Abstract
Phosphoinositide (PI) lipids are essential components of eukaryotic cell membranes. They are produced by mono-, bis- and trisphosphorylation of the inositol headgroup of phosphatidylinositol (PtdIns) and are concentrated in separate pools of cytosolic membranes. PIs serve as markers of the cell compartments and form unique docking sites for protein effectors. Collectively, seven known PIs, the protein effectors that bind them and enzymes that generate or modify PIs compose a remarkably complex protein-lipid signaling network. A number of cytosolic proteins contain one or several effector modules capable of recognizing individual PIs and recruiting the host proteins to distinct intracellular compartment. The recently determined atomic-resolution structures and membrane-targeting mechanisms of a dozen PI effectors have provided insights into the molecular basis for regulation of endocytic membrane trafficking and signaling. In this review, I highlight the structural aspects of the deciphering of the ‘PI code’ by the most common PI-recognizing effectors and discuss the mechanistic details of their membrane anchoring.
Phosphoinositides (PIs), phosphorylated derivatives of phosphatidylinositol (PtdIns), regulate fundamental biological processes including cell growth and survival, membrane trafficking and cytoskeletal dynamics (reviewed in refs. 1–4). PIs comprise less than 1% of cell lipids, yet they play very important roles in major signal transduction pathways, serving as docking sites for signaling effectors and as precursors of secondary messengers. The inositol headgroup of PI can be reversibly phosphorylated at three positions, 3, 4 and 5, and all seven PI isoforms, including three monophosphorylated (PtdIns(3)P, PtdIns(4)P and PtdIns(5)P), three bisphosphorylated (PtdIns(3,4)P2, PtdIns(3,5)P2 and PtdIns(4,5)P2) and one trisphosphorylated (PtdIns(3,4,5)P3) species have been identified in eukaryotic cells.
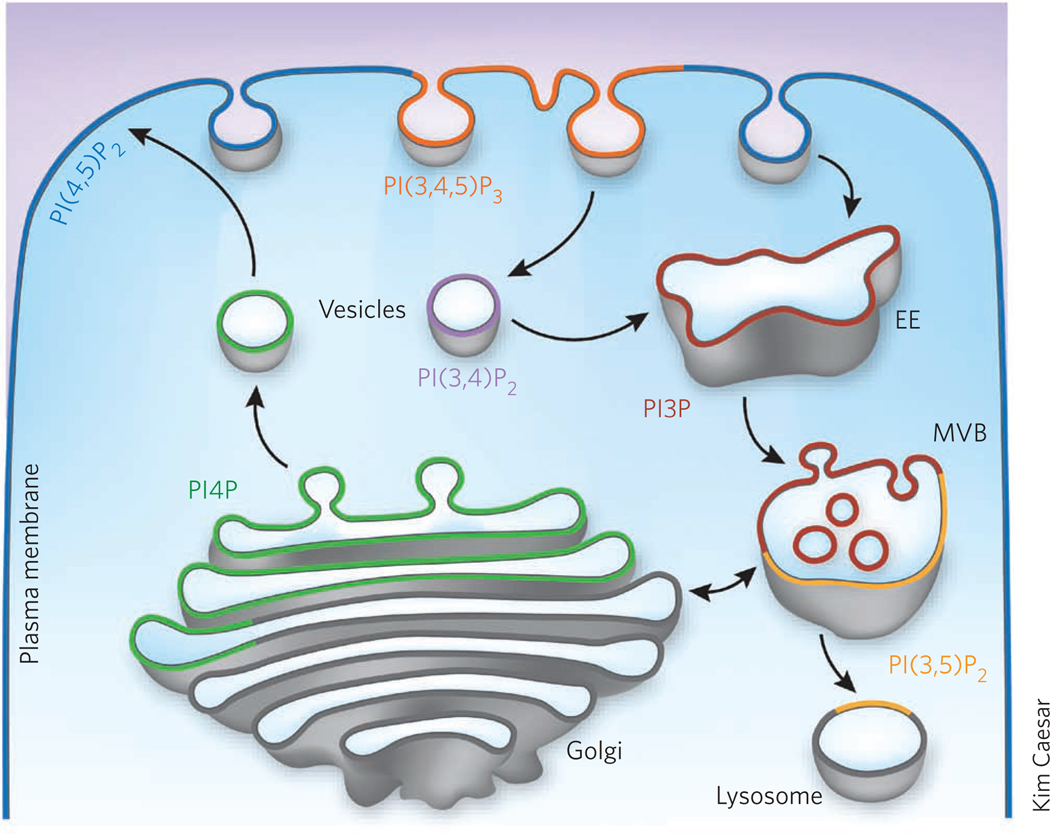
The amount and the spatial and temporal distribution of PIs in the cell vary substantially5. The largest pool of these lipids is composed of PtdIns(4)P and PtdIns(4,5)P2, whereas PtdIns(3,4,5)P3 appears to be undetectable in unstimulated cells. While some PIs, such as PtdIns(4)P and PtdIns(4,5)P2, are constitutively present in membranes, others are rapidly and transiently produced in response to the activation of cell surface receptors and other stimuli. The levels and turnover of PIs are tightly controlled by a large set of PI-specific enzymes. Found exclusively on the cytosolic side of membrane bilayers, PIs are readily accessible to PI kinases and phosphatases capable of attaching and removing phosphate groups, respectively, and to phospholipases that cleave the lipids. Because the PI-modifying enzymes are heterogeneously localized in the cell, PIs are clustered in distinct intracellular membranes and thus each form of PI essentially serves as a marker of an organelle. For example, the plasma membrane is enriched in PtdIns(4,5)P2, whereas PtdIns(4)P and PtdIns(3)P are detected primarily in the Golgi and early endosomes, respectively (Fig. 1). The unique distribution of PIs may provide the mechanism for fine-tuning the membrane trafficking flow and controlling the proper sequence of signaling events.
