Abstract
Transport proteins of the neurotransmitter sodium symporter (NSS) family regulate the extracellular concentration of several neurotransmitters in the central nervous system. The only member of this family for which atomic-resolution structural data are available is the prokaryotic homologue LeuT. This protein has been used as a model system to study the molecular mechanism of transport of the NSS family. In this Journal Club, we discuss two strikingly different LeuT transport mechanisms: one involving a single high-affinity substrate binding site and one recently proposed alternative involving two high-affinity substrate binding sites that are allosterically coupled.
Summary of the problem
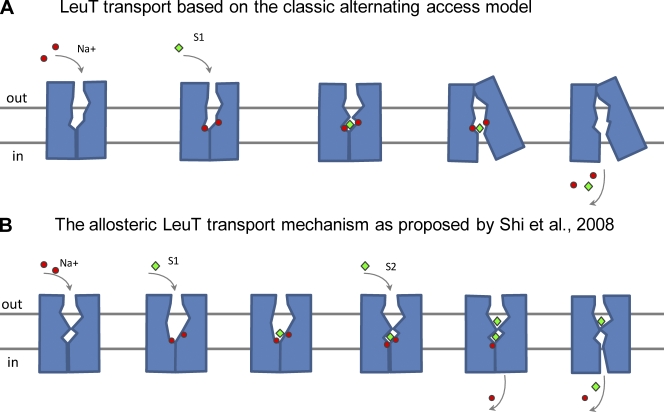
Active transporters are integral membrane proteins that move their substrates from one side of a membrane to the other against their electrochemical gradients. The first “molecular cartoons” for the mechanism of active transporters appeared in the literature around 50 years ago (Jardetzky, 1966) and represented a very elegant and general idea: To move their substrates across the membrane, the transporters alternate between two conformational states—outward- and inward-facing states—in which an aqueous pathway leads to a substrate-binding site from the extracellular space and the cytoplasm, respectively (Fig. 1 A). Today, we are just beginning to understand the molecular details of this concept in different families of active transporters, as highlighted by the recent debate on the transport mechanism of the neurotransmitter sodium symporter (NSS) family.
Figure 1.
Putative mechanisms for substrate transport by LeuT. (A) Classical alternating access mechanism for LeuT. Sodium ions (red spheres) and the amino acid substrate (green rhombus) bind to a central binding pocket to form a complex with the transporter. A conformational change closes external access and opens a path to the inside. After dissociation of substrates, the empty transporter undergoes a conformational change to regenerate the outward facing conformation. (B) The mechanism by Shi et al. (2008) requiring binding of two substrate molecules. This cartoon is strictly based on the model proposed by Shi et al. (2008; Fig. 7 is adapted with permission from Molecular Cell).
NSS are secondary active transporters essential to both brain physiology and pathology and the main targets for antidepressants, psychostimulants, and drugs of abuse (Murphy et al., 2004; Gether et al., 2006). They transport several neurotransmitters into the cytoplasm of neurons, glia, and other cells using the energy stored in transmembrane ionic gradients. In 2005, the laboratory of E. Gouaux published the first x-ray crystal structure of an NSS family member, LeuT, at the enviable resolution of 1.6 Å (Yamashita et al., 2005). LeuT is a Na+/amino acid symporter from the thermophilic bacterium Aquifex aeolicus, and it is currently a model system to understand the molecular mechanism of transport of the NSS family. The crystal structure of LeuT showed the transporter in an outward-facing state with a single leucine molecule and two sodium ions in a binding pocket, right underneath an extracellular aqueous vestibule and occluded from the extracellular solution by residues Y108 and F253 (Fig. 2). Shortly after, the crystal structure of LeuT was solved in complex with a variety of nonpolar amino acid substrates (Singh et al., 2008). All these structures showed two sodium ions bound and a single molecule of substrate in the same binding pocket as leucine. These structural data were in excellent agreement with the transport stoichiometry of two Na+ ions to one substrate molecule, measured for other NSS proteins (Krause and Schwartz, 2005).
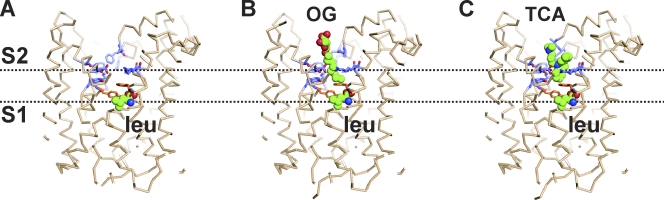
Figure 2.
