Abstract
The CS-RDC-NOE Rosetta program was used to generate the solution structure of a 27 kDa fragment of the E. coli BamC protein from a limited set of NMR data. The BamC protein is a component of the essential five-protein β-barrel assembly machinery in E. coli. The first 100 residues in BamC were disordered in solution. The Rosetta calculations showed that BamC101-344 forms two well-defined domains connected by an ~18 residue linker, where the relative orientation of the domains was not defined. Both domains adopt a helix-grip fold, previously observed in the Bet v I superfamily. 15N relaxation data indicated a high degree of conformational flexibility for the linker connecting the N- and C-terminal domains in BamC. The results here show that CS-RDC-NOE Rosetta is robust and has a high tolerance for misassigned NOE restraints, which greatly simplifies NMR structure determinations.
Keywords: β-barrel assembly machinery, helix-grip motif, NMR structure, Rosetta, outer membrane protein
Introduction
The traditional NMR solution structure determination of a protein involves three steps: assignment of 1H, 13C and 15N resonances, measurement of structural information such as 1H-1H nuclear Overhauser effect (NOE) distance data and residual dipolar coupling (RDC) orientational data, followed by computational methods to generate an ensemble of conformations consistent with the NMR-derived structural restraints.1 This approach is routinely applied to small proteins but as the size of the protein increases (> 15 kDa) additional methods such as deuteration, relaxation-enhanced experiments (such as TROSY, CRIPT, CRINEPT) or specific labeling (such as 13C-labeling of ILV methyl groups) are employed to improve spectral resolution and sensitivity.2, 3 A major challenge in the structure determination of larger proteins is the time-consuming step of resonance assignment of NOEs involving side chain protons. Thus, various procedures are being developed to assign NOEs automatically as part of the structure refinement process.4, 5, 6, 7, 8
An alternate strategy for determining solution structures of proteins is to include a sparse set of experimental NMR data in the Rosetta protein structure-prediction program.9, 10 Rosetta was originally developed as a de novo method for generating protein structures from the sequence.11 It has since been extended to a suite of programs that perform homology modeling, protein-protein docking, protein-ligand docking, protein design or structure determinations using limited experimental data.12, 13 For de novo structure determinations, Rosetta starts by assembling fragments derived from a database of known protein structures using a low-resolution energy function.11 The low energy conformations are then subjected to further sampling using an all-atom representation. Experimental data can dramatically increase the efficiency with which near-native conformations are sampled. For example, CS-Rosetta incorporates backbone NMR chemical shift data to help guide fragment selection and conformational searching, which enables atomic-resolution structure determination for small proteins.9, 14 However, for proteins larger than ~12 kDa, chemical shifts may not provide enough information to guide the conformational search.10 Thus, additional experimental data such as RDCs and NOEs have been incorporated into iterative CS-RDC-NOE Rosetta and shown to increase its ability to generate accurate protein structures.10 For proteins above ~20 kDa a significant fraction of residues did not converge to well-defined conformations using only a sparse set of amide 1H-1H NOEs (from 21 to 52) in CS-RDC-NOE Rosetta.10 Thus, improvements in conformational sampling or additional experimental data are required to generate models for even larger proteins using Rosetta.
This iterative CS-RDC-NOE Rosetta program was used here to generate structures of the 27 kDa E. coli periplasmic protein BamC by including chemical shift data and a limited set of NOE restraints (149 NOEs). The lipoprotein BamC is a component of the essential five-protein β-barrel assembly machine, BAM.15 This complex is involved in the folding and insertion of β-barrel proteins into the outer membranes of gram-negative bacteria. Although bamC null E. coli strains are viable, the mutants display outer membrane permeability defects and reduced levels of β-barrels in the outer membrane.16, 17 The CS-RDC-NOE Rosetta calculations show that BamC consists of two helix-grip type domains connected by an ~18 amino acid linker. Additional backbone-backbone and methyl-methyl NOE data, not used in the structure calculations, validated the BamC structures generated by Rosetta. A set of 15N NMR relaxation data was collected for BamC and demonstrated a high degree of conformational dynamics for the backbone in the linker region. Analysis of the 15N relaxation data and 1H-15N RDCs indicate that the two domains in BamC do not have a fixed orientation in solution. The studies here also showed that CS-RDC-NOERosetta is quite robust to including inconsistent NOE distance restraints in the experimental data.
Results and Discussion
The first 100 N-terminal residues are not structured in BamC
The first 24 residues of E. coli BamC contain a periplasmic localization signal, which is cleaved in vivo, and were not included in the protein constructs studied here. In addition, the N-terminal Cys that is normally modified with a lipid anchor was mutated to Ala to prevent intermolecular disulfide crosslinking. The previously reported chemical shifts for BamC26-344 indicated that the next N-terminal ~75 residues had no regular secondary structure.18 To confirm this hypothesis, the 35 kDa BamC26-344 was subjected to limited aminopeptidase digestion, resulting in a stable fragment ~27 kDa (data not shown). To test whether the first 75 amino acids alter the structure of the rest of the protein, hydrogen-deuterium exchange NMR experiments were carried out on BamC26-344 and BamC101-344. The amide protons of the N-terminal 75 amino acids in the longer construct exchanged by the end of the first 2D HSQC experiment (~11 min), consistent with a disordered N-terminus (Fig. S1). The exchange profiles of the remaining amide protons were essentially unchanged for the two constructs, indicating that the rest of the protein is unaffected by the presence of the 75 amino acid N-terminal tail. Thus, a shorter construct, BamC101-344, was used for the structure determination and most of the NMR studies here (Fig. S2). The backbone and Cβ chemical shifts of BamC101-344 were then assigned using conventional heteronuclear triple resonance NMR experiments, followed by partial assignment of side chain resonances, as described in Methods.1 Except for residues near the N-terminus, no significant differences were observed when comparing the backbone chemical shifts in the previously assigned BamC26-344.18
The structure of BamC generated by CS-RDC-NOE Rosetta reveals a two-domain protein connected by a flexible linker
CS-RDC-NOE Rosetta calculations10 on BamC101-344 were carried out as described in Methods. The input consisted of the backbone (HN, N, C’, Cα, and Cβ) chemical shifts for all residues (excluding the terminal and Pro amides), 156 1H-15N RDCs and two different sets of NOE restraints. A preliminary calculation was performed with 62 1H-1H amide-amide NOE distance restraints, but the low-scoring models consistently violated 10 NOEs. Analysis of the NOE spectra showed that these 10 NOEs were misassigned or had ambiguous assignments (discussed below). Thus, a calculation (BamC_I) was carried out with the remaining 52 amide-amide NOE restraints. This greatly improved convergence in both domains and reduced the overall Rosetta energy of the 10 lowest scoring models (the scores were computed without NOE and RDC restraints) by 60 score units to a range of -556 to -565. An additional set of 97 1H-1H NOE restraints (for a total of 149 NOEs) involving backbone and sidechain protons, which were consistent with the BamC_I structures, were included in the final calculations (BamC_II) where the 10 lowest energy structures had Rosetta energies ranging from -590 to -596.