Figure 1. Subcellular localization of Pls.
PIs are concentrated in distinct pools of cytosolic membranes and serve as markers of various cell compartments and regions. Only predominant PIs are shown for simplicity. A heterogeneous distribution of PIs within a given membrane has also been observed. EE, early endosome; MVB, multivesicular bodies; PI3P, PtdIns(3)P; PI4P, PtdIns(4)P; PI(3,4)P2, PtdIns(3,4)P2; PI(3,5)P2, PtdIns(3,5)P2; PI(4,5)P2, PtdIns(4,5)P2; PI(3,4,5)P3; PtdIns(3,4,5)P3.
A major breakthrough in understanding the significance of PI signaling was the identification of protein effectors able to recognize individual PIs (Fig. 2). The pleckstrin homology (PH) domain was the first effector found to associate with PIs6. The list of PI-binding domains has since grown rapidly and at present contains 11 modules that display a wide range of affinities and selectivities for lipid membranes. It includes the ANTH (AP180 N-terminal homology), C2 (conserved region-2 of protein kinase C), ENTH (epsin N-terminal homology), FERM (4.1, ezrin, radixin, moiesin), FYVE (Fab1, YOTB, Vac1 and EEA1), GOLPH3 (Golgi phosphoprotein 3), PDZ (postsynaptic density 95, disk large, zonula occludens), PROPPINs (β-propellers that bind PIs), PTB (phosphotyrosine binding), PX (Phox homology) and Tubby modules. The recognition of a unique arrangement of phosphate groups around the inositol ring, which can be referred to as a ‘PI code’, by these domains results in the recruitment of the host proteins to specific intracellular compartments. Many of these proteins are modular in architecture and contain other lipid- and protein-binding domains or possess catalytic activities. These in turn trigger phosphorylation/dephosphorylation of other membrane-associated complexes and adaptors and promote the interconversion of PIs, subsequently leading to the activation or termination of signaling cascades. The cross-talk between PIs, the PI-modifying enzymes and effectors capable of ‘reading’ the PI code constitutes one of the most intriguing and complex signaling networks in the cell, which we have only begun to understand. This review focuses on the structural and mechanistic aspects of the deciphering of the PI code by the most common PI-binding effectors.
Figure 2. PI-recognizing effectors.
Signaling domains and their target PIs are colored as in Figure 1, according to their predominant distribution in intracellular membranes. PI3P, PtdIns(3)P; PI4P, PtdIns(4)P; PI5P, PtdIns(5)P; PI(3,4)P2, PtdIns(3,4)P2; PI(3,5)P2, PtdIns(3,5)P2; PI(4,5)P2, PtdIns(4,5)P2; PI(3,4,5)P3; PtdIns(3,4,5)P3.
The FYVE domain is an effector of PtdIns(3)P
The FYVE domain is a ~70-residue zinc-binding finger, found in 28 human proteins (reviewed in ref. 7). It binds PtdIns(3)P with high specificity and affinity and bridges a number of cytosolic proteins with PtdIns(3)P-enriched early endosomes, multivesicular bodies and phagosomes8–10. A small fraction of PtdIns(3)P has been identified in the nucleus and the Golgi apparatus, and FYVE domain–containing proteins DFCP1 and Alfy preferentially localize to these sites11,12. The FYVE domain is defined by three conserved sequences—the WxxD, RR/KHHCR and RVC motifs—that form a highly positively charged binding site for PtdIns(3)P. Although topologically, the FYVE domain belongs to a larger family of zinc-coordinating RING fingers, it can be distinguished from other DNA- and protein-binding members of this family by the presence of the three signature motifs.
FYVE domain–containing proteins have diverse biological functions. One of the largest subsets of FYVE proteins is involved in the regulation of endocytic trafficking and the fusion of endosomal membranes with transport vesicles and other organelles13. This subset includes the well-characterized mammalian proteins EEA1, Endofin, FENS-1, FYCO1, Hrs, Rabenosyn-5, Rabip4 and WDFY2 and yeast proteins Vac1p and Vps27p. Another fast-growing subset plays a critical role in signal transduction and TGFβ-Smad activation (Hrs and SARA), adipocyte differentiation (ProF), leukocyte signaling (FGD2), cytoskeletal reorganization (EhFP), autophagosome formation (DFCP1) and apoptosis (Phafin1 and 2). A number of enzymes such as kinases (Fab1 and PIKfyve), phosphatases (MTMR3 and MTMR4) and ubiquitin ligases (Pib1p) contain the FYVE domain, and their catalytic activities and localization to endosomal membranes require binding to PtdIns(3)P.