LeuT substrate and inhibitor binding sites. x-ray crystal structures of LeuT in complex with leucine (PDB accession no. 2A65; A), leucine and OG (3GJC; B), and leucine and clomipramine, a TCA (2Q6H; C). The ligands in the structures are represented as spheres and the residues forming the S2 site are shown in blue. Residues Y108 and F253, which are occluding leucine bound to S1 from the extracellular solution, are shown in orange. The broken lines pass through S1 and S2 to indicate the position in the binding sites.
Remarkably, a very different mechanism for NSS was proposed by J. Javitch’s laboratory (Shi et al., 2008). The authors used measurements of radiolabeled ligand binding to propose an allosteric transport mechanism for LeuT in which the occupancy of two high-affinity substrate binding sites is required to achieve transport (Shi et al., 2008). Specifically, binding of a second substrate molecule at a secondary site was proposed to be an essential trigger for the transition of the protein to the inward facing state and for the release of the substrate from the primary site into the cytoplasm. This proposed mechanism created an ongoing discussion in the field; since its inception, there have been studies both supporting and contesting it.
Key results
The LeuT mechanism proposed by Shi et al. (2008) was based on radiolabeled ligand binding measurements using a scintillation proximity assay (SPA). SPA is a method to measure binding of a radio-labeled ligand to a protein attached to a scintillant-containing bead. When the authors measured the binding of leucine to wild-type (WT) LeuT, they found a binding stoichiometry of approximately two substrate molecules to one LeuT molecule. Molecular dynamic simulations suggested that a second substrate-binding site, called S2, was located in the extracellular vestibule of LeuT, ∼10 Å away from the central substrate binding site (S1), as determined by crystallography. Mutants at the S2 site abolished transport as well as binding of substrate to S2, and yielded a ∼1:1 substrate binding stoichiometry. From these experiments, they concluded that the substrate can simultaneously bind to S1 and S2 in WT LeuT, but only to S1 in the S2 mutants.
To address the functional role of the proposed S2 binding site, Shi et al. (2008) measured the dissociation of leucine from LeuT under different conditions. First, they found that prolonged preincubation (at least 3–5 h) of detergent-solubilized LeuT with radiolabeled leucine led to trapping of approximately half the bound leucine. Using the S2 mutant or a tricyclic antidepressant (TCA) to preclude substrate binding to S2, they suggested that leucine was trapped to the S1 site after the prolonged preincubation. The complete dissociation of leucine was only achieved upon addition of unlabeled substrate and Na+ removal. Based on this finding, Shi et al. (2008) proposed that binding to S2 triggered the release of substrate from S1 in an allosteric manner. However, it is intriguing that the time needed for the formation of the trapped state was much longer than the overall turnover rate of leucine transport (Kcat for leucine was ∼1–2/h; Singh et al., 2008). Shorter preincubations (30 min) with leucine, within the overall transport cycle of LeuT, or overnight incubations with alanine, a different substrate that is transported at a faster rate than leucine, didn’t lead to the formation of the trapped complex (Shi et al., 2008).
Quick et al. (2009) also measured the Kd of leucine binding and found it to be similar for S1 and S2, and <100 nM. Based on these binding data, it would be expected to see substrate bound to both S1 and S2 sites in the LeuT crystal structures. However, the structures of LeuT in complex with leucine and other substrates showed one molecule of substrate bound to S1, but no substrate bound to S2 (Fig. 2 A), even when the crystals were grown in a solution containing 30 mM leucine (Yamashita et al., 2005; Singh et al., 2008). Quick et al. (2009) investigated this discrepancy using functional and structural approaches. They showed that the detergent used for crystallization, n-octyl-β-d-glucopyranoside (OG), can act as an inhibitor and binds to S2, precluding the binding of leucine to that site (Fig. 2 B). It is worth noting that in contrast to the crystallographic experiments, all the LeuT functional studies to date have been performed in the presence of a larger and milder detergent, n-dodecyl-β-d-maltopyranoside (DDM). Additionally, the authors solved the crystal structure of a LeuT mutant (E290S) at 2.8-Å resolution, and found distinct electron density in S2 consistent with the presence of an OG molecule at this site (Quick et al., 2009). Triggered by this discovery, they solved the structure of WT LeuT at 2.0 Å resolution and found weaker electron density in S2 that could correspond to the aliphatic chain of the detergent molecule (Quick et al., 2009). Interestingly, electron density for an OG molecule at that position was not seen in the first structure of LeuT solved at 1.6 Å (Yamashita et al., 2005).
