activity by multiple steps including transcriptional control of Ccnd1 (81, 82). Furthermore, Myc and Ccnd1 are known to be up-regulated by gonadotropins (83–85). Thus, the decreasing numbers of preantral and antral follicles in Ctgf cKO (Amhr2cre/+) females may be explained by the down-regulation of Myc and Ccnd1 expression. On the other hand, Myc and Ccnd1 are also known as apoptosis-related genes. Overexpression of Myc can trigger apoptosis (85, 86). Ccnd1 activates a large number of gene transcriptions, leading to cell cycle progression, and down-regulation of Ccnd1 can induce cell cycle arrest at G1 (87–89). Therefore, down-regulation of Ccnd1 may underlie granulosa cell apoptosis in Ctgf cKO ovaries.
In addition to abnormal follicle development, Ctgf cKO ovaries have defective ovulatory function. The ovulation rate of PMSG/hCG-stimulated Ctgf cKO females decreases by approximately 60%. This was found in mature Ctgf cKO female mice (4–5 months of age), but not in immature Ctgf cKO female mice (3 wk of age). One of the reasons for no ovulatory differences between 3 wk-old control and both Ctgf cKO mice may be due to the correlation of Cre recombination with age, as previously reported in mice expressing Amhr2-cre (90) and Pgr-cre (49).
Despite the observation that Lhcgr is up-regulated in both Ctgf cKO mice, ovulation rates are decreased in both Ctgf cKO strains compared with control mice. Furthermore, Cyp17a1 is down-regulated; however, Hsd3b is up-regulated in granulosa cells from Ctgf cKO (Amhr2cre/+) female mice. These results indicate that the synthesis of progesterone should be significantly induced in Ctgf cKO (Amhr2cre/+) female mice; however, the ovulation rate is decreased in Ctgf cKO (Amhr2cre/+) female mice. Furthermore, Ctgf cKO mice have defects in ovulation after cumulus expansion. Thus, we speculated that the ovulatory dysfunction in Ctgf cKO mice may be due to the defects of PGR-regulated genes. This was confirmed by the finding that Ctgf cKO female ovaries exhibit reduced expression of Adamts1, which is one of the PGR-regulated genes critical for ovulation. Additionally, we show that Adamts1 acts downstream of Ctgf in the granulosa cells of preovulatory follicles. Thus, it is likely that Ctgf is a potential candidate gene for PGR regulation during preovulatory period.
Versican is a proteoglycan secreted abundantly by granulosa cells during the ovulatory process and is thought to be essential for the remodeling of the extracellular matrix surrounding granulosa cells of preovulatory follicles during the ovulation period (69, 91, 92). In the preovulatory period, ADAMTS1 mediates cleavage of versican into the 70-kDa V1 form, which is thought to be the functionally active-form. ADAMTS1 and versican are secreted by mural granulosa cells around the time of ovulation (93, 94). In addition, Adamts1-deficient female mice have impairments in ovulation, decreasing their fertility (91, 93, 95). Furthermore, CTGF is also required for the remodeling of the extracellular matrix in many tissues and has been shown to regulate the expression of multiple matrix metalloproteinases and their inhibitors (22, 40, 70, 96–99). Thus, CTGF may regulate ADAMTS1 expression in the ovary, impacting the cleavage and activation of versican, which is necessary for the remodeling of follicle wall in preovulatory follicles for ovulatory process.
Versican and the V1 form have also shown to be secreted by the matrix of COC around the time of ovulation (99). Adamts1-deficient female mice have a decreased cumulus expansion rate (91, 93). However, cumulus expansion in Ctgf cKO ovaries appears to be normal. There are no defects in cumulus expansion in Ctgf cKO females. The expression of Adamts1 decreases by approximately 20% in Ctgf cKO (Amhr2cre/+) and 45% in Ctgf cKO (Pgrcre/+) mice compared with control mice. Therefore, down-regulation of Adamts1 may not have been sufficient to cause a defect of cumulus expansion in Ctgf cKO females.
The expression of Adamts1 continues to decrease at 11 h after hCG in Ctgf cKO mice, indicating that ADAMTS1 is induced in a CTGF-dependent manner at the time of ovulation. Therefore, the ovulatory dysfunction of Ctgf cKO mice may be caused by decreased ADAMTS1 expression and subsequent inhibition of versican cleavage. In addition, PGR regulation of CTGF indicates that CTGF is a critical intermediate for follicle wall remodeling of periovulatory follicles. Furthermore, our study suggests that CTGF is the missing link in the PGR regulation of ADAMTS1 during ovulation, thereby unveiling a new ovarian signaling pathway comprising PGR, CTGF, and ADAMTS1 (Fig. 9).
Collectively, the present study demonstrates that CTGF is functionally required for normal follicle development, ovulation, and maintenance of female fertility. Our results provide genetic evidence that CTGF functions in inhibition of granulosa cell apoptosis, corpus luteum development, and follicle wall degradation, which are essential for normal fertility.
Materials and Methods
Generation of Ctgf cKO female mice
Mice carrying the Ctgf-null allele (Ctgf+/−) have been described previously (40). Mice carrying the Ctgf conditional allele (Ctgfflox/flox) were created in the laboratory of Dr. Karen Lyons and will be described elsewhere. The Ctgf+/− female mice were bred to male mice carrying anti-Müllerian hormone receptor type 2-cre knockin (Amhr2cre/+) (45) or progesterone receptor-cre knockin (Pgrcre/+) (48) alleles to generate Ctgf+/− Amhr2cre/+ male mice and Ctgf+/−Pgrcre/+ male mice. These mice were then bred to Ctgfflox/flox female mice to generate Ctgfflox/− Amhr2cre/+ and Ctgfflox/−Pgrcre/+ female mice, which were designated as Ctgf cKO (Amhr2cre/+) and Ctgf cKO (Pgrcre/+), respectively. Ctgfflox/− female mice were used as controls. Mice were genotyped by PCR analyses of genomic tail DNA using specific primers (Table 3). The primers for the null allele have been described previously (40), and the details of the primers for the conditional allele will be described elsewhere. Analyses of DNA recombination in the granulosa cell compartment were performed using DNA isolated from preovulatory stage granulosa cells as described previously (55, 100). Experiments were performed with 3- to 35-wk-old female mice. All mouse lines used in the present study were maintained on a hybrid C57BL/6J and 129S5 genetic mixed background. Animals were maintained according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Table 3.