Molecular mechanism of membrane association by FYVE
While specific recognition of PtdIns(3)P is a major characteristic of the FYVE finger, it localizes to membranes through a multivalent mechanism that also involves nonspecific electrostatic contacts with acidic lipids other than PtdIns(3)P14–16, activation of a histidine switch17–19, hydrophobic insertion into the bilayer14–16,20–24 and, in some cases, dimerization25–28. Each of these components uniquely contributes to the FYVE domain specificity and increases the binding affinity for PtdIns(3)P-enriched membranes to the low nanomolar level11,14,22.
The FYVE domain contains a variable-length loop next to the PtdIns(3)P binding pocket. Upon binding to PtdIns(3)P, the hydrophobic residues at the tip of this loop (termed the turret loop or membrane insertion/interaction loop (MIL)) insert into the bilayer. The MIL is flanked by a set of basic and polar residues that are positioned at the level of the lipid headgroups when the protein penetrates the membrane. These residues make nonspecific electrostatic contacts with acidic phospholipids, such as phosphatidylserine (PS) or phosphatidic acid (PA). The strong positive potential around the MIL also drives the initial membrane docking and facilitates association with PtdIns(3)P. It has recently been shown that interaction of the FYVE domain with PtdIns(3)P exhibits pH dependence and is regulated by a histidine switch composed of a pair of adjacent histidine residues in the RR/KHHCR motif. The FYVE domain binds PtdIns(3)P when both histidine residues are positively charged and releases the lipid upon their deprotonation. Membrane association of FYVE domain–containing proteins can be further enhanced by bivalent or multivalent interactions with PtdIns(3)P. For example, EEA1 forms a parallel coiled-coil homodimer that juxtaposes two FYVE domains, allowing for simultaneous interaction with two PtdIns(3)P headgroups26.
Structural basis of PtdIns(3)P recognition by FYVE
The three-dimensional structures of the FYVE domain of human EEA1 (Protein Data Bank: 1JOC and 1HYI) bound to inositol 1,3-bisphosphate26 and dibutanoyl PtdIns(3)P21, and the ligand-free FYVE domains of EEA1 (1HYJ)21, fly Hrs (1DVP)29, Leishmania major Lm5-1 (1Z2Q)19, human RUFY (2YW8 and 2YQM) and yeast Vps27p (1VFY)20, have been determined by X-ray crystallography and NMR spectroscopy. All structures reveal a similar overall fold that consists of two double-stranded antiparallel β-sheets and a C-terminal α-helix (Fig. 3). An additional N-terminal α-helical turn is seen in the EEA1 FYVE domain structure, and a short α-helix connecting β2 and β3 is present in the structures of EEA1, Lm5-1 and RUFY. The functionally important β1 strand spans three residues of the RR/KHHCR motif and pairs with the β2 strand, which links two zinc-binding clusters. The zinc ions are bound by four CXXC motifs in a cross-braced topology. One zinc ion is coordinated by the first and third cysteine motifs, whereas another zinc ion is bound by the second and fourth motifs in all human proteins. In yeast Vps27p, the fourth cysteine residue is replaced by a histidine.
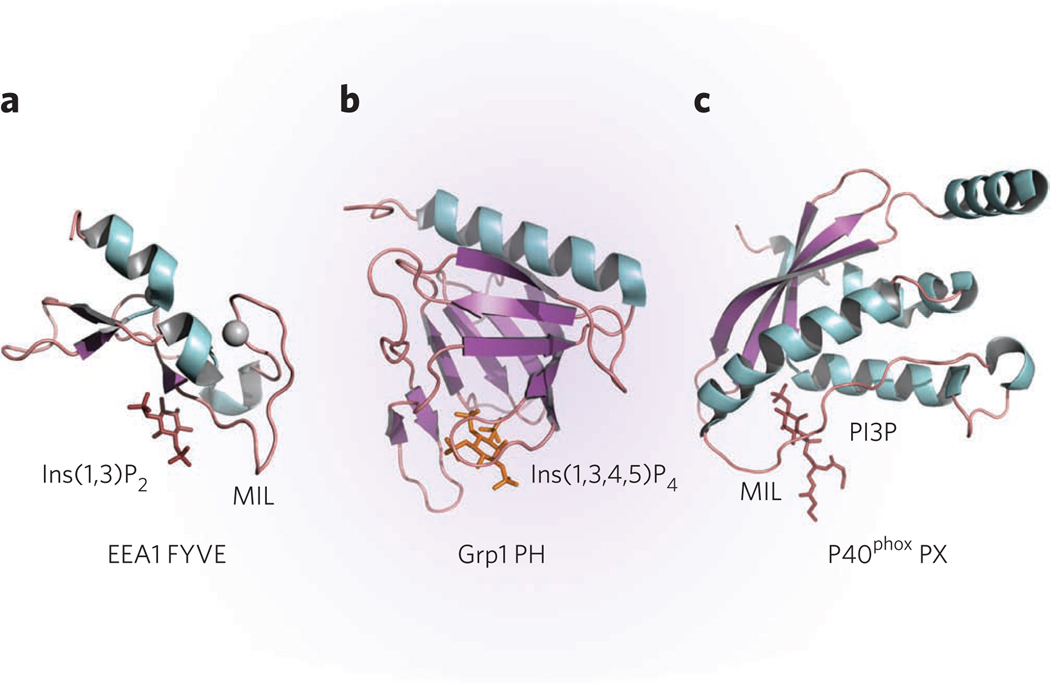
Figure 3. Molecular mechanism of PI recognition.