The site occupied by OG appears to be similar to the one where TCAs bind to inhibit LeuT function (Fig. 2 B). The crystal structures of LeuT in complex with different TCAs (Singh et al., 2007; Zhou et al., 2007) are, overall, very similar to the structure of LeuT in complex with leucine (Yamashita et al., 2005). Although these structures were also obtained in the presence of OG, they showed a molecule of leucine bound to S1 and a molecule of TCA in a binding pocket overlapping with S2, where the OG molecule was proposed to bind (Fig. 2 C). Considering that the TCAs inhibit LeuT transport with micromolar affinity (Singh et al., 2007; Zhou et al., 2007), it is still unclear how TCA molecules can displace OG from the S2 site but substrates with nanomolar affinity for LeuT, such as leucine, would fail to do so. Moreover, although there is agreement that the TCAs inhibit LeuT transport by binding to its extracellular vestibule and precluding the formation of the inward-facing state, the effect of the TCAs on the substrate binding is yet under discussion. In Shi et al. (2008) and Quick et al. (2009), Javitch’s group used the SPA method and found ∼50% displacement of substrate bound to WT LeuT by the TCA clomipramine, but no effect of the drug on the substrate bound to mutants designed to disrupt S2, which is consistent with the existence of two high-affinity substrate binding sites. The authors proposed a model of inhibition in which the TCAs, similarly to OG, compete with substrate for S2 and inhibit transport by disrupting the allosteric coupling between the two binding sites. In contrast, Singh et al. (2007) combined steady-state kinetics and radioactive substrate binding to propose a noncompetitive model of inhibition and showed that the TCAs do not displace substrate bound to LeuT, which is consistent with the existence of a single high-affinity substrate binding site (Singh et al., 2007).
Very recently, in Piscitelli et al. (2010), Gouaux’s laboratory directly tested the model of a single high-affinity site using several measurements of substrate binding stoichiometry. Strikingly, their results were in direct contradiction to those of Shi et al. (2008) and Quick et al. (2009). They used three different techniques to measure substrate binding stoichiometry to LeuT—SPA, isothermal titration calorimetry (ITC), and equilibrium dialysis—and found consistently that the binding stoichiometry of leucine was near unity. Moreover, using SPA, they found no difference in the binding stoichiometry of leucine between WT LeuT and the mutants designed to disrupt the S2 site. From these experiments, Piscitelli et al. (2010) concluded that there is a single high-affinity biding site (S1), although they suggested that substrate may bind weakly to other sites on its way from the extracellular medium to S1.
Discussion and conclusions
Despite the fact that both Gouaux’s (Yamashita et al., 2005; Singh et al., 2007; Piscitelli et al., 2010) and Javitch’s (Shi et al., 2008; Quick et al., 2009) groups have used structural and functional approaches to clarify the debate on the LeuT substrate stoichiometry and elucidate the mechanism of LeuT transport and inhibition, these approaches have not been enough to establish a consensus. Even more surprising is the fact that the same or very similar techniques have produced very different results. Crystallography has shown mixed results. One group detected density in the proposed second binding site corresponding to a detergent molecule (Quick et al., 2009), whereas the other group modeled structural water molecules to account for the excess of electron density in the extracellular vestibule of LeuT (Yamashita et al., 2005). From a crystallographic perspective, there could be two different ways to shed light on these intriguing results. The first one would be to screen for other detergent–lipid systems in which LeuT can be crystallized and that do not interfere with substrate transport and binding. This would allow detection of the substrate bound to S2, if it binds to this site with high affinity. The second way is to prove unequivocally that the density seen in S2 corresponds to OG. This could be achieved using OG with a heavy atom in its chemical structure, like sulfur or selenium. The advantage of these detergents is that the exact position of the sulfur or selenium atom can be accurately determined by measuring the x-ray anomalous scattering of the heavy atoms.