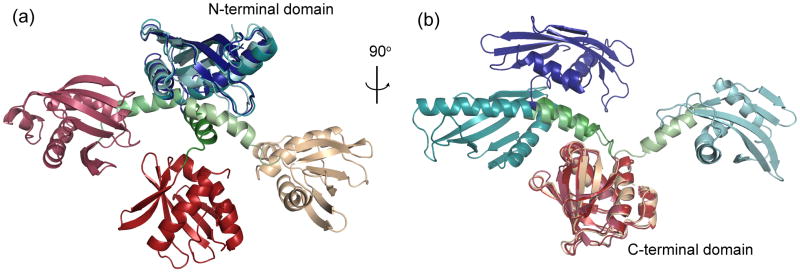
The final calculations indicated that BamC has two well-defined domains, an N-terminal domain from residues 101 to 210, a C-terminal domain from residues 229 to 346 and a linker from residues 211 to 228. However, the relative orientation of the two domains was not defined by Rosetta (Fig. 1). One possibility for this conformational heterogeneity is that the N- and C-terminal domains do not interact and the residues between these regions are flexible. A second possibility is that BamC101-344 does have a well-defined orientation of the two domains in solution, but the calculations did not find the global minimum due to incomplete sampling of conformational space. To try to observe long-range NOEs between the N- and C-terminal domains, a 3D (13C, 13C, 1H) HMQC-NOESY-HMQC spectrum was acquired on an Ile, Leu and Val (ILV) methyl-protonated 2H, 15N, 13C-labeled sample of BamC26-344. Previous studies have shown that methyl-methyl distances over 8 Å can be observed in NOESY spectra.19 A total of 100 new ILV methyl-methyl NOEs were observed, but none were between residues in the N- and C-terminal domains. The absence of interdomain NOEs alone does not prove the N- and C-terminal domains are not interacting, but it is a strong indication that there is no stable interaction between the two domains in isolated BamC.
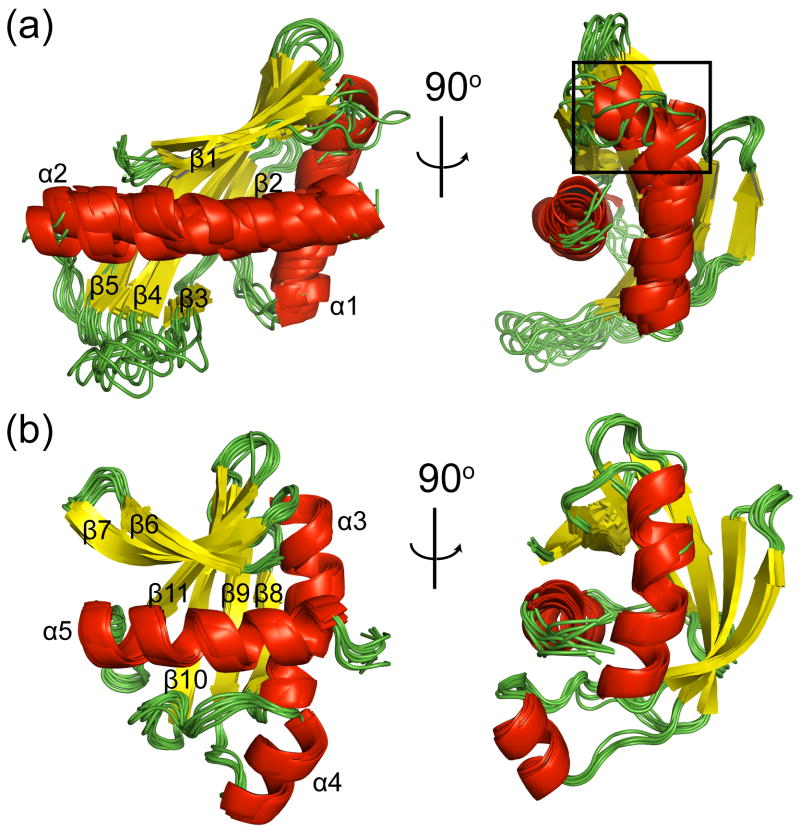
Figure 1.
Three low energy structures for BamC101-344 shown as superimpositions of (a) their N- terminal domain (cool colors) and (b) C-terminal domain (warm colors), illustrating that the folds of the N- and C-terminal domains converged in the Rosetta calculations, but the relative orientation of the domains did not converge. These three structures represent the range of orientations for the two domains in the set of low energy Rosetta structures.
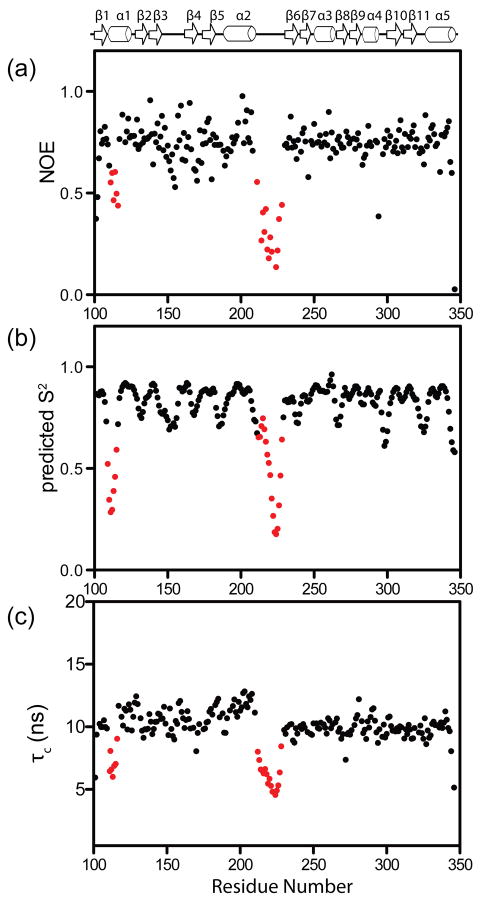
Since the size of a 1H-1H NOE is a function of both the distance and dynamics between the nuclei, 15N relaxation NMR experiments were performed to directly probe backbone dynamics in BamC. Heteronuclear 15N{1H} NOEs were measured for the backbone amides in BamC101-344 and as seen in Fig. 2a, residues in the N- and C-terminal domains generally had NOE values above 0.7, whereas residues 214 to 227 in the linker had significantly lower 15N{1H} NOE values (0.13 to 0.42), indicating a flexible linker. A similar conclusion is obtained by predicting the order parameter of backbone amide group, S2, from the chemical shifts.20 As seen in Figure 2b, residues 217 to 228 in BamC show lower predicted S2 values, supporting flexibility for the linker region.
Figure 2.
The 15N relaxation data on BamC101-344 show regions of increased backbone flexibility. Plots of (a) 15N{1H} NOE, (b) the predicted backbone order parameter, S2, (c) the calculated residue-specific rotational correlation time, τc, as a function of residue number. The S2 is predicted from the experimental chemical shifts using a random coil index.20 The red circles highlight two regions of the molecule that exhibit increased dynamics (see text). The regions of regular secondary structure are illustrated at the top of the figure.