(a–c) Crystal structures of (a) the EEA1 FYVE domain in complex with inositol 1,3-bisphosphate (Ins(1,3)P2), a headgroup of PtdIns(3)P (1JOC)26, (b) the Grp1 PH domain in complex with inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4), a headgroup of PtdIns(3,4,5)P3 (1FGY)43 and (c) the p40phox PX domain in complex with dibutanoyl PtdIns(3)P (PI3P) (1H6H)67.
Structural insight into PtdIns(3)P recognition by the FYVE domain is provided by the crystal structure of the EEA1 FYVE domain in complex with inositol 1,3-bisphosphate26 (Fig. 4). The structure shows that the WXXD, RR/KHHCR and RVC motifs are centrally involved in coordination of the inositol headgroup (Fig. 4a). Critical hydrogen bonds are formed between the 3-phosphate group of PtdIns(3)P and the RR/KHHCR motif, particularly the guanidino moiety of the last arginine (Arg1375), the imidazole ring of the first histidine (His1372) and the backbone amide of the second histidine (His1373). The 1-phosphate is bound by the first arginine of the motif (Arg1370) and through water-mediated contact with the backbone carboxyl group of the second arginine (Arg1371). Arg1400 of the RVC motif, positioned between His1372 and Arg1375, forms another water-mediated hydrogen bond to the 3-phosphate group. Coordination of the 4-, 5- and 6-hydroxyl groups of the inositol ring is crucial for stereospecificity and the exclusion of alternatively phosphorylated phosphoinositides. The 4- and 5-hydroxyl groups are hydrogen bonded to the imidazole ring of His1373, whereas the carboxylate of Asp1352 in the N-terminal WXXD motif makes contacts with the hydroxyl groups at the 5 and 6 positions.
Figure 4. The structural basis for deciphering the PI code.
(a–e) Schematic diagrams showing PtdIns(3)P headgroup coordination by (a) EEA1 FYVE (1JOC) and (c) p40phox PX (1H6H) domains; PtdIns(3,4,5)P3 headgroup coordination by (b) Grp1 PH domain (1FGY); and PtdIns(4,5)P2 headgroup coordination by (d) CALM ANTH (1HFA) and (e) Epsin1 ENTH (1H0A) domains. Only charged residues of the proteins are depicted for clarity.
The PH domain reads various PI codes
The PH domains comprise one of the largest families of signaling modules and are the most thoroughly characterized among the PI-binding domains (reviewed in refs. 30,31). The PH domain was identified within a set of human proteins in 1993 and derives its name from the two homologous regions of pleckstrin, the major protein kinase C substrate in platelets32,33. Since then, this module has been found in 275 human proteins involved in intracellular signaling, membrane trafficking, cytoskeletal transformations and lipid metabolism. The PH domain contains ~120 residues that are folded in a highly conserved three-dimensional structure despite little sequence similarity between the family members. As a result of the high sequence variability, the PH domains have diverse functions and interact with numerous ligands, including proteins, acidic phospholipids, inositol polyphosphates and PIs. Many of those able to recognize PIs do so weakly and promiscuously, however a subset of PH domains (about 10–20% of all PH modules) binds individual PIs specifically and strongly, most commonly PtdIns(3,4,5)P3 and PtdIns(4,5)P2, as well as PtdIns(3,4)P2. The binding affinity of the PH domains for PIs varies significantly, ranging from low nanomolar to low micromolar31. Among the best-characterized PtdIns(3,4,5)P3 effectors are the PH domains of ARNO, Btk, Gap1, Grp1 and cytohesin-1 (refs. 34–36). The PLC™1 PH domain is specific for PtdIns(4,5)P2, whereas PH domains of TAPP1, centaurin β2, PEPP1 and FAPP1 prefer PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(3)P and PtdIns(4)P, respectively, and DAPP1, PDK1 and PKB (Akt) bind both PtdIns(3,4)P2 and PtdIns(3,4,5)P3 (refs. 30,31,37).
PH domains are present in GTP-GDP exchange factors (ARNO, cytohesin-1, FGD2, Grp1), GTPase-activating proteins (centaurin β2, Gap1), lipid-metabolizing (PLC™1, PLD1 and 2), lipid-transport (FAPP2) and cytoskeletal (dynamin, β-spectrin) proteins, kinases (Btk (Itk), CERK, PKB), phosphatases (PHLPP1) and other macromolecules that are implicated in vital biological processes including growth, proliferation, metabolism, cell polarization and migration, receptor endo- and exocytosis, membrane budding and trafficking, actin rearrangement, immune responses and apoptosis.