Regarding the binding assays, the results are even more dissimilar: Piscitelli et al. (2010) measured a substrate stoichiometry of ∼1:1 (one substrate molecule to one LeuT molecule) and Shi et al. (2008) measured a stoichiometry of ∼2:1, even when the two groups assayed binding with the same technique, the SPA. However, their experiments were performed in very different ways. Shi et al. (2008) used a concentration of LeuT (∼5 nM) approximately one order of magnitude below the Kd for leucine (40–70 nM). Under this condition, determination of the binding stoichiometry requires a precise knowledge of how the radioactivity, measured in counts per minute (cpm), translates into moles of bound radio-ligand. In other words, the system has to be calibrated. To do so, Shi et al. (2008) used a scintillation liquid mixture and measured the cpm for the known total amount of radio-ligand (Quick and Javitch, 2007; Shi et al., 2008). However, it is not clear that the counting efficiency in the scintillation liquid is similar to the one in the scintillating beads used in SPA. In the former, both bound and unbound radio-ligands react with an isotropic scintillating medium, whereas in the latter only the bound radio-ligand reacts with an anisotropic one. This can introduce errors in the calculation of the amount of substrate bound to the protein.
In contrast, the approach of Piscitelli et al. (2010) was to use an excess of LeuT over the Kd for leucine in their SPA experiments. Under these conditions, knowledge of the detection efficiency is not required, and the stoichiometry of the binding reaction can be estimated from the intersection abscissa of the linear regions of the binding curve at low and high substrate concentrations. The accuracy of the binding stoichiometry calculation depends on how much excess of protein over Kd is used. In the experiments of Piscitelli et al. (2010), this excess was ∼20-fold, although ideally at least 100-fold is required for accurate determination of the stoichiometry (Beckett, 2011). However, the limitation in these experiments is the amount of radio-ligand used to saturate the protein (already 1.2 µM in their assay) because the transporter is at a relatively high concentration. Noticeably, Piscitelli et al. (2010) also performed binding experiments using isothermal titration calorimetry, which does not require radio-labeled ligand, using LeuT in ∼400-fold excess over leucine Kd, and the leucine binding stoichiometry was still near unity.
There are two more important differences in the way the two groups performed their binding experiments. One is the determination of the background radioactivity in the SPA experiments: Shi et al. (2008) measured it using high imidazole concentration (400 mM) to detach the His-tagged protein from the scintillating beads. This approach could carry errors if the scintillating beads bind protein by means other than through the histidine tag. In contrast, Piscitelli et al. (2010) estimated the background radioactivity using 5 mM unlabeled alanine in their binding assays and determined the counts from 2 to 60 h after adding the substrate in the presence of sodium. This method could also introduce errors if the substrate gets kinetically trapped in the transporter, but alanine does not, as has been proposed (Shi et al., 2008). Nevertheless, it is unclear if the two approaches yielded similar background levels of radioactivity. The second consideration applies to all stoichiometric assays: the determination of the protein concentration, an essential parameter in these experiments. Again, the two groups used different methods to calculate the protein concentration. Shi et al. (2008) used colorimetric assays based on the absorbance shift of a dye that binds to the protein (Bradford, 1976). Piscitelli et al. (2010) used protein absorbance at 280 nm, but they corrected the theoretical extinction coefficient of LeuT, calculated based on its primary sequence, using quantitative amino acid analysis (QAAA). In our view, the standard methods to calculate protein concentration, like the Bradford assay and protein absorbance at 280 nm, are in general not accurate when applied to membrane proteins, and other techniques such as QAAA are required for this purpose. Accordingly, Piscitelli et al. (2010) found an ∼20% difference between the theoretical and the corrected extinction coefficients using QAAA.
Nevertheless, the differences in the way the two groups performed their experiments does not explain why Shi et al. (2008) measured a substrate binding stoichiometry of 2:1 for WT and 1:1 for the mutant designed to disrupt S2 (L400C), and Piscitelli et al. (2010) measured a 1:1 stoichiometry for both WT and L400C. In other words, the errors in protein concentration and nonspecific radioactivity determinations should be the same for the WT and mutant binding measurements, and should not affect the relative stoichiometry between the two proteins.