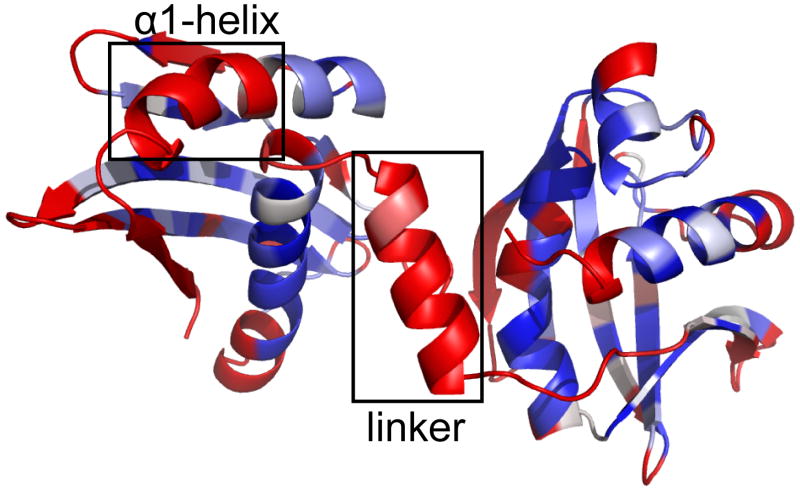
H-D exchange experiments provided additional evidence for N- and C-terminal domains being connected by a conformationally dynamic linker in BamC101-344, where slowly exchanging amide protons were observed for many residues in the N- and C-terminal domains but rapid exchange was observed for all the amide protons in the linker (Fig. 3). Moreover, it was recently shown that BamC101-344 is susceptible to cleavage by subtilisin in the linker region, yielding two stable fragments of 12.2 and 14.5 kDa.21 All these results support a model where BamC101-344 contains well-ordered N- and C-terminal domains connected by a flexible linker. The CS-RDC-NOE Rosetta calculations on BamC101-344 consistently generated an α-helical conformation for this linker region (Figs. 1 and 3). However, the NMR data demonstrate that the linker is flexible. This indicates that Rosetta has a tendency to ‘over fold’ dynamic regions, which likely results from backbone fragments being selected from a database of known well-ordered protein structures. Methods are currently being developed in Rosetta to reduce over folding of regions that are experimentally identified as dynamic (D. Baker, unpublished results).
Figure 3.
Hydrogen-deuterium exchange profiles are mapped onto a low energy structure of BamC101-344 with residues colored in red, white and blue indicating rapid (< 12 min), medium (12 min to 24 hr) and slow exchange (> 24 hr) at 30 °C, a nd gray for prolines or residues where exchange could not be determined due to spectral overlap. Black boxes highlight the rapid exchange for part of the α1 helix and the linker connecting the domains. Note that Rosetta modeled most of the linker region as a helix even though the 15N relaxation and deuterium exchange data indicate that this region is conformationally dynamic (see text).
High flexibility for the linker does not rule out a stable interaction between the N- and C-terminal domains in full-length BamC. However, if the two domains are not tumbling as a rigid single species in solution then the individual domains should have smaller rotational correlation times than predicted for the full protein. Residue-specific rotational correlation times, τc, were calculated from 15N R1 and R2 measurements (data not shown) on BamC (Fig. 2c).1, 22 The residues in the N- and C-terminal domains have average τc values of 10.8 and 9.8 ns, respectively, whereas the linker has lower τc values (ranging from 4.6 to 7.4 ns for residues 214 to 227). The values for the N- and C-terminal domains are ~35% smaller than what would be predicted for a 27 kDa spherical protein, but larger than predicted if the N- and C-terminal domains were tumbling independently as ~13 kDa spheres (~7.7 ns). This pattern of 15N{1H} NOE and τc values is very similar to what was previously observed in Ca2+-loaded calmodulin, which has a flexible linker connecting its N- and C-terminal domains.23
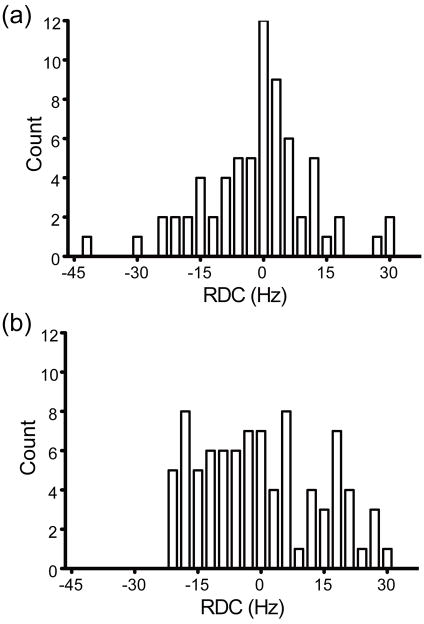
RDC data are routinely used to determine the relative orientation of well-defined domains in proteins or nucleic acids.24, 25 Thus, we wanted to address whether the 1H-15N RDCs could be used to define the orientation of the N- and C-terminal domains in BamC. The first step in domain orientation using RDC data is to assess whether the two domains have a fixed orientation and therefore the same alignment tensor. One indication of a rigid orientation for the two domains is if the individual domains have similar values for the magnitude, Da, and rhombicity, R, of their alignment tensors.24 A structure-independent method was used here for comparing Da and R in the two domains, which involves analysis of the shape of histograms of the RDCs in a molecule.26 If the N- and C-terminal domains in BamC had a fixed orientation, the histograms of the individual domains would have similar shapes, indicating similar values for Da and R (assuming that the set of RDCs adequately sample all orientations of bond vectors). The histograms of the 86 and 68 1H-15N RDCs for the N- and C-terminal domains have quite different shapes (see Fig. 4). These data are consistent with the conclusion obtained from the τc data, that N- and C-terminal domains in BamC do not have a fixed orientation in solution and are not tumbling as a rigid species.
Figure 4.
Histograms of the 86 and 68 1H-15N RDCs for the (a) N- and (b) C-terminal domains in BamC101-344. The shape of the histogram provides information on the magnitude, Da, and rhombicity, R, of the alignment tensor.24 The differences in the shapes of the two histograms indicate that the N- and C-terminal domains have different alignment tensors and therefore do not have a fixed orientation in solution (assuming that these RDCs adequately sample all orientations of 1H-15N bond vectors).
The CS-RDC-NOE Rosetta calculations on the full-length BamC assumed a single alignment tensor for the whole molecule, but this is not correct if domains do not have a fixed orientation. Thus, separate CS-RDC-NOE Rosetta calculations were performed for the N- (101 - 212) and C-terminal (229 - 344) domains (Fig. 5 and Table 1). There was no significant difference in the overall folds of the individual domains in the Rosetta calculations performed on the full protein or the separate domains. Thus, all further analyses were performed on the structures generated by the calculations for the individual domains.
Figure 5.
Ensemble of structures of the N- and C-terminal domains of BamC generated by separate CS-RDC-NOE Rosetta calculations for the two domains. Superimposition of 9 low energy structures of the (a) N- and (b) C-terminal domains of BamC. Part of the α1 helix in the N-terminal domain, which shows conformational flexibility in the 15N relaxation data (see text), is highlighted by a black box.
TABLE 1.