Primer sequences for genotyping PCR
| Gene | Primer sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| Ctgf WT | TGTGTAGGACTTCATTCAGTTCT | GTTGGTGTCTGGACGCTCCA |
| Ctgf null | TGTGTAGGACTTCATTCAGTTCT | TCGCCTTCTTGACGAGTTCTTC |
| Ctgf flox | GCCATTTGGTCTGAACTCAT | GGGCTGTAGTCTCTTGGTG |
| Amhr2-cre | CGCATTGTCTGAGTAGGTGT | AGAGAGGCTGCGTTGAGTGT |
| Pgr-cre | TATACCGATCTCCCTGGACG | ATGTTTAGCTGGCCCAAATG |
Fertility analysis
To examine the fertility of female Ctgf cKO mice, Ctgf cKO (Amhr2cre/+) and Ctgf cKO (Pgrcre/+) females were mated independently with WT male mice for a 6-month period and 10 cages each. Cages were monitored daily, and the numbers of litters and pups were recorded over.
Histological analysis and follicle count
Ovaries and uteri were dissected and fixed in 10% neutral buffered formalin for histology. Tissue processing and embedding were performed in the Baylor College of Medicine Department of Pathology Core laboratory. Paraffin sections, cut at 5 μm, were stained with periodic acid-Schiff-hematoxylin and hematoxylin and eosin to examine the morphology of ovaries and uteri.
Follicles were classified according to the morphological criteria described by Pedersen and Peters (101). Follicle counting was performed as reported elsewhere (102, 103). Briefly, at least six ovaries of each genotype were serially sectioned at 8 μm, and every tenth section was examined. Follicles were counted from six of the largest sections, and the numbers of follicles were normalized by the total area of the section. The measurements were obtained by using ImageJ 1.44v software (http://rsb.info.nih.gov/ij), and the results are reported as average numbers of follicles/mm2.
Immunohistochemistry
Paraffin sections of ovaries were deparaffinized in xylene and subsequently hydrated by decreasing concentrations of ethanol (100%, 100%, 100%, 95%, 70%, 50%) before being hydrated in tap water. For antigen retrieval, these sections were then boiled in 10 mm citrate buffer (pH 6.0) in a microwave oven at 600W for 4 × 5 min. After washing in PBS, endogenous peroxidase activity was blocked by incubating sections in 3% hydrogen peroxide for 10 min. Endogenous avidin and biotin were also blocked by incubating the sections in avidin, followed by biotin (Blocking Kit SP-2001; Vector Laboratories, Burlingame, CA). To block nonspecific binding, sections were incubated in 3% normal serum (Vector)/PBS for 1 h at room temperature. The sections were then incubated overnight at 4 C with the primary antibody against 3ß-HSD [1:300 dilution (no.sc-30820); Santa Cruz Biotechnology, Inc., Santa Cruz, CA] in 3% goat serum/PBS. After washing in PBS, the sections were then incubated with a biotinylated secondary antibody (Vector). This secondary antibody was detected by streptoavidin-conjugated peroxidase (Vectastain ABC Kit Standard, PK-4000; Vector). For immunostaining, liquid diaminobenzide tetrahydrochloride (PK-4100; Vector) was used as a substrate for color development. Finally, the sections were briefly counterstained with Mayer's hematoxylin, dehydrated through increasing concentrations of ethanol, followed by xylene, and mounted. A section without the primary antibody was used as a negative control.
Western blot analysis
The whole uterine tissue cells and ovarian granulosa cells collected from WT, control, and both Ctgf cKO female mice (at least three animals were pooled for each genotype) were lysed with 200 μl cold lysis buffer [20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1% Na-deoxycholate, 0.1% sodium dodecyl sulfate, 1 mm Na3VO4, 50 mm NaF, and 1 mm Na2MoO4] containing protease inhibitor cocktail (Roche Molecular Biochemicals, Mannheim, Germany). After brief sonication, the lysates were centrifuged at 17,000 × g for 10 min at 4 C. The protein concentration was measured by BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). The lysates were stored at −80 C until electrophoresis. In a typical experiment, 25 μg lysates were mixed with lysis buffer. After the addition of 2× sodium dodecyl sulfate sample buffer (Invitrogen Corp., Carlsbad, CA), each sample was heated at 70 C for 10 min. The heated samples were separated on a NuPAGE 4 to 12% Bis-Tris gel electrophoresis (Invitrogen) at 200 V for 35 min and subsequently transferred onto a nitrocellulose membranes at 30 V for 60 min. The membrane was incubated in TBS-T [20 mm Tris-HCl, 100 mm NaCl (pH 7.6), and 0.1% Tween 20] containing 3% milk for 1 h at room temperature to block nonspecific binding sites. After three 10-min washes in TBS-T, the membranes were incubated overnight with primary antibodies against CTGF [1:500 dilution (no. PAB9497); Abnova, Taipei, Taiwan] in TBS-T containing 3% milk at 4 C. After overnight incubation and three 10-min washes in TBS-T, the membranes were incubated with horseradish peroxidase-conjugated goat antirabbit antibodies (1:10,000 dilution; The Jackson Laboratory, Bar Harbor, ME) in TBS-T containing 3% milk for 1 h at room temperature. After washing three times for 10 min each in TBS-T, the antibody-bound enzymes were detected by using SuperSignal West Pico detection kit (Thermo Fisher Scientific, Pittsburgh, PA) according to the manufacturer's instructions and exposed to x-ray films (Eastman Kodak, Rochester, NY). Immunoblots were stripped by using stripping buffer (Thermo Fisher) and reprobed with antibody against Actin [1:500 dilution (no. sc-1616); Santa Cruz Biotechnology] in TBS-T containing 3% milk.