The two current models of transport by the Gouaux (Piscitelli et al., 2010) and Javitch (Shi et al., 2008) groups greatly differ on how binding of substrate is coupled to the conformational change that leads to its release into the cytoplasm. But do they predict different substrate dependence of the transport rate? Experimentally, both groups have determined a simple hyperbolic Michaelis-Menten–type substrate dependence of the rate of transport using protein reconstituted into liposomes. This is consistent with the hyperbolic substrate binding isotherms that the two groups have measured using detergent-solubilized protein. Such simple binding and transport behaviors are clearly expected for a single substrate-binding site model. However, they can also be obtained with a two-binding site model if there is no cooperativity between the two sites and the substrate affinities, for the sites are similar. Interestingly, the SPA results by Quick et al. (2009) are consistent with similar leucine Kd for S1 and S2, although the nature of the coupling between the substrate binding events in the two proposed binding sites is unclear in this model. One way to shed light on this important question would be to study the sodium dependence of leucine binding. Because leucine in S1 helps to coordinate Na1 and is in very close proximity to Na2, the leucine Kd for this site is expected to be strongly dependent on the sodium concentration. In contrast, binding of leucine to S2 would occur after the sodium ions are bound, and it is expected to be sodium independent. Therefore, studies of leucine binding at different sodium concentrations can help to understand how changes in the leucine Kd for S1 affect the Kd for S2, and the nature of the coupling between the two sites.
Elucidating the mechanism of LeuT transport will be a significant advance, as it might, apart from the NSS family, also have implications in other transporter families that have been recently identified to have similar structural fold to that of LeuT (Forrest and Rudnick 2009; Krishnamurthy et al., 2009). The clarification of this debate will require the use of similar techniques and experimental conditions, and most likely the development of new binding measurements using fluorescence or other binding methods.
Please participate in a discussion of this Journal Club article on the JGP Facebook page (www.facebook.com/JGenPhysiol).
Acknowledgments
We are indebted to Dr. J. A. Mindell for critical and thoughtful reading of the manuscript. We would like to thank Drs. G. Rudnick and O. Boudker for encouragement and discussions, and Dr. E. Gouaux for sharing results ahead of publication.
Joseph A. Mindell served as faculty advisor.
Christopher Miller served as editor.
Footnotes
Abbreviations used in this paper:
- NSS
- neurotransmitter sodium symporter
- OG
- n-octyl-β-d-glucopyranoside
- QAAA
- quantitative amino acid analysis
- SPA
- scintillation proximity assay
- TCA
- tricyclic antidepressant
- WT
- wild type
References
- Beckett D. 2011. Measurement and analysis of equilibrium binding titrations: A beginner’s guide. Methods Enzymol. 488:1–16 10.1016/B978-0-12-381268-1.00001-X [DOI] [PubMed] [Google Scholar]
- Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Forrest L.R., Rudnick G. 2009. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda). 24:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U., Andersen P.H., Larsson O.M., Schousboe A. 2006. Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol. Sci. 27:375–383 10.1016/j.tips.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Jardetzky O. 1966. Simple allosteric model for membrane pumps. Nature. 211:969–970 10.1038/211969a0 [DOI] [PubMed] [Google Scholar]
- Krause S., Schwarz W. 2005. Identification and selective inhibition of the channel mode of the neuronal GABA transporter 1. Mol. Pharmacol. 68:1728–1735 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy H., Piscitelli C.L., Gouaux E. 2009. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 459:347–355 10.1038/nature08143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.L., Lerner A., Rudnick G., Lesch K.P. 2004. Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol. Interv. 4:109–123 10.1124/mi.4.2.8 [DOI] [PubMed] [Google Scholar]
- Piscitelli C.L., Krishnamurthy H., Gouaux E. 2010. Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature. 468:1129–1132 10.1038/nature09581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M., Javitch J.A. 2007. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. USA. 104:3603–3608 10.1073/pnas.0609573104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M., Winther A.M., Shi L., Nissen P., Weinstein H., Javitch J.A. 2009. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl. Acad. Sci. USA. 106:5563–5568 10.1073/pnas.0811322106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Quick M., Zhao Y., Weinstein H., Javitch J.A. 2008. The mechanism of a neurotransmitter:sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell. 30:667–677 10.1016/j.molcel.2008.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Yamashita A., Gouaux E. 2007. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 448:952–956 10.1038/nature06038 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Piscitelli C.L., Yamashita A., Gouaux E. 2008. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 322:1655–1661 10.1126/science.1166777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Singh S.K., Kawate T., Jin Y., Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 437:215–223 10.1038/nature03978 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Zhen J., Karpowich N.K., Goetz R.M., Law C.J., Reith M.E., Wang D.N. 2007. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 317:1390–1393 10.1126/science.1147614 [DOI] [PMC free article] [PubMed] [Google Scholar]