Structural statistics of the BamC N- and C-terminal domainsa
| N-terminal domain | C-terminal domain | |
|---|---|---|
| Number of residues | 112 | 118 |
|
| ||
| NOE-based distance restraints | ||
| NOE distance restraints (viol. ≥ 0.5 Å)b | 69 (16 ± 1) | 78 (30 ± 2) |
| Number of restraints per residue | 0.62 | 0.66 |
|
| ||
| Other restraints | ||
| ϕ + ψ dihedral angle restraints (viol. ≥ 5 °)c | 193 (11 ± 3) | 202 (11 ± 4) |
| Residual dipolar coupling restraints (viol. ≥ 5 Hz) | 60 (24 ± 3) | 82 (20 ± 4) |
|
| ||
| Average RMSD to the average structure | ||
| Backbone (Å) | 1.39 ± 0.33 | 0.54 ± 0.06 |
| Heavy atom (Å) | 1.88 ± 0.30 | 0.95 ± 0.11 |
|
| ||
| Ramachandran plotd | ||
| Most favored regions | 92.2% | 94.3% |
| Allowed regions | 7.8% | 4.8% |
| Generously allowed regions | 0.0% | 0.0% |
| Disallowed regions | 0.0% | 0.9% |
Statistics are given for the 9 lowest energy structures after all-atom refinement with CS-RDC-NOE Rosetta.
All NOEs are long range (|i-j| ≥ 5). Two of the 149 NOEs were between the amino acids in the linker region and N-terminal domain and were not included in the calculations of the individual domains.
Torsion angle restraints were derived from TALOS+.46
PROCHECK was used to calculate these data.48
The N-terminal domain of BamC is composed of two α helices packed against a five-stranded anti-parallel β sheet (Fig. 5a), reminiscent of the helix-grip fold.27 The C-terminal domain closely resembles the N-terminal domain with two additional structural elements (Fig. 5b): i) a short β-strand before β1 and ii) a seven-residue helix inserted between strands β3 and β4 (where the corresponding residues in the N-terminal domain are an extended loop). The N-terminal domain superimposes on the C-terminal domain with a pairwise RMSD of 1.7 Å for the Cα backbone for 60 residues in structurally similar regions identified using LSQMAN.28
Figure 2 indicates there is another region of flexibility in the N-terminal domain, where residues 112 – 117 show lower 15N{1H} NOE, predicted S2 and τc values than residues in the domains. The amide protons for these residues exchanged rapidly in the H-D exchange experiments (Fig. 3). This region is not uniquely defined in the Rosetta calculations where some structures show a kink in the helix near Pro 117 and others exhibit fraying of the N-terminal residues in this helix (boxed region in Fig. 5a). Thus, the 15N relaxation and H-D exchange data support a model where the N-terminal part of the α1 helix is conformationally dynamic, consistent with the conformational variation of this region in the Rosetta structures. These results demonstrate how NMR relaxation data can be used to determine whether non-converged regions observed in the Rosetta calculations reflect true conformational heterogeneity of the protein.
Validation of the BamC structure generated by Rosetta
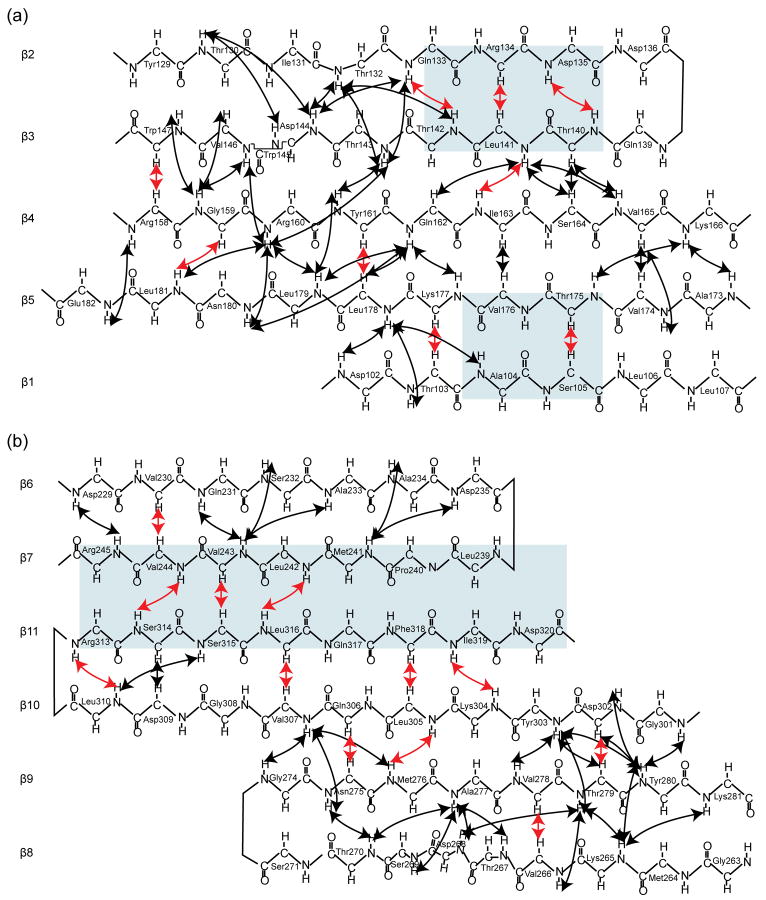
Previous studies on a number of known structures showed that iterative CS-RDC-NOE Rosetta yields accurate conformations if the calculations converge.10 The N- and C-terminal regions in BamC converged well (Fig. 5), thus based on previous experience it is expected that Rosetta generated accurate structures for these domains. Nevertheless, it is advisable to validate the Rosetta structures with independent experimental data. Thus, we analyzed how additional backbone-backbone and methyl-methyl 1H-1H NOEs, not included in the calculations, fit to the Rosetta models for BamC101-344. The input NOE data set used for the Rosetta calculations comprised 149 readily assigned long-range 1H-1H NOEs (between residues i and j where | i − j | > 5). These NOEs were not evenly distributed over the structure, where regions have clusters of NOEs and others have no NOEs. The gray boxes in Figure 6 highlight regions of antiparallel β sheet-secondary structure where no cross-strand NOEs were included in the Rosetta calculations. For example, β7 and β11 had no cross-strand NOE restraints, which could lead to ambiguity in the register or orientation of the two strands. This lack of NOE restraints provided a valuable opportunity to directly validate the Rosetta models. Further analysis of the original NOESY spectra yielded three cross-strand backbone-backbone NOEs in this region (HN V244 and HN S314, Hα V243 and Hα Ser315, and HN L316 and HN L242), which unambiguously confirmed the strand orientation and register between β7 and β11 generated by Rosetta (Fig. 6). The cross-strand Hα- Hα NOE is especially diagnostic of an anti-parallel β sheet because the Hα-Hα distance is ~2.2 − 2.5 Å. Strong Hα- Hα cross peaks, as well as other cross strand 1H-1H NOEs, were also observed for the other two boxed regions in Figure 6, validating the β-sheet secondary structure generated by Rosetta.
Figure 6.