TUNEL labeling assay of apoptotic cell
The TUNEL assay was performed to analyze DNA fragmentation associated with apoptosis using the ApopTag Plus Fluorescein In Situ Apoptosis Detection Kit (Millipore Corp., Billerica, MA). Ovaries were first fixed in 10% neutral-buffered formalin, embedded in paraffin, and sectioned at 5 μm. Five sections from each of three independent control and Ctgf cKO (Amhr2cre/+) ovaries were analyzed in parallel. TUNEL labeling assay was performed according to the manufacturer's instructions, and slides were mounted in Vectashield (Vector) containing propidium iodine to visualize chromatin. The total numbers of TUNEL-positive follicles per section were counted and normalized to the total numbers of follicles. Data are presented as the average percentage of TUNEL-positive follicles per section ± sem.
Superovulation and oocyte counts
To examine the ovulation ability of Ctgf cKO mice, at least five immature (age 21–23 d) and mature (age 4–5 months) female mice of control and both Ctgf cKO were superovulated: these mice were given ip injections with 5 IU PMSG (Calbiochem, San Diego, CA) for 46 h, followed by ip injections with 5 IU hCG (Calbiochem) for an additional 18 h. COC were isolated from oviducts and collected into M2 medium (Sigma-Aldrich Co., St. Louis, MO). After treating with 1 mg/ml hyaluronidase (Sigma-Aldrich) to dissociate cumulus cells from oocytes for 10 min, the number of oocytes per female was recorded.
Cumulus expansion analysis
To study in vivo cumulus expansion, adult (age 4–5 months) female mice, including control (n = 3) and both Ctgf cKO (n = 3 for each genotype), were given ip injections with 5 IU PMSG, followed by ip injections with 5 IU hCG 46 h later. The ovaries were collected 6 h after hCG injection and fixed in 10% neutral buffered formalin. The ovaries were then processed for periodic acid-Schiff-hematoxylin staining and histologically examined. In vitro cumulus expansion assay was performed according to a previously described protocol with slight modifications (55). In brief, adult (age 4–5 months) control (n = 3) and both Ctgf cKO female mice (n = 3 for each genotype) were treated with 5 IU PMSG. After 44 h of PMSG treatment, to assess cumulus expansion in vitro, COC were dissected from large antral follicles. These intact COC were then cultured in droplets of DMEM/F12 (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 0.25 mm sodium pyruvate (Invitrogen), 3 mm l-glutamine (Invitrogen), and 100 U of penicillin-streptomycin (Invitrogen)/ml with or without 10 ng/ml of EGF (Becton Dickinson, BD, Franklin Lakes, NJ).
Uterine hormonal stimulation
Adult (age 4–5 months) control and both Ctgf cKO female mice were unstimulated or treated with 5 IU PMSG for 46 h, followed by 5 IU hCG for an additional 18 h. Body weights and uterine weights were measured, and the average uterine weight/body weight ± sem was calculated for each group. For each genotype, fold changes between the PMSG/hCG and unstimulated groups were calculated.
Artificial induction of decidualization
The artificial decidualization of the uterus was performed as previously described (52). Briefly, 10 d after ovariectomy, ovariectomized female mice were treated with sc injections of 100 ng 17ß-estradiol, once a day for 3 d. After 2 d rest, mice were then treated with daily injections of progesterone (1 mg, sc) and 17ß-estradiol (6.7ng, sc), once a day for 3 d. One uterine horn was traumatized by a needle scratch on the antimesometrial lumen at 6 h after the last injection. The contralateral horn was not traumatized and served as a control. Mice were then given sc injections of progesterone (1 mg, sc) and 17ß-estradiol (6.7ng, sc), once a day. The mice were killed 5 d after trauma, and the wet weight of the traumatized and control horns was recorded. Uterine tissues were then placed in 10% neutral buffered formalin for histology.
Alkaline phosphatase staining
Isolated uterine tissues were fixed at 4 C in 4% paraformaldehyde for 2 h by a sucrose gradient in PBS (10%, 20%, 30%, respectively) and frozen in Tissue-Tek optimal cutting temperature embedding medium (Sakura Finetek, Torrance, CA) for sectioning. The sections were postfixed in 0.2% glutaraldehyde, rinsed in PBS, and incubated with a 100 mm Tris buffer (pH 9.5) containing 5-bromo-4-chloro-3-indolyl phosphate and Nitro blue tetrazolium chloride (Roche Molecular Biochemicals). Nuclear Fast Red (Vector) was used for counterstaining.
Hormone assays
Blood was collected from adult (age 4–5 months) female mice by cardiac puncture under isoflurane inhalation (Abbott Laboratories, Chicago, IL). The serum was separated by centrifugation (5 min, 17,000 × g) in microtainer tubes (Becton Dickinson, Franklin Lakes, NJ) and stored at −20 C until further use. RIA for estradiol and progesterone were performed by the Center for Research in Reproduction Ligand Assay and Analysis Core in University of Virginia (Specialized Cooperative Centers Program in Reproductive Research NICHD/NIH U54 HD28934). The limit of detection of the assays is as follows: 17β-estradiol (5 pg/ml) and progesterone (0.1 ng/ml). Samples that are below the assay threshold were assigned with the threshold value. Detailed information on the hormone analyses is available at http://www.medicine.virginia.edu/research/institutes-and-programs/crr/ligand-page.
Granulosa cell isolation and RNA extraction
Granulosa cells were isolated from immature female mice (age 21–23 d) or mature female mice (age 6–8 months) as described previously (55, 57, 104). Briefly, Ctgf cKO (Amhr2cre/+), Ctgf cKO (Pgrcre/+), Pgr +/−, and Pgr −/− female mice were treated with 5 IU PMSG (46 h) or 5 IU PMSG (46 h) and 5 IU hCG (6 h or 11 h). After treatment, granulosa cells were collected for RNA isolation. At least three mice were used in each group. Large antral follicles were punctured with needles (104), and granulosa cells were collected into DMEM/F12 medium (Invitrogen) containing 0.3% fetal bovine serum, 10 mm HEPES, and 100 U/ml penicillin and streptomycin. After filtration through a 40-μm pore-size nylon mesh filter (Nalgene, Rochester, NY) for removing ovarian debris and oocytes, the granulosa cells were collected by centrifugation and subjected to RNA extraction by using the RNeasy minikit (QIAGEN, Valencia, CA). To remove possible DNA contamination, on-column digestion with ribonuclease-free deoxyribonuclease (QIAGEN) was performed during RNA extraction.