Molecular mechanism of membrane association by PH
The PI-binding site of the PH domain is formed by three variable loops flanking the open end of a β-barrel (described below and in Fig. 3b). Like the binding sites of all other PI-binding modules, it contains a cluster of basic lysine and arginine residues that make direct contacts to the phosphate groups of the lipid. As a result, the PH domain is electrostatically polarized and has a strong positive electrostatic potential around the binding site that contributes to both specific PI binding and nonspecific electrostatic interactions with other anionic lipids in membranes30,31,38–40. Single-molecule fluorescence studies reveal that the Grp1 PH domain can interact with PS via one or more secondary binding sites and that the electrostatic search mechanism speeds its association with PtdIns(3,4,5)P3 (ref. 41). PtdIns(3,4,5)P3 binding of this domain can be further enhanced by acidification of the medium and consequent protonation of the histidine residue40 that forms a critical hydrogen bond to the 4-phosphate group of the PI42,43. The pH dependence is less pronounced for the PH domain than for the FYVE domain. Unlike FYVE domains, which all contain two invariable histidine residues in the binding pocket, a single histidine is present in only a small set of PH domains40. Additionally, PH domains of ARNO, DAPP1, Fapp1, Grp1, PLC™1 and TAPP1 have been shown to penetrate PI-containing monolayers to various degrees39,40,44,45, and, at least in the case of Grp1, the membrane insertion is triggered by specific recognition of the inositol headgroup and is increased in an acidic environment40.
Structural basis of PtdIns(3,4,5)P3 recognition by PH
Over 100 three-dimensional crystal and NMR structures of various canonical PH domains have been deposited in the PDB. Of them, two are complexes with PtdIns(3,4,5)P3 (Btk (2Z0P) and PDK1 (1W1G)46), 12 are complexes of seven PH domains with inositol 1,3,4,5-tetrakisphosphate (IP4) (ARNO (1U27)36, Btk (1B55)47, DAPP1 (PHISH or 1FAO)42, Grp1 (1FHX, 1FGY, 2R0D and 2R09)42,43,48, PDK1 (1W1D)46, PEPP1 (1UPR), and PKB (1UNQ, 1H10 and 2UZS)49–51), two are complexes with inositol 1,4,5-trisphosphate (IP3) (ARNO (1U29)36 and PLC™1 (1MAI)52), two are complexes with inositol 1,2,3,5,6-pentakisphosphate (pleckstrin (2I5C and 2I5F)53) and one is a complex with inositol 1,3,4,5,6-pentakisphosphate (Grp1 (1FHW)42). Additionally, PH domains of ArhGAP9 (2P0D, 2P0F and 2P0H)54 and β-spectrin (1BTN)55 have been shown to a typically interact with IPs. The canonical PH domain folds into a seven-stranded β-barrel, capped by an amphipathic αhelix at one open end, whereas the opposite end is framed by three variable loops (Fig. 3b). The variable loops form a large PI-binding pocket, and their length and primary sequence define the specificity of the PH domain.
Details of how the PH domain recognizes the pattern of sequential 3-, 4- and 5-phosphate groups in PtdIns(3,4,5)P3 is provided by the crystal structure of the Grp1 PH domain in complex with IP4, an isolated headgroup of PtdIns(3,4,5)P3 (ref. 42,43) (Figs. 3b and 4b). The inositol ring lies in the center of a deep, positively charged pocket formed by the β1–β2, β3–β4 and β6–β7 loops and the strands they connect. The distal phosphates are buried deepest in the pocket, whereas the 1-phosphate group is positioned near the tips of the loops. A network of hydrogen bonds, formed between conserved lysine and arginine residues in β2, β3, β4 and β7 and all four phosphate groups of IP4, efficiently restrains the inositol molecule. A unique β-hairpin in the long β6–β7 loop of Grp1 is involved in additional hydrogen bonding contacts with the 5-phosphate group of IP4. These additional hydrogen bonds with the 5-phosphate account for the high specificity of the Grp1 PH domain toward PtdIns(3,4,5)P3. In general, the β1–β2 loop of PH domains functions as a platform for the interaction with PIs1. It contains the sequence motif Kxn(K/R)xR, which makes the most critical contacts with the phosphate groups of the lipid1.
The PX domain prefers PtdIns(3)P
The PX domain consists of ~130 residues and is found in 43 human signaling and regulatory proteins (reviewed in ref. 56). It is named after the two phagocyte NADPH oxidase (phox) subunits p40phox and p47phox, in which it was first identified in 199657. Of all PIs, PtdIns(3)P appears to be the primary target of PX domain–containing proteins, as the majority of them associate with PtdIns(3)P-enriched endosomes and vacuoles (KIF16B, p40phox, PXK, sorting nexins (SNXs), Vam7p), although binding to other PIs has also been reported for Bem1, CISK, CPK, FISH, NOXO1, p47phox, PI3K-C2α, PLD1 and SNXs. All yeast PX domains bind PtdIns(3)P, though only four with relatively high affinity (Kd ~2–3 µM)58.
PX domain–containing proteins play essential roles in endocytosis, protein sorting, membrane trafficking, transcription, cell polarity and signaling56. SNXs, found in both yeast and mammalian cells, comprise the largest subset of proteins harboring this domain. Human SNXs are involved in endosomal sorting and recycling, in internalization, transport and lysosomal degradation of epidermal growth factor receptor, and in membrane tubulation. Yeast SNXs are required for regulation of protein retrieval and recycling traffic from prevacuolar and late endosome compartments to the late Golgi. The t-SNARE Vam7p mediates fusion of multiple transport intermediates with the vacuole. The subunits of neutrophilic NADPH oxidase complex, p40phox and p47phox, are implicated in phagocyte-mediated destruction of ingested microbes. The cytokine-independent survival kinase (CISK), PI 3-kinases and the adaptor protein FISH play roles in cell signaling.