Backbone-backbone 1H-1H NOEs confirm the β-sheet secondary structure in BamC generated by CS-RDC-NOE Rosetta. Schematics for the β-sheet secondary structures in the (a) N- and (b) C-terminal domains showing NOEs included (black) or not included (red) in the CS-RDC-NOE Rosetta calculations. The NOEs in red were used to help validate the Rosetta structures.
One of Rosetta’s strengths is that in cases of high backbone convergence (< 2 Å) it can generate accurate side chain packing.10 The ILV methyl-protonated NOESY spectra on BamC were initially collected to search for long-range interdomain methyl-methyl NOEs. However, these spectra also provide data on packing of the Ile, Leu and Val side chains. A set of 103 NOEs were observed between the ILV methyl groups in BamC, but only 3 of these were included as restraints in the Rosetta calculations. In all cases, the range of distances predicted from these methyl-methyl NOEs were consistent with the hydrophobic packing observed in the BamC structures generated by Rosetta.
CS-RDC-NOE Rosetta is very robust to errors in an NOE restraint list
A valuable feature of using CS-RDC-NOE Rosetta to generate protein structures from NMR data is its ability to deal with incorrectly assigned NOEs, which can lead to distance restraints that are inconsistent with the correct structure. This can be a major problem when generating structures from NMR data, where inconsistent distance restraints lead to high energies and can drive the conformation away from the correctly folded structure during refinement. Rosetta is not as susceptible to such problems because, as previously discussed, there is generally a low probability that inaccurate experimental restraints will also yield low energy structures.10 Furthermore, as noted in Methods, the weightings for the NOE and RDC restraints are reduced from 5.0 to 0.1 in the all-atom refinement and for large distance violations Rosetta uses a linear potential as opposed to the quadratic potential commonly used for NOE restraints. The results here demonstrate that CS-RDC-NOE Rosetta is very robust in dealing with inconsistent NOEs. As seen in Figure 7, a preliminary calculation that included 10 misassigned NOEs generated similar structures for the N- and C-terminal domains as the final structures, even though 8 of the NOEs violated their restraint by > 12 Å. Rosetta is able to handle inaccurate data because the experimental data is primarily used to increase the efficiency of searching for low energy conformations.10 To test the effect of having this level of misassigned NOEs on a standard structure calculation, XPLOR-NIH29 simulations were performed using the N-terminal domain of one of the final Rosetta structures as the target (Supplemental Material). As expected, when no misassignments were introduced in the simulated data, the calculations generated well-defined structures with low NOE energies and small RMSDs to the target structures (average RMSD 0.34 ± 0.14 Å). However when 16% of ~1700 simulated NOEs were misassigned, mimicking the 10 of 62 misassignments in the preliminary CS-RDC-NOE Rosetta calculations, the ensemble of structure had huge NOE energies and did not converge to the target (average RMSD 15.5 ± 2.0 Å)(see Supplemental Material). These results demonstrate that incorrect NOE assignments have much less influence on structures generated by CS-RDC-NOE Rosetta than a standard NMR structure determination. On the other hand, as seen in Table 1, the percentage of NOE and RDC violations are much greater than observed in standard NMR structure determination. This results from reducing the weights of the NOE and RDC restraints during the all-atom refinement (see Methods). Therefore, the CS-RDC-NOE Rosetta ensemble will generally be lower resolution than an NMR ensemble generated with thousands of NOE restraints. However, proteins greater than 15 kDa generally required extensive deuteration, which drastically reduces the number of NOEs. Hence, the approach used by CS-RDC-NOE Rosetta, which combines limited experimental data with efficient computational methods for predicting protein folding, will have a clear advantage when applied to large proteins.
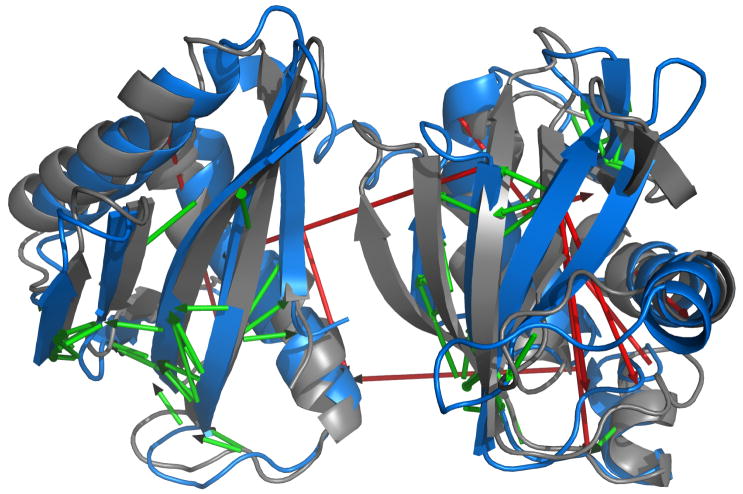
Figure 7.
CS-RDC-NOE Rosetta modeling is tolerant of misassigned NOEs. The best scoring model in a preliminary calculation that included 10 misassigned NOEs is shown in blue. Superimposed in gray are the individual domains of the best scoring model of the final calculation, BamC_II. Amide HN-HN NOEs used as restraints in the preliminary calculation are shown as arrows. Arrows are colored in green if the NOE is within the distance bound of the restraint and in red if it violates the restraint. The red arrows are between residues: 53-15, 92-113, 92-191, 101-174, 142-155,149-194, 183-207, 197-207, 198-207 and 242-237.
Comparison of BamC with other helix-grip motifs
Independent queries of the N- and C-terminal domains to the Dali server30 resulted in similar hits of helix-grip motifs that belong to the Bet v I superfamily of structural homologues. Figure 8a shows the superimposition of the N-terminal domain with the major latex protein, At1g24000.1.27 Proteins with the Bet v 1 motif are known to bind a diverse set of hydrophobic ligands such as membrane lipids, plant hormones and steroids.31 Structurally, these proteins are characterized by a hydrophobic cavity between the β-sheet and a long α-helix that accommodates the ligand. However, this cavity is not present in the Rosetta structures of either domain of BamC. Consistent with this observation, preliminary lipid binding studies performed with 1H, 15N BamC101-344 and palmitic acid (40 μM) or an extract of E. coli polar lipids (120 μM) showed no changes of the amide chemical shifts in BamC (data not shown).
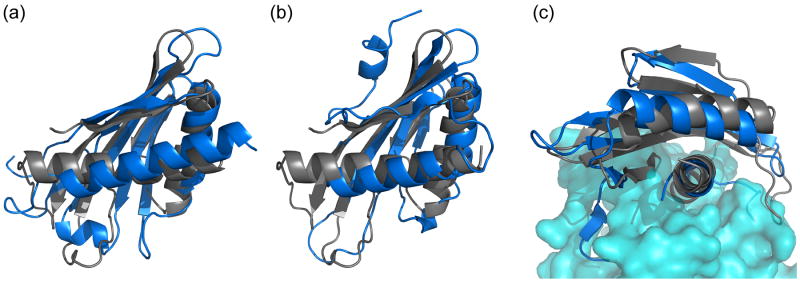
Figure 8.