Real-time qPCR
Gene expressions were analyzed by real-time qPCR. Briefly, total RNA (200 ng) was first converted to cDNA using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)12–18 primers (Invitrogen). The cDNAs were then amplified using gene-specific primers and ABI Prism 7500 Sequence Detection System [Applied Biosystems (ABI), Foster City, CA] using standardized cDNA and TaqMan Assays-On-Demand PCR primer and probe sets (ABI). In addition, custom gene-specific primers were designed by Primer Express software (ABI). The following TaqMan assays were used: Ctgf, Mm00439093; Cdkn1b, Mm00438168; Cdkn2b, Mm00483241; Fshr, Mm00442819; Lhcgr, Mm00442931; Cyp11a1, Mm00490735; Cyp17a1, Mm00484040; Cyp19a1, Mm00484049; Star, Mm00441558; Grem1, Mm00488615; Kitl, Mm00442972; Myc, Mm00487803; Ghr, Mm00439093; and Gapdh, Pre-Developed TaqMan Assay Reagents Control Kits. For the following genes, custom primers were designed using Primer Express software, and primer sequences are listed in Table 4: Ccnd1; Ccnd2; Hsd17b7; Hsd3b; Ptgfr; Sfrp4; Taf4b; Adamts1; Edn2; Il6; Prkg2; Caspase-3; and Gapdh. TaqMan PCR was performed using TaqMan Universal PCR Master Mix (ABI), and PCR with custom primers was performed using SYBR Green PCR Master Mix (ABI). The reaction conditions were as follows: 2 min at 50 C, 10 min at 95 C, followed by 40 cycles of 15 sec at 95 C (denaturation) and 1 min at 60 C (annealing/extension). The assay was performed in a 20-μl reaction volume with a 96-well plate. Minimums of five animals per genotype were used, and each sample was analyzed in duplicate. Two nontemplate controls (nuclease-free water) were included in each plate for each primer-probe set. The relative quantity of transcript was calculated by the 2−ΔΔC method as described elsewhere (105), normalized to the endogenous reference (Gapdh), and plotted as means ± sem using Excel software (Microsoft Corp., Redmond, WA).
Table 4.
Primer sequences for real-time quantitative PCR
| Gene symbol | RefSeq Acc. no. | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|---|
| Ccnd1 | NM_007631 | CATCAAGTGTGACCCGGACTG | CCTCCTCCTCAGTGGCCTTG |
| Ccnd2 | NM_009829 | AGACCTTCATCGCTCTGTGC | TAGCAGATGACGAACACGCC |
| Hsd17b7 | NM_010476 | CCTGTGCTCAGTCCGTTTTT | CCAAGGCCCTGAATTCAATA |
| Hsd3b | NM_008293 | AGCCGAATGTGTCCATAAGC | TAGTGGCTGACGTGGAACTG |
| Ptgfr | NM_008966 | CTGGCCATAATGTGCGTCTC | TGTCGTTTCACAGGTCACTGG |
| Sfrp4 | NM_016687 | CCTGCCAGTGTCCACATATCC | GCAATTTTCAAGAAGCATCATCCT |
| Taf4b | NM_001100449 | TCACAAGAATCTGCCTCAGG | GCCACAAAGACAAGACGTAGC |
| Adamts1 | NM_009621 | TGCTCCAAGACATGCGGCTCAG | TGGTACTGGCTGGCTTCACTTCC |
| versican | NM_001081249 | TCCTGATTGGCATTAGTGAAG | CTGGTCTCCGCTGTATCC |
| Edn2 | NM_007902 | CCTGTGCTACCTTCTGCCATC | CCCTCAGCAGTCCACATCTTG |
| Il6 | NM_031168 | CCGCTATGAAGTTCCTCTCTGC | AGGGAAGGCCGTGGTTGTC |
| Prkg2 | NM_008926 | GCCCGATTCTCCTCAACCTCCC | TCCACTCTTCCGAACCCACCAAC |
| Caspase-3 | NM_009810 | CGATCTGGTACAGACGTG | GCCATGTCATCCTCA |
| Gapdh | NM_008084 | CAATGTGTCCGTCGTGGATCT | GCCTGCTTCACCACCTTCTT |
Acc. no., Accession no.
Statistical analysis
All experiments were repeated at least three times independently. Differences among groups were assessed by using Student's t test or one-way ANOVA, and the mean between individual groups was further compared using Tukey's HSD test or Dunnett's test. The hormone data were log transformed before statistical analysis. The data are shown as means ± sem, and a P value of <0.05 was considered statistically significant.
Acknowledgments
We thank the Matzuk laboratory members for helpful advice and suggestions, Ms. Shirley Baker (Baylor College of Medicine) for aid with manuscript formatting, and Dr. Richard Behringer (M.D. Anderson Cancer Center) for the generous gift of the Amhr2-Cre mice.
This research is supported by National Institute of Health grants HD32067 (to M.M.M.) and R01HD042311 (to F.J.D.) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, R01AR052686 (to K.M.L.) from the National Institute of Musculoskeletal and Skin Disease, and CA077530 (to J.P.L.) from the National Cancer Institute.
Present address for Q.L.: Department of Veterinary Integrative Biosciences, College of Veterinary Medicine & Biomedical Sciences, Texas A&M University, College Station, Texas 77843.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CDK
- Cyclin-dependent kinase
- cKO
- conditional knockout
- COC
- cumulus-oocyte complexes
- CTGF
- connective tissue growth factor
- EGF
- epidermal growth factor
- hCG
- human chorionic gonadotropin
- HSD
- hydroxysteroid dehydrogenase
- PGR
- progesterone receptor
- PMSG
- pregnant mare serum gonadotropin (PPAR, peroxisome proliferator activated receptor
- qPCR
- quantitative PCR
- TBS-T
- 20 mm Tris-HCl, 100 mm NaCl (pH 7.6), and 0.1% Tween 20
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling
- WT
- wild type.