Molecular mechanism of membrane association by PX
Similar to the FYVE and PH modules, the PX domain associates with membranes that contain PtdIns(3)P or other PIs via multiple interactions. Specific recognition of the inositol headgroup is often facilitated by nonspecific electrostatic contacts with acidic membrane surfaces and is accompanied by a hydrophobic insertion into the bilayer. The PX domain was identified as a PI-binding module independently by several groups in 200158–63. The p40phox, SNX3 and Vam7p PX domains were found to specifically recognize PtdIns(3)P, whereas the p47phox PX domain was shown to prefer PtdIns(3,4)P2.
Several mechanistic studies have demonstrated that PI binding induces membrane penetration of hydrophobic residues in the MIL (a variable α1–α2 loop). X-ray reflectivity experiments show that the p40phox PX domain penetrates 9 Å into the lipid layer, with the side chains of a tyrosine and valine inserted most deeply64. Corresponding hydrophobic residues in the p47phox and Vam7p PX domains also penetrate membrane mimetics61,65,66. Alignment of the PX domain sequences reveals some conservation of the hydrophobic residues despite the fact that overall the MIL is highly variable. Bem1, CISK, CPK, FISH, Grd19p, p40phox, p47phox and SNX3 contain VPYV, IFG, MVLG, VYVGV, ILF, ILL, WFDG and LPF sequences, respectively, in place of the hydrophobic residues in p40phox, p47phox and Vam7p, and the MIL occupies analogous conformations in the PX domain structures67–73.
Basic residues located in and around the PI binding pocket and the MIL of the p40phox, p47phox and Vam7p PX domains are involved in nonspecific electrostatic contacts with the negatively charged lipids65,66,68. A separate, well-defined binding site for PS or PA, aside from the PtdIns(3,4)P2 binding pocket, is identified in the p47phox PX domain65,68. Electrostatic interactions of the PX domains were shown to alleviate recognition of the PI lipid, enhance affinity and induce hydrophobic insertion64,65,74,75.
Structural basis of PtdIns(3)P recognition by PX
The atomic resolution crystal and solution structures of three PX domains bound to PtdIns(3)P (Grd19p (1OCU)69, p40phox (1H6H)67 and SNX9 (2RAK)76) and 15 PX domains in the ligand-free form (Bem1p (2V6V and 2CZO)45, CISK (1XTE and 1XTN)72, Grd19p (1OCS)69, KIF16B (2V14)77, p40phox (2DYB)78, p47phox (1O7K, 1KQ6 and 1GD5)68,71, PI3K-C2α (2IWL, 2AR5, 2REA and 2RED)75,79, PI3K-C2β (2WWE), SNX1 (2I4K)80, SNX7 (3IQ2), SNX9 (2RAI and 2RAJ)76, SNX12 (2CSK), SNX17 (3FOG and 3LUI), SNX22 (2ETT)81 and Vam7p (1KMD)70) have been determined. The structures show a similar fold that consists of a three-stranded β-sheet, packed against a helical subdomain composed of 3–4 α-helices (Fig. 3c). An additional 310 helix is present in the structures of the Bem1p, CISK, p40phox and PI3K-C2αPX domains, and another helix α0 is formed by the residues N-terminal to the β-sheet in p40phox 45,67,72,75. The α1- and α2-helices are connected by a long variable loop (MIL), which in Bem1p, CISK, p40phox, p47phox and PI3K-C2α contains a type II polyproline helix45,67,68,72,75. The β1 strand has a β-bulge that twists the β-sheet, forming one wall of the lipid binding pocket.
In the p40phox PX complex, PtdIns(3)P is bound in a relatively narrow and deep (7Å) groove formed by three elements, the loop connecting β3 and α1, a part of MIL closest to α2, and the N-terminal halves of β2 and α267 (Figs. 3c and 4c). The 3-phosphate group of the lipid is restrained through the formation of hydrogen bonds with the guanidino moiety of Arg58 in the β3-α1 loop and with backbone amides of Tyr59 and Arg60. The side chains of Lys92 and Arg60 are involved in the hydrogen bonding contacts with the 1-phosphate, whereas the 4- and 5-hydroxyl groups of PtdIns(3)P are hydrogen bonded to Arg105. The three motifs essential for PI binding, RRYx2Fx2Lx3L of β3-α1, Px2PxK of the MIL, and RR/Kx2L of α2, are present in the majority of PX domain sequences.