The N-terminal domain of BamC (gray) is shown superimposed with structurally similar proteins (blue) identified in a DALI search. (a) The major latex protein, At1g24000.1 (PDB ID:1VJH), (b) the yeast septin-associated Kcc4p (PDB ID:3OSM) and c) the helix-grip domain of the α subunit of AMPK (PDB ID:2V8Q) all share similar helix-grip motifs with the BamC domains. The superimposition in (c) is rotated to highlight the protein-protein interaction surface between the α subunit and the βγ subunits of AMPK (cyan, in surface representation). The central helix in the α subunit of AMPK (red) is important for protein-protein interaction. The residues used to align the proteins were defined by LSQMAN.28
The Dali search also identified KA-1 domains as structurally related to both the N- and C-terminal domains of BamC (Fig. 8b). It was recently shown that in some kinases, such as human MARK/Par1 and the yeast septin-associated Kcc4p, the KA-1 domains bind acidic phospholipids and are responsible for targeting the kinase to the plasma membrane.32 Phospholipid binding is not accommodated by a hydrophobic cavity as in Bet v 1 proteins. Instead, binding is mediated by electrostatic interactions between positively charged residues on the protein surface and negatively charged phosphates in the phospholipid head groups, consistent with their membrane-targeting role. The electrostatic surface potentials for both domains of BamC display more negative character than the KA-1 domains in these kinases (not shown). These results, together with the lack of changes in chemical shifts upon addition of E. coli phospholipids, suggest that BamC is not involved in membrane binding.
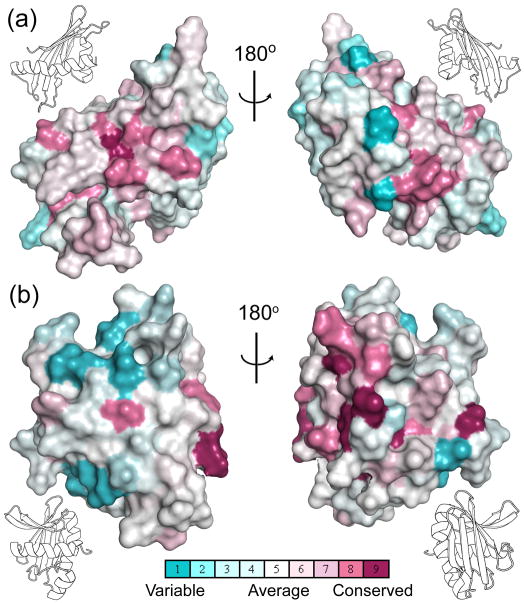
In multisubunit kinases, such as AMPK and Kcc4p, the KA-1 domains mediate intersubunit contacts important for the integrity of the kinase complex.33, 34 The long C-terminal α-helix in these KA-1 domains contains most of the residues that participate in the intersubunit contacts (see Fig. 8c). A superimposition of the BamC N-terminal domain with AMPK is shown in Figure 8c, illustrating how this region in BamC could be involved in binding to other BAM components. Residues on the surface of the corresponding helix (α2) of the N-terminal BamC domain have relatively high phylogenetic conservation (Fig. 9). A second region of surface-exposed highly conserved residues is also observed on the α3-helix in the C-terminal domain of BamC. Previous studies have shown that BamC interacts directly with the C-terminus of BamD and helps stabilize the BAM complex.35, 36 One possibility is that, analogous to the KA-1 domain of AMPK, each domain of BamC mediates intersubunit contacts. The presence of two structurally similar domains in BamC could thus serve as a scaffold to stabilize the structure of the multisubunit BAM complex.
Figure 9.
The conserved amino acids in BamC are clustered in sites potentially important for protein-protein interactions. Conserved sequences are mapped onto surface representations of the (a) N- and (b) C-terminal domains of BamC (the insets show the secondary structure representation of the domain in the same orientation). The color scale is shown where darker blue indicates higher than average level of sequence variability, darker red indicates a higher than average level of sequence conservation and white indicates an average level of sequence changes.
Conclusions
The lipoprotein BamC is one of five proteins in the β-barrel assembly machine in E. coli, but its specific role in outer membrane protein folding and insertion is not known. The CS-RDC-NOE Rosetta calculations using a limited set of NMR data showed that BamC is made up of two well-defined helix-grip domains. This helix-grip motif has been previously observed in the Bet v I superfamily, where this fold serves as either a ligand-binding domain or protein-protein interaction domain. The helix-grip domains in BamC do not have the hydrophobic binding pocket observed in Bet v 1 or the positively charged loop used to interact with phospholipids observed in Kcc4p. Therefore, we speculate that helix-grip motifs in BamC function as protein-protein interaction domains to help stabilize interactions between the subunits of the BAM complex. Specifically the long helix in the helix-grip domains may mediate the interactions with the other components of the BAM complex, similar to the protein-protein interaction domain observed in AMPK.34
The studies here also showed that CS-RDC-NOE Rosetta is tolerant of including some incorrect NOE assignments in the NMR restraints, even ones with large distance violations. For some structure generation programs, this level of inconsistent NOE restraints could trap molecules in high-energy conformations, making it difficult to refine the structures. Rosetta is quite robust to such inconsistencies because it identifies correctly folded structures based on low energies for its scoring function where the experimental data have low weights and serve primarily to guide the conformational search. This greatly simplifies analysis of NOE spectra and makes CS-RDC-NOE Rosetta an attractive alternative to restrained molecular mechanics/dynamics methods for generating solution structures of proteins from NMR data.
Materials and Methods
Cloning of BamC constructs
The gene for E. coli BamC (residues 26-344) was obtained by PCR from genomic E. coli DNA using primers that introduce unique NcoI and XmaI restriction sites. This gene was introduced into the pMS174 vector (an engineered variant of the pET28 vector that generates an N-terminal His-tag fusion that can be specifically cleaved with TEV protease) to yield pMS282. A second construct, shorter by 75 amino acids at the N-terminus, BamC101-344 was PCR amplified from pMS282 and ligated into pMS174 with the NcoI/Xma restriction site to yield pMS639. Plasmids pMS282 (BamC26-344) and pMS639 (BamC101-344) were sequenced to confirm the absence of mutations. To facilitate cloning, three non-native amino acids were included at the N-terminus (AGM) and two non-native amino acids at the C-teriminus (PG) in BamC26-344 and BamC101-344 constructs, but these amino acids are not included in the numbering system used to refer to the constructs.