References
- 1. Rodgers RJ, van Wezel IL, Irving-Rodgers HF, Lavranos TC, Irvine CM, Krupa M. 1999. Roles of extracellular matrix in follicular development. J Reprod Fertil Suppl 54:343–352 [PubMed] [Google Scholar]
- 2. Smith MF, McIntush EW, Ricke WA, Kojima FN, Smith GW. 1999. Regulation of ovarian extracellular matrix remodelling by metalloproteinases and their tissue inhibitors: effects on follicular development, ovulation and luteal function. J Reprod Fertil Suppl 54:367–381 [PubMed] [Google Scholar]
- 3. Werb Z, Chin JR. 1998. Extracellular matrix remodeling during morphogenesis. Ann NY Acad Sci 857:110–118 [DOI] [PubMed] [Google Scholar]
- 4. Yada H, Hosokawa K, Tajima K, Hasegawa Y, Kotsuji F. 1999. Role of ovarian theca and granulosa cell interaction in hormone productionand cell growth during the bovine follicular maturation process. Biol Reprod 61:1480–1486 [DOI] [PubMed] [Google Scholar]
- 5. Slee RB, Hillier SG, Largue P, Harlow CR, Miele G, Clinton M. 2001. Differentiation-dependent expression of connective tissue growth factor and lysyl oxidase messenger ribonucleic acids in rat granulosa cells. Endocrinology 142:1082–1089 [DOI] [PubMed] [Google Scholar]
- 6. Bradham DM, Igarashi A, Potter RL, Grotendorst GR. 1991. Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol 114:1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grotendorst GR. 1997. Connective tissue growth factor: a mediator of TGF-β action on fibroblasts. Cytokine Growth Factor Rev 8:171–179 [DOI] [PubMed] [Google Scholar]
- 8. Hashimoto Y, Shindo-Okada N, Tani M, Nagamachi Y, Takeuchi K, Shiroishi T, Toma H, Yokota J. 1998. Expression of the Elm1 gene, a novel gene of the CCN (connective tissue growth factor, Cyr61/Cef10, and neuroblastoma overexpressed gene) family, suppresses In vivo tumor growth and metastasis of K-1735 murine melanoma cells. J Exp Med 187:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. 1992. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol 12:10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Brien TP, Lau LF. 1992. Expression of the growth factor-inducible immediate early gene cyr61 correlates with chondrogenesis during mouse embryonic development. Cell Growth Differ 3:645–654 [PubMed] [Google Scholar]
- 11. O'Brien TP, Yang GP, Sanders L, Lau LF. 1990. Expression of cyr61, a growth factor-inducible immediate-early gene. Mol Cell Biol 10:3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, Watanabe C, Cohen RL, Melhem MF, Finley GG, Quirke P, Goddard AD, Hillan KJ, Gurney AL, Botstein D, Levine AJ. 1998. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA 95:14717–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang R, Averboukh L, Zhu W, Zhang H, Jo H, Dempsey PJ, Coffey RJ, Pardee AB, Liang P. 1998. Identification of rCop-1, a new member of the CCN protein family, as a negative regulator for cell transformation. Mol Cell Biol 18:6131–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bork P. 1993. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett 327:125–130 [DOI] [PubMed] [Google Scholar]
- 15. Hadjiargyrou M, Ahrens W, Rubin CT. 2000. Temporal expression of the chondrogenic and angiogenic growth factor CYR61 during fracture repair. J Bone Miner Res 15:1014–1023 [DOI] [PubMed] [Google Scholar]
- 16. Nakanishi T, Nishida T, Shimo T, Kobayashi K, Kubo T, Tamatani T, Tezuka K, Takigawa M. 2000. Effects of CTGF/Hcs24, a product of a hypertrophic chondrocyte-specific gene, on the proliferation and differentiation of chondrocytes in culture. Endocrinology 141:264–273 [DOI] [PubMed] [Google Scholar]
- 17. Nishida T, Nakanishi T, Shimo T, Asano M, Hattori T, Tamatani T, Tezuka K, Takigawa M. 1998. Demonstration of receptors specific for connective tissue growth factor on a human chondrocytic cell line (HCS-2/8). Biochem Biophys Res Commun 247:905–909 [DOI] [PubMed] [Google Scholar]
- 18. Shakunaga T, Ozaki T, Ohara N, Asaumi K, Doi T, Nishida K, Kawai A, Nakanishi T, Takigawa M, Inoue H. 2000. Expression of connective tissue growth factor in cartilaginous tumors. Cancer 89:1466–1473 [PubMed] [Google Scholar]
- 19. Chen CC, Chen N, Lau LF. 2001. The angiogenic factors Cyr61 and connective tissue growth factor induce adhesive signaling in primary human skin fibroblasts. J Biol Chem 276:10443–10452 [DOI] [PubMed] [Google Scholar]
- 20. Igarashi A, Okochi H, Bradham DM, Grotendorst GR. 1993. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell 4:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Babic AM, Chen CC, Lau LF. 1999. Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin αvβ3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol Cell Biol 19:2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimo T, Nakanishi T, Kimura Y, Nishida T, Ishizeki K, Matsumura T, Takigawa M. 1998. Inhibition of endogenous expression of connective tissue growth factor by its antisense oligonucleotide and antisense RNA suppresses proliferation and migration of vascular endothelial cells. J Biochem 124:130–140 [DOI] [PubMed] [Google Scholar]
- 23. Shimo T, Nakanishi T, Nishida T, Asano M, Kanyama M, Kuboki T, Tamatani T, Tezuka K, Takemura M, Matsumura T, Takigawa M. 1999. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J Biochem 126:137–145 [DOI] [PubMed] [Google Scholar]
- 24. Pawar S, Kartha S, Toback FG. 1995. Differential gene expression in migrating renal epithelial cells after wounding. J Cell Physiol 165:556–565 [DOI] [PubMed] [Google Scholar]