Other PI effectors
The number of PI effectors is increasing rapidly and, in addition to the FYVE, PH and PX domains described above, includes the ANTH, C2, ENTH, FERM, GOLPH3, PDZ, PROPPINs, PTB and Tubby modules. The ANTH domain and its structural relative ENTH bind strongly and specifically to PtdIns(4,5)P2 in the plasma membrane82. Whereas the majority of C2 domains associate with the most common anionic and zwitterionic lipids such as PS and phosphocholine (PC), some show preference for PtdIns(3,4,5)P3 and PtdIns(4,5)P2 (ref. 83). Another specific effector of PtdIns(4,5)P2 is the FERM domain84. GOLPH3 targets PtdIns(4)P in the Golgi apparatus85,86. PDZ recognizes PtdIns(4,5)P2 in the plasma membrane87. Human and yeast PROPPINs bind PtdIns(3,5)P2 in membranes of endosomes, lysosomes and vacuoles88, and the fly PROPPIN Dm3 associates with both PtdIns(3,5)P2 and PtdIns(3)P (ref. 1). Interactions with PtdIns(4,5)P2 and PtdIns(4)P have been reported for the PTB domain that normally binds phosphotyrosine peptides31. The Tubby domain localizes to the plasma membrane through binding to PtdIns(4,5)P2 and to a lesser degree to PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (refs. 89,90). Several distinct proteins—for example, AKAP79, CAP23, GAP43, MARKS, profilin and WASP—recognize PIs via clusters of basic residues. However, the structural details of these interactions have not been characterized91–95.
Although PI effectors have diverse and unrelated structures, their membrane docking and PI-binding mechanisms share many similarities. First, all effectors possess a highly basic binding site composed of at least three positively charged residues (three basic residues and a histidine in CALM ANTH96) and as many as six positively charged residues (six basic residues and a histidine in Epsin1 ENTH97 and Grp1 PH42,43) (Fig. 4). Second, membrane binding involves some or all components of multiple anchoring. Association with PI-containing membranes can be augmented by nonspecific electrostatic interactions, hydrophobic insertion, protonation of a histidine switch and increased avidity, with each component substantially contributing to binding energetics. Cooperation of multiple interactions is particularly essential for the recruitment of less selective PI-binding modules, including many PH domains. The affinity and specificity can be further increased because of cooperative binding of some of the PI-recognizing domains or the adjacent regions to other membrane elements, including certain lipid headgroups and membrane-attached proteins, such as small GTPases (reviewed in refs. 1,3).
Concluding remarks
Phosphoinositide-binding domains have recently emerged as a family of ‘PI code’ readers, and a considerable effort has been put forth by many groups to determine their role in mediating acute and constitutive membrane signaling. This review focuses on the mechanistic aspects of single PI effectors. However, these domains are often found next to other PI-binding modules and PI-modifying catalytic domains. Of those discussed here, pleckstrin and centaurin γ3 contain two and five PH domains, respectively; FGD1 contains two PH domains separated by a FYVE finger; PLCγ1 has two PH domains, a catalytic phospholipase module and a C2 domain; PLD1C contains PX, PH and phospholipase domains; DFCP1 contains tandem FYVE fingers; and PDZ and PX domains are present in SNX27. Depending on the specificities, these modules may act either in concert or compete for targeting their host proteins to particular subcellular membranes and regions. The location and duration of membrane association by proteins containing multiple distinct PI effectors can be mediated by activities of organelle-specific PI kinases, phosphatases and lipases. The cross-talk between PIs can provide a mechanism for the regulation of temporal and spatial membrane localization of these proteins and may be essential for controlling signaling cascades. The link between dysregulation of the PI signaling network and numerous diseases suggests a strong therapeutic potential98–100, and further mechanistic studies will be essential to fully understand and exploit this potential.
Acknowledgments
The research in T.G.K.’s laboratory is supported by the US National Institutes of Health grants GM 071424 and CA 113472.
Footnotes
Competing financial interests
The author declares no competing financial interests.
References
- 1.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH. Membrane binding domains. Biochim Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 4.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 6.Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 7.Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta. 2006;1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 9.Gaullier JM, et al. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 10.Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla AA. functional PtdIns(3)P-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 11.Ridley SH, et al. FENS-1 and DFCP1 are FYVE domain–containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 12.Simonsen A, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 13.Gillooly DJ, Simonsen A, Stenmark H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001;355:249–258. doi: 10.1042/0264-6021:3550249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahelin RV, et al. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 15.Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J. Mol. Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 16.Kutateladze TG, et al. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 17.Lee SA, et al. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 2005;102:13052–13057. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J, et al. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 2009;76:852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertens HD, Callaghan JM, Swarbrick JD, McConville MJ, Gooley PR. A high-resolution solution structure of a trypanosomatid FYVE domain. Protein Sci. 2007;16:2552–2559. doi: 10.1110/ps.073009807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 21.Kutateladze T, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 22.Blatner NR, et al. The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]
- 23.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 24.Brunecky R, et al. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 2005;44:16064–16071. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H. The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem. J. 1999;338:539–543. [PMC free article] [PubMed] [Google Scholar]
- 26.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 27.Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 28.Hayakawa A, et al. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 2004;279:5958–5966. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y, et al. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 30.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 31.DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta. 2006;1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 33.Mayer BJ, Ren R, Clark KL, Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993;73:629–630. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda M, Kojima T, Kabayama H, Mikoshiba K. Mutation of the pleckstrin homology domain of Bruton’s tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J. Biol. Chem. 1996;271:30303–30306. doi: 10.1074/jbc.271.48.30303. [DOI] [PubMed] [Google Scholar]