Protein expression and purification of BamC constructs
Plasmids pMS282 and pMS639 were transformed into E. coli Rosetta (DE3) cells (Novagen) and small-scale growths from single colonies were used to inoculate 100 mL LB supplemented with 50 μg/mL kanamycin. Cultures were grown overnight, spun down and resuspended in 3 L of M9 minimal medium supplemented with 50 μg/mL kanamycin, 1.5 g/L 13C-glucose and 1 g/L 15NH4Cl (Sigma/Isotec). Cultures were grown at 37 °C to OD600 of 0.6 and cooled on ice for 10 min. Expression was induced with 1.0 mM IPTG (Gold Bio Technology Inc.) and grown overnight at 20 °C. Cells were harvested by centrifugation, and the cell pellet was re-suspended in lysis buffer containing 25 mM Tris-Cl, 300 mM NaCl pH 8.0 and Complete EDTA-free protease inhibitor (Roche), then sonicated on ice. Cell debris was removed by centrifugation and the supernatant was applied to a Ni-NTA column (Qiagen) pre-equilibrated with buffer A (25 mM, Tris pH 8, 150 mM NaCl). The Ni-NTA beads were washed with 2 column volumes of buffer A, followed by 5 column volumes of buffer A containing 25 mM imidazole. The protein was eluted with buffer A containing 200 mM imidazole. Fractions containing the protein were incubated with His-tagged TEV protease for 24 hr at 4 °C by dialysis against buffer A supplemented with 10 mM DTT to cleave the His tag and then dialyzed overnight at 4 °C against buffer A. The TEV protease was removed using Ni-NTA beads and the protein was loaded on a size exclusion column (HiLoad 26/60 Superdex 200, Amersham Pharmacia Biotech) pre-equilibrated with buffer A and eluted in the same buffer. Protein was concentrated to 1.6 mM for BamC26-344 and 1.8 mM for BamC101-344 and stored at -70 °C.
For the ILV methyl protonated labeled sample, pMS282 was transformed into E. coli Rosetta (DE3) cells (Novagen) and plated on LB-agarose with 50 μg/mL kanamycin. Single colonies were used to inoculate small cultures grown in rich medium for 3 hr at 37 °C. Cells were first conditioned to minimal medium in solutions containing H2O and finally transferred to 2 L of M9 minimal medium prepared in 99.8% 2H2O with 2 g/L 2H, 13C glucose and 1 g/L 15NH4Cl (Isotec). Cells were grown at 37 °C until an OD600 of 0.6 at which point 100 mg/L of 2-keto-3-(methyl-d3)- butyric acid-1,2,3,4-13C4,3-d1 sodium salt and 50 mg/L of 2-ketobutyic acid-13C4,3,3-d2 sodium salt (Isotec) were added to the media. Cells were cold shocked, followed by induction with 0.4 mM IPTG and grown overnight at 20 °C. Purification was as described above.
NMR spectroscopy
For resonance assignments the 13C, 15N-labeled BamC101-344 sample was exchanged into NMR buffer containing 10% 2H2O, 50 mM sodium phosphate, pH 6.0, 50 mM NaCl, 0.02% NaN3 and a protease inhibitor cocktail (1X HALT from Pierce), to a concentration of ~1.0 mM. The following 3D spectra were used to generate backbone and side chain assignments of BamC101-344: HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HNHAHB, HBHA(CO)NH, H(CCO)NH, (H)C(CO)NH and (H)CCH-TOCSY.1 The backbone assignments for BamC101-344 are very similar to the previously published assignments for BamC26-344.18 The 15N- and 13C-edited 3D NOESY spectra were collected with a 150 ms mixing time on the 13C, 15N-labeled BamC101-344 sample. For the 13C-edited 3D NOESY, the 13C, 15N-labeled BamC101-344 sample was exchanged into NMR buffer with 99.8% 2H2O. All NMR spectra were collected at 30 °C on VNMRS 900MHz, VNMRS 800MHz, or Inova 600 MHz spectrometers equipped with HCN z-axis gradient cold probes.
The amide 1H-15N RDCs were measured using a 1.1 mM 13C, 15N-labeled BamC101-344 sample with no Pf1 phage and a 0.14 mM 13C, 15N-labeled BamC101-344 sample in 21 mg/mL liquid crystalline Pf1 phage prepared as described.37 2D HSQC sensitivity-enhanced 15N-IPAP spectra38 were collected on the isotropic (no Pf1 phage) and aligned (with Pf1 phage) samples. For hydrogen-deuterium exchange experiments, 0.2 mM 13C, 15N-labeled BamC26-344 and 1.0 mM 13C, 15N-labeled BamC101-344 were rapidly exchanged into NMR buffer with 99.8% 2H2O using buffer exchange spin columns (Pierce). The SOFAST 15N HMQC sequence39 was used to collect 2D 1H, 15N spectra, each with a total experiment time of 4 min. Time points were collected every 4 mins for the first 4 hours, then every 3 hours for the following 24 hours, then once a day for 7 days. The peak volumes from the spectra were fit to an exponential decay function. The data for determining 15N R1 and R2 relaxations rates and 15N{1H} heteronuclear NOE data were collected as described.1, 22 Residue specific τc values were calculated from the 15N R1 and R2 relaxations rates using .40 Values for τc for the N- and C- terminal domains were averaged using R1 and R2 relaxations rates from residues with 15N{1H} heteronuclear NOE values greater than 0.7. Estimates for τc values of the individual domains were made using an empirical relation between molecular weight and τc values (http://www.nmr2.buffalo.edu/nesg.wiki/NMR_determined_Rotational_correlation_time). The methyl groups were assigned in the ILV methyl protonated 2H, 13C, 15N-labeled sample of BamC26-344 using a spectrum collected with an HMCMCGCBCA pulse sequence.3 Methyl-methyl NOEs were measured using a 3D 13C,13C,1H NOESY pulse sequence41 recorded with a 240 ms mixing time. TSP was used as internal chemical shift reference and spectra were referenced to DSS by correcting for the pH dependence of TSP chemical shift.42 All spectra were processed with NMRPipe43 and analyzed with either Sparky44 or CCPNMR Analysis.45
Structure generation with iterative CS-RDC-NOE Rosetta
The backbone (HN, N, C’, Cα, and Cβ) chemical shifts of the construct BamC101-344 were used in CS-Rosetta 3.X to select 25 and 200 fragments of 9 and 3 residues length, respectively.9 Since TALOS+46 predicted dynamic or disordered regions at the termini of the BamC101-344 construct to be less than 5 residues of length, no tails were removed for the modeling procedure. However, sequence regions that are at least 3 residues of length or are at the termini (residues 110-114, 218-228, 296-301, 345, and 346, respectively) and have TALOS+ predicted order parameters46 less than 0.7 were excluded from Rosetta score computation although they were explicitly modeled. Residues 345 and 346 were included to facilitate the cloning of the BamC construct used in the NMR studies but are not part of wt E. coli BamC. Models for the full-length construct, residues 101-346, as well as for independent domains, residues 101-212 and residues 229-346, for the N-terminus and the C-terminus, respectively were generated with the iterative CS-RDC-NOE Rosetta protocol as described previously.10 A set of 50,000 structures was produced in the initial low-resolution sampling and 15,000 of these were chosen for all-atom refinement. The NOE and RDC data had weights of 5 for the overall scoring in the low-resolution sampling stage and a weight of 0.1 in the all-atom sampling stage. To select the pool of structures that are iterated further, weights of 5 were used for NOE and RDC data for the low-resolution and all-atom structures. The standard Rosetta all-atom energy function was used with weight set, score13_env_hb.47 We note that this weight set has recently been found to hold no improvement over the more established score12,47 which therefore is recommended for use in future. The amide 1H-1H NOE restraints were modeled with a flat bottom potential where no score penalty is introduced if the proton-proton distance is between 1.5 and 6.0 Å. Violations of the bounds are penalized quadratically until 0.5 Å above the upper bound where the potential switches smoothly to a linear function. The iterative CS-RDC-NOE Rosetta calculations on the full-length BamC required ~40 h on 512 nodes of a Blue Gene/P computer (~20,000 node hours).