- 25. Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Lüscher TF, Fujii T. 1999. Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J Biol Chem 274:37461–37466 [DOI] [PubMed] [Google Scholar]
- 26. Wenger C, Ellenrieder V, Alber B, Lacher U, Menke A, Hameister H, Wilda M, Iwamura T, Beger HG, Adler G, Gress TM. 1999. Expression and differential regulation of connective tissue growth factor in pancreatic cancer cells. Oncogene 18:1073–1080 [DOI] [PubMed] [Google Scholar]
- 27. Lin SL, Chen RH, Chen YM, Chiang WC, Lai CF, Wu KD, Tsai TJ. 2005. Pentoxifylline attenuates tubulointerstitial fibrosis by blocking Smad3/4-activated transcription and profibrogenic effects of connective tissue growth factor. J Am Soc Nephrol 16:2702–2713 [DOI] [PubMed] [Google Scholar]
- 28. Albrecht C, von Der Kammer H, Mayhaus M, Klaudiny J, Schweizer M, Nitsch RM. 2000. Muscarinic acetylcholine receptors induce the expression of the immediate early growth regulatory gene CYR61. J Biol Chem 275:28929–28936 [DOI] [PubMed] [Google Scholar]
- 29. Kondo Y, Nakanishi T, Takigawa M, Ogawa N. 1999. Immunohistochemical localization of connective tissue growth factor in the rat central nervous system. Brain Res 834:146–151 [DOI] [PubMed] [Google Scholar]
- 30. Schwab JM, Postler E, Nguyen TD, Mittelbronn M, Meyermann R, Schluesener HJ. 2000. Connective tissue growth factor is expressed by a subset of reactive astrocytes in human cerebral infarction. Neuropathol Appl Neurobiol 26:434–440 [DOI] [PubMed] [Google Scholar]
- 31. Ryseck RP, Macdonald-Bravo H, Mattéi MG, Bravo R. 1991. Structure, mapping, and expression of fisp-12, a growth factor-inducible gene encoding a secreted cysteine-rich protein. Cell Growth Differ 2:225–233 [PubMed] [Google Scholar]
- 32. Brigstock DR. 1999. The connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed (CCN) family. Endocr Rev 20:189–206 [DOI] [PubMed] [Google Scholar]
- 33. Lau LF, Lam SC. 1999. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res 248:44–57 [DOI] [PubMed] [Google Scholar]
- 34. Surveyor GA, Wilson AK, Brigstock DR. 1998. Localization of connective tissue growth factor during the period of embryo implantation in the mouse. Biol Reprod 59:1207–1213 [DOI] [PubMed] [Google Scholar]
- 35. Harlow CR, Hillier SG. 2002. Connective tissue growth factor in the ovarian paracrine system. Mol Cell Endocrinol 187:23–27 [DOI] [PubMed] [Google Scholar]
- 36. Wandji SA, Gadsby JE, Barber JA, Hammond JM. 2000. Messenger ribonucleic acids for MAC25 and connective tissue growth factor (CTGF) are inversely regulated during folliculogenesis and early luteogenesis. Endocrinology 141:2648–2657 [DOI] [PubMed] [Google Scholar]
- 37. Harlow CR, Davidson L, Burns KH, Yan C, Matzuk MM, Hillier SG. 2002. FSH and TGF-β superfamily members regulate granulosa cell connective tissue growth factor gene expression in vitro and in vivo. Endocrinology 143:3316–3325 [DOI] [PubMed] [Google Scholar]
- 38. Schindler R, Nilsson E, Skinner MK. 2010. Induction of ovarian primordial follicle assembly by connective tissue growth factor CTGF. PLoS One 5:e12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duncan WC, Hillier SG, Gay E, Bell J, Fraser HM. 2005. Connective tissue growth factor expression in the human corpus luteum: paracrine regulation by human chorionic gonadotropin. J Clin Endocrinol Metab 90:5366–5376 [DOI] [PubMed] [Google Scholar]
- 40. Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. 2003. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mo FE, Lau LF. 2006. The matricellular protein CCN1 is essential for cardiac development. Circ Res 99:961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. 2002. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol 22:8709–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arango NA, Kobayashi A, Wang Y, Jamin SP, Lee HH, Orvis GD, Behringer RR. 2008. A mesenchymal perspective of Mullerian duct differentiation and regression in Amhr2-lacZ mice. Mol Reprod Dev 75:1154–1162 [DOI] [PubMed] [Google Scholar]
- 44. Deutscher E, Hung-Chang Yao H. 2007. Essential roles of mesenchyme-derived β-catenin in mouse Mullerian duct morphogenesis. Dev Biol 307:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. 2002. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet 32:408–410 [DOI] [PubMed] [Google Scholar]
- 46. Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, Behringer RR, Parker KL. 2004. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol 18:1610–1619 [DOI] [PubMed] [Google Scholar]
- 47. Robker RL, Akison LK, Russell DL. 2009. Control of oocyte release by progesterone receptor-regulated gene expression. Nucl Recept Signal 7:e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP. 2005. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 41:58–66 [DOI] [PubMed] [Google Scholar]
- 49. Ismail PM, Li J, DeMayo FJ, O'Malley BW, Lydon JP. 2002. A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol Endocrinol 16:2475–2489 [DOI] [PubMed] [Google Scholar]
- 50. Jorgez CJ, Klysik M, Jamin SP, Behringer RR, Matzuk MM. 2004. Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol Endocrinol 18:953–967 [DOI] [PubMed] [Google Scholar]
- 51. Ma X, Dong Y, Matzuk MM, Kumar TR. 2004. Targeted disruption of luteinizing hormone β-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility. Proc Natl Acad Sci USA 101:17294–17299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Finn CA, Martin L. 1972. Endocrine control of the timing of endometrial sensitivity to a decidual stimulus. Biol Reprod 7:82–86 [DOI] [PubMed] [Google Scholar]