- 35.Klarlund JK, et al. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 36.Cronin TC, DiNitto JP, Czech MP, Lambright DG. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 2004;23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 38.Singh SM, Murray D. Molecular modeling of the membrane targeting of phospholipase C pleckstrin homology domains. Protein Sci. 2003;12:1934–1953. doi: 10.1110/ps.0358803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 40.He J, et al. Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson KM, et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 43.Lietzke SE, et al. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 44.Flesch FM, Yu JW, Lemmon MA, Burger KN. Membrane activity of the phospholipase C-delta1 pleckstrin homology (PH) domain. Biochem. J. 2005;389:435–441. doi: 10.1042/BJ20041721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J. Biol. Chem. 2007;282:25737–25747. doi: 10.1074/jbc.M702861200. [DOI] [PubMed] [Google Scholar]
- 46.Komander D, et al. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baraldi E, et al. Structure of the PH domain from Bruton s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure. 1999;7:449–460. doi: 10.1016/s0969-2126(99)80057-4. [DOI] [PubMed] [Google Scholar]
- 48.DiNitto JP, et al. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 50.Milburn CC, et al. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem. J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 52.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 53.Jackson SG, Zhang Y, Haslam RJ, Junop MS. Structural analysis of the carboxy terminal PH domain of pleckstrin bound to d-myo-inositol 1,2,3,5,6-pentakisphosphate. BMC Struct. Biol. 2007;7:80. doi: 10.1186/1472-6807-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ceccarelli DF, et al. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- 55.Hyvönen M, et al. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 1995;14:4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Ponting CP, et al. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol-3-phosphate. J. Biol. Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 59.Kanai F, et al. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 61.Cheever ML, et al. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 62.Ellson CD, et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat. Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 63.Song X, et al. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry. 2001;40:8940–8944. doi: 10.1021/bi0155100. [DOI] [PubMed] [Google Scholar]
- 64.Málková S, Stahelin RV, Pingali SV, Cho W, Schlossman ML. Orientation and penetration depth of monolayer-bound p40phox-PX. Biochemistry. 2006;45:13566–13575. doi: 10.1021/bi061133l. [DOI] [PubMed] [Google Scholar]
- 65.Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J. Biol. Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 66.Lee SA, et al. Molecular mechanism of membrane docking by the Vam7p PX domain. J. Biol. Chem. 2006;281:37091–37101. doi: 10.1074/jbc.M608610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bravo J, et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 68.Karathanassis D, et al. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou CZ, et al. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 2003;278:50371–50376. doi: 10.1074/jbc.M304392200. [DOI] [PubMed] [Google Scholar]
- 70.Lu J, Garcia J, Dulubova I, Sudhof TC, Rizo J. Solution structure of the Vam7p PX domain. Biochemistry. 2002;41:5956–5962. doi: 10.1021/bi020050b. [DOI] [PubMed] [Google Scholar]
- 71.Hiroaki H, Ago T, Ito T, Sumimoto H, Kohda D. Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 2001;8:526–530. doi: 10.1038/88591. [DOI] [PubMed] [Google Scholar]
- 72.Xing Y, et al. Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX) J. Biol. Chem. 2004;279:30662–30669. doi: 10.1074/jbc.M404107200. [DOI] [PubMed] [Google Scholar]
- 73.Kutateladze TG. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol 3-phosphate containing membranes. Prog. Lipid Res. 2007;46:315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stahelin RV, et al. Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 2004;279:54918–54926. doi: 10.1074/jbc.M407798200. [DOI] [PubMed] [Google Scholar]
- 75.Stahelin RV, et al. Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J. Biol. Chem. 2006;281:39396–39406. doi: 10.1074/jbc.M607079200. [DOI] [PubMed] [Google Scholar]
- 76.Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blatner NR, et al. The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 2007;26:3709–3719. doi: 10.1038/sj.emboj.7601800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Honbou K, et al. Full-length p40phox structure suggests a basis for regulation mechanism of its membrane binding. EMBO J. 2007;26:1176–1186. doi: 10.1038/sj.emboj.7601561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parkinson GN, Vines D, Driscoll PC, Djordjevic S. Crystal structures of PI3K-C2alpha PX domain indicate conformational change associated with ligand binding. BMC Struct. Biol. 2008;8:13. doi: 10.1186/1472-6807-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong Q, et al. Determinants of the endosomal localization of sorting nexin 1. Mol. Biol. Cell. 2005;16:2049–2057. doi: 10.1091/mbc.E04-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song J, Zhao KQ, Newman CL, Vinarov DA, Markley JL. Solution structure of human sorting nexin 22. Protein Sci. 2007;16:807–814. doi: 10.1110/ps.072752407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 83.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 84.Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19:4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wood CS, et al. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zimmermann P. The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim. Biophys. Acta. 2006;1761:947–956. doi: 10.1016/j.bbalip.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 88.Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem. J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 89.Santagata S, et al. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 90.Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 2009;10:67. doi: 10.1186/1471-2121-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Arbuzova A, Hangyas-Mihalyne G, McLaughlin S. The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2001;276:5012–5019. doi: 10.1074/jbc.M008355200. [DOI] [PubMed] [Google Scholar]
- 92.Caroni P. New EMBO members’ review: actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 2001;20:4332–4336. doi: 10.1093/emboj/20.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. EMBO J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- 95.Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ford MG, et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 97.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 98.McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prestwich GD. Phosphoinositide signaling; from affinity probes to pharmaceutical targets. Chem. Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 100.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]