Supplementary Material
Structure of BamC was determined using CS-RDC-NOE Rosetta and limited NMR data
BamC consists of two domains that consist of helix-grip type fold
CS-RDC-NOE Rosetta is robust to misassigned nuclear Overhauser effect data
Acknowledgments
This research was supported by NIH grant AI080709 to MCS, GM092802 to DB and NIH training grant T32 GM08759 to LRW. The NMR instrumentation was purchased with partial support from NIH Grants GM068928, RR11969 and RR16649, NSF Grants 9602941 and 0230966 and the W. M. Keck Foundation. We also thank Dr. Geoff Armstrong for help in NMR data collection.
Abbreviations
- BAM
beta-barrel assembly machine
- CS
chemical shift
- DSS
2,2-dimethyl- 2-silapentane-5-sulfonic acid
- H-D
hydrogen-deuterium
- KA-1
kinase associated
- ILV
isoleucine, leucine, valine
- MARK
microtubule affinity regulating kinase
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- Par1
partitioning-defective 1 kinase
- PCR
polymerase chain reaction
- RDC
residual dipolar coupling
- RMSD
root mean squared deviation
- TSP
2,2,3,3-tetradeutero-3-(trimethylsilyl)-propionic acid
Footnotes
Accession numbers Atomic coordinates have been deposited in the PDB under accession codes, PDB ID: 2LAF (N-terminal domain) and PDB ID: 2LAE (C-terminal domain). The chemical shifts and structural data have been deposited in the BRMB under accession code 17521.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2. Academic Press; San Diego: 2007. [Google Scholar]
- 2.Wider G. NMR techniques used with very large biological macromolecules in solution. Methods Enzymol. 2005;394:382–398. doi: 10.1016/S0076-6879(05)94015-9. [DOI] [PubMed] [Google Scholar]
- 3.Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125:13868–78. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 4.Lemak A, Gutmanas A, Chitayat S, Karra M, Fares C, Sunnerhagen M, Arrowsmith CH. A novel strategy for NMR resonance assignment and protein structure determination. J Biomol NMR. 2011;49:27–38. doi: 10.1007/s10858-010-9458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guntert P. Automated structure determination from NMR spectra. Eur Biophys J Biophy. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 6.Altieri AS, Byrd RA. Automation of NMR structure determination of proteins. Curr Opin Struct Biol. 2004;14:547–553. doi: 10.1016/j.sbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Raman S, Huang YJP, Mao BC, Rossi P, Aramini JM, Liu GH, Montelione GT, Baker D. Accurate Automated Protein NMR Structure Determination Using Unassigned NOESY Data. J Am Chem Soc. 2010;132:202–207. doi: 10.1021/ja905934c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Schneider WM, Shen Y, Raman S, Inouye M, Baker D, Roth MJ, Montelione GT. Fully automated high-quality NMR structure determination of small (2)H-enriched proteins. J Struct Funct Genomics. 2010;11:223–32. doi: 10.1007/s10969-010-9095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–90. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327:1014–8. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 12.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–98. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–14. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 16.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–45. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol. 2005;187:4552–61. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowles TJ, McClelland DM, Rajesh S, Henderson IR, Overduin M. Secondary structure and (1)H, (13)C and (15)N backbone resonance assignments of BamC, a component of the outer membrane protein assembly machinery in Escherichia coli. Biomol NMR Assign. 2009;3:203–6. doi: 10.1007/s12104-009-9175-3. [DOI] [PubMed] [Google Scholar]
- 19.Mueller GA, Choy WY, Yang D, Forman-Kay JD, Venters RA, Kay LE. Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein. J Mol Biol. 2000;300:197–212. doi: 10.1006/jmbi.2000.3842. [DOI] [PubMed] [Google Scholar]
- 20.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970–1. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht R, Zeth K. Crystallization and preliminary X-ray data collection of the Escherichia coli lipoproteins BamC, BamD and BamE. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1586–90. doi: 10.1107/S1744309110034160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrow NA, Zhang OW, Formankay JD, Kay LE. Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 23.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–78. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 24.Fischer MW, Losonczi JA, Weaver JL, Prestegard JH. Domain orientation and dynamics in multidomain proteins from residual dipolar couplings. Biochemistry. 1999;38:9013–22. doi: 10.1021/bi9905213. [DOI] [PubMed] [Google Scholar]
- 25.Mollova ET, Hansen MR, Pardi A. Global Structure of RNA Determined with Residual Dipolar Couplings. J Am Chem Soc. 2000;122:11561–11562. [Google Scholar]
- 26.Clore GM, Gronenborn AM, Bax A. A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J Magn Res. 1998;133:216–221. doi: 10.1006/jmre.1998.1419. [DOI] [PubMed] [Google Scholar]
- 27.Lytle BL, Song J, de la Cruz NB, Peterson FC, Johnson KA, Bingman CA, Phillips GN, Jr, Volkman BF. Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds. Proteins Struct Funct Bioinformat. 2009;76:237–43. doi: 10.1002/prot.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–45. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters CD, Kuszewski JJ, Clore GM. Using XPLOR-NIH for NMR molecular structure determination. Progr NMR Spectroscopy. 2006;48:47–62. [Google Scholar]
- 30.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase Associated-1 Domains Drive MARK/PAR1 Kinases to Membrane Targets by Binding Acidic Phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–U14. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 34.Oakhill JS, Scott JW, Kemp BE. Structure and function of AMP-activated protein kinase. Acta Physiol. 2009;196:3–14. doi: 10.1111/j.1748-1716.2009.01977.x. [DOI] [PubMed] [Google Scholar]
- 35.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 37.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 38.Ding KY, Gronenborn AM. Sensitivity-enhanced 2D IPAP, TROSY-anti-TROSY, and E.COSY experiments: alternatives for measuring dipolar N-15-H-1(N) couplings. J Magn Res. 2003;163:208–214. doi: 10.1016/s1090-7807(03)00081-8. [DOI] [PubMed] [Google Scholar]
- 39.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 40.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–9. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 41.Zwahlen C, Gardner KH, Sarma SP, Horita DA, Byrd RA, Kay LE. An NMR experiment for measuring methyl-methyl NOEs in C-13-labeled proteins with high resolution. J Am Chem Soc. 1998;120:7617–7625. [Google Scholar]
- 42.Tynkkynen T, Tiainen M, Soininen P, Laatikainen R. From proton nuclear magnetic resonance spectra to pH. Assessment of 1H NMR pH indicator compound set for deuterium oxide solutions. Anal Chim Acta. 2009;648:105–12. doi: 10.1016/j.aca.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. SPARKY 3. University of California, San Francisco [Google Scholar]
- 45.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Bioinformat. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyka MD, Keedy DA, Andre I, Dimaio F, Song Y, Richardson DC, Richardson JS, Baker D. Alternate states of proteins revealed by detailed energy landscape mapping. J Mol Biol. 2011;405:607–18. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.