- 53. Sugimoto Y, Hasumoto K, Namba T, Irie A, Katsuyama M, Negishi M, Kakizuka A, Narumiya S, Ichikawa A. 1994. Cloning and expression of a cDNA for mouse prostaglandin F receptor. J Biol Chem 269:1356–1360 [PubMed] [Google Scholar]
- 54. Hsieh M, Mulders SM, Friis RR, Dharmarajan A, Richards JS. 2003. Expression and localization of secreted frizzled-related protein-4 in the rodent ovary: evidence for selective up-regulation in luteinized granulosa cells. Endocrinology 144:4597–4606 [DOI] [PubMed] [Google Scholar]
- 55. Pangas SA, Li X, Robertson EJ, Matzuk MM. 2006. Premature luteinization and cumulus cell defects in ovarian-specific Smad4 knockout mice. Mol Endocrinol 20:1406–1422 [DOI] [PubMed] [Google Scholar]
- 56. Yu J, Prado GN, Schreiber B, Polgar P, Polgar P, Taylor L. 2002. Role of prostaglandin E(2) EP receptors and cAMP in the expression of connective tissue growth factor. Arch Biochem Biophys 404:302–308 [DOI] [PubMed] [Google Scholar]
- 57. Pangas SA, Jorgez CJ, Tran M, Agno J, Li X, Brown CW, Kumar TR, Matzuk MM. 2007. Intraovarian activins are required for female fertility. Mol Endocrinol 21:2458–2471 [DOI] [PubMed] [Google Scholar]
- 58. Thomas FH, Vanderhyden BC. 2006. Oocyte-granulosa cell interactions during mouse follicular development: regulation of kit ligand expression and its role in oocyte growth. Reprod Biol Endocrinol 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Otsuka F, Shimasaki S. 2002. A negative feedback system between oocyte bone morphogenetic protein 15 and granulosa cell kit ligand: its role in regulating granulosa cell mitosis. Proc Natl Acad Sci USA 99:8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kumar TR, Wang Y, Lu N, Matzuk MM. 1997. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet 15:201–204 [DOI] [PubMed] [Google Scholar]
- 61. Richards JS. 1994. Hormonal control of gene expression in the ovary. Endocr Rev 15:725–751 [DOI] [PubMed] [Google Scholar]
- 62. Loutradis D, Bletsa R, Aravantinos L, Kallianidis K, Michalas S, Psychoyos A. 1991. Preovulatory effects of the progesterone antagonist mifepristone (RU486) in mice. Hum Reprod 6:1238–1240 [DOI] [PubMed] [Google Scholar]
- 63. Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 9:2266–2278 [DOI] [PubMed] [Google Scholar]
- 64. Park OK, Mayo KE. 1991. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol 5:967–978 [DOI] [PubMed] [Google Scholar]
- 65. Chen Q, Sun X, Chen J, Cheng L, Wang J, Wang Y, Sun Z. 2009. Direct rosiglitazone action on steroidogenesis and proinflammatory factor production in human granulosa-lutein cells. Reprod Biol Endocrinol 7:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chinetti G, Fruchart JC, Staels B. 2000. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res 49:497–505 [DOI] [PubMed] [Google Scholar]
- 67. Kim J, Sato M, Li Q, Lydon JP, Demayo FJ, Bagchi IC, Bagchi MK. 2008. Peroxisome proliferator-activated receptor γ is a target of progesterone regulation in the preovulatory follicles and controls ovulation in mice. Mol Cell Biol 28:1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Palanisamy GS, Cheon YP, Kim J, Kannan A, Li Q, Sato M, Mantena SR, Sitruk-Ware RL, Bagchi MK, Bagchi IC. 2006. A novel pathway involving progesterone receptor, endothelin-2, and endothelin receptor B controls ovulation in mice. Mol Endocrinol 20:2784–2795 [DOI] [PubMed] [Google Scholar]
- 69. Sriraman V, Rudd MD, Lohmann SM, Mulders SM, Richards JS. 2006. Cyclic guanosine 5′-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol Endocrinol 20:348–361 [DOI] [PubMed] [Google Scholar]
- 70. Boerboom D, Russell DL, Richards JS, Sirois J. 2003. Regulation of transcripts encoding ADAMTS-1 (a disintegrin and metalloproteinase with thrombospondin-like motifs-1) and progesterone receptor by human chorionic gonadotropin in equine preovulatory follicles. J Mol Endocrinol 31:473–485 [DOI] [PubMed] [Google Scholar]
- 71. Harlow CR, Bradshaw AC, Rae MT, Shearer KD, Hillier SG. 2007. Oestrogen formation and connective tissue growth factor expression in rat granulosa cells. J Endocrinol 192:41–52 [DOI] [PubMed] [Google Scholar]
- 72. Wandji SA, Gadsby JE, Simmen FA, Barber JA, Hammond JM. 2000. Porcine ovarian cells express messenger ribonucleic acids for the acid-labile subunit and insulin-like growth factor binding protein-3 during follicular and luteal phases of the estrous cycle. Endocrinology 141:2638–2647 [DOI] [PubMed] [Google Scholar]
- 73. Cameron VA, Nishimura E, Mathews LS, Lewis KA, Sawchenko PE, Vale WW. 1994. Hybridization histochemical localization of activin receptor subtypes in rat brain, pituitary, ovary, and testis. Endocrinology 134:799–808 [DOI] [PubMed] [Google Scholar]
- 74. Myers M, Gay E, McNeilly AS, Fraser HM, Duncan WC. 2007. In vitro evidence suggests activin-A may promote tissue remodeling associated with human luteolysis. Endocrinology 148:3730–3739 [DOI] [PubMed] [Google Scholar]
- 75. van den Hurk R, Van de Pavert SA. 2001. Localization of an activin/activin receptor system in the porcine ovary. Mol Reprod Dev 60:463–471 [DOI] [PubMed] [Google Scholar]
- 76. Erickson GF, Shimasaki S. 2003. The spatiotemporal expression pattern of the bone morphogenetic protein family in rat ovary cell types during the estrous cycle. Reprod Biol Endocrinol 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Di Simone N, Lanzone A, Petraglia F, Ronsisvalle E, Caruso A, Mancuso S. 1994. Effect of activin-A on progesterone synthesis in human luteal cells. Fertil Steril 62:1157–1161 [DOI] [PubMed] [Google Scholar]
- 78. Brannian JD, Woodruff TK, Mather JP, Stouffer RL. 1992. Activin-A inhibits progesterone production by macaque luteal cells in culture. J Clin Endocrinol Metab 75:756–761 [DOI] [PubMed] [Google Scholar]
- 79. Thomas FH, Ethier JF, Shimasaki S, Vanderhyden BC. 2005. Follicle-stimulating hormone regulates oocyte growth by modulation of expression of oocyte and granulosa cell factors. Endocrinology 146:941–949 [DOI] [PubMed] [Google Scholar]
- 80. Yoshida H, Takakura N, Kataoka H, Kunisada T, Okamura H, Nishikawa SI. 1997. Stepwise requirement of c-kit tyrosine kinase in mouse ovarian follicle development. Dev Biol 184:122–137 [DOI] [PubMed] [Google Scholar]
- 81. Nasi S, Ciarapica R, Jucker R, Rosati J, Soucek L. 2001. Making decisions through Myc. FEBS Lett 490:153–162 [DOI] [PubMed] [Google Scholar]
- 82. Sears RC, Nevins JR. 2002. Signaling networks that link cell proliferation and cell fate. J Biol Chem 277:11617–11620 [DOI] [PubMed] [Google Scholar]
- 83. Piontkewitz Y, Sundfeldt K, Hedin L. 1997. The expression of c-myc during follicular growth and luteal formation in the rat ovary in vivo. J Endocrinol 152:395–406 [DOI] [PubMed] [Google Scholar]
- 84. Delidow BC, White BA, Peluso JJ. 1990. Gonadotropin induction of c-fos and c-myc expression and deoxyribonucleic acid synthesis in rat granulosa cells. Endocrinology 126:2302–2306 [DOI] [PubMed] [Google Scholar]
- 85. Matsumura I, Tanaka H, Kanakura Y. 2003. E2F1 and c-Myc in cell growth and death. Cell Cycle 2:333–338 [PubMed] [Google Scholar]
- 86. Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, Bertino JR, Wahl GM. 1995. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci USA 92:5436–5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev 12:2245–2262 [DOI] [PubMed] [Google Scholar]
- 88. Nevins JR. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ 9:585–593 [PubMed] [Google Scholar]
- 89. Harbour JW, Dean DC. 2000. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2:E65–67 [DOI] [PubMed] [Google Scholar]
- 90. Boerboom D, Paquet M, Hsieh M, Liu J, Jamin SP, Behringer RR, Sirois J, Taketo MM, Richards JS. 2005. Misregulated Wnt/β-catenin signaling leads to ovarian granulosa cell tumor development. Cancer Res 65:9206–9215 [DOI] [PubMed] [Google Scholar]
- 91. Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, Russell DL. 2010. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod 83:549–557 [DOI] [PubMed] [Google Scholar]
- 92. Richards JS, Hernandez-Gonzalez I, Gonzalez-Robayna I, Teuling E, Lo Y, Boerboom D, Falender AE, Doyle KH, LeBaron RG, Thompson V, Sandy JD. 2005. Regulated expression of ADAMTS family members in follicles and cumulus oocyte complexes: evidence for specific and redundant patterns during ovulation. Biol Reprod 72:1241–1255 [DOI] [PubMed] [Google Scholar]
- 93. Mittaz L, Russell DL, Wilson T, Brasted M, Tkalcevic J, Salamonsen LA, Hertzog PJ, Pritchard MA. 2004. Adamts-1 is essential for the development and function of the urogenital system. Biol Reprod 70:1096–1105 [DOI] [PubMed] [Google Scholar]
- 94. Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley BW, Richards JS. 2000. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci USA 97:4689–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai T, Wang Y, Ogata M, Nishimatsu H, Moriyama N, Oh-hashi Y, Morita H, Ishikawa T, Nagai R, Yazaki Y, Matsushima K. 2000. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest 105:1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst GR. 1999. Connective tissue growth factor mediates transforming growth factor β-induced collagen synthesis: down-regulation by cAMP. FASEB J 13:1774–1786 [PubMed] [Google Scholar]
- 97. Eguchi T, Kubota S, Kondo S, Shimo T, Hattori T, Nakanishi T, Kuboki T, Yatani H, Takigawa M. 2001. Regulatory mechanism of human connective tissue growth factor (CTGF/Hcs24) gene expression in a human chondrocytic cell line, HCS-2/8. J Biochem 130:79–87 [DOI] [PubMed] [Google Scholar]
- 98. Fan WH, Pech M, Karnovsky MJ. 2000. Connective tissue growth factor (CTGF) stimulates vascular smooth muscle cell growth and migration in vitro. Eur J Cell Biol 79:915–923 [DOI] [PubMed] [Google Scholar]
- 99. Russell DL, Doyle KM, Ochsner SA, Sandy JD, Richards JS. 2003. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem 278:42330–42339 [DOI] [PubMed] [Google Scholar]
- 100. Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, Gutierrez C, Wang D, Martin JF, Jamin SP, Behringer RR, Robertson EJ, Matzuk MM. 2008. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol 28:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pedersen T, Peters H. 1968. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil 17:555–557 [DOI] [PubMed] [Google Scholar]
- 102. Cheng G, Weihua Z, Mäkinen S, Mäkelä S, Saji S, Warner M, Gustafsson JA, Hovatta O. 2002. A role for the androgen receptor in follicular atresia of estrogen receptor β knockout mouse ovary. Biol Reprod 66:77–84 [DOI] [PubMed] [Google Scholar]
- 103. Hu YC, Wang PH, Yeh S, Wang RS, Xie C, Xu Q, Zhou X, Chao HT, Tsai MY, Chang C. 2004. Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc Natl Acad Sci USA 101:11209–11214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pangas SA, Jorgez CJ, Matzuk MM. 2004. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J Biol Chem 279:32281–32286 [DOI] [PubMed] [Google Scholar]
- 105. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time qPCR and the 2(−Δ Δ C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]