Summary
Previous experiments suggest a connection between the N-alpha-acetylation of proteins and the sensitivity of cells to apoptotic signals. Here, we describe a novel biochemical assay to detect the acetylation status of proteins and demonstrate that protein N-alpha-acetylation is regulated by the availability of acetyl-CoA. Because the anti-apoptotic protein Bcl-xL is known to influence mitochondrial metabolism, we reasoned that Bcl-xL may provide a link between protein N-alpha-acetylation and apoptosis. Indeed, Bcl-xL overexpression leads to a reduction in levels of acetyl-CoA and N-alpha-acetylated proteins in the cell. This effect is independent of Bax and Bak, the known binding partners of Bcl-xL. Increasing cellular levels of acetyl-CoA by addition of acetate or citrate restores protein N-alpha-acetylation in Bcl-xL-expressing cells and confers sensitivity to apoptotic stimuli. We conclude that acetyl-CoA serves as a signaling molecule that couples apoptotic sensitivity to metabolism by regulating protein N-alpha-acetylation.
Introduction
Increasing evidence suggest that specific metabolic alterations associated with cancer cells may not be ancillary to their transformation but instrumental to their tumorigenic potential by mediating cell proliferation, growth and survival (Vander Heiden et al., 2009). Many oncogenes and tumor suppressor genes known to promote excess cell proliferation also alter biosynthetic (or anabolic) processes. For example, Akt expression stimulates glucose uptake and glycolysis, the pentose phosphate pathway and fatty acid synthesis. c-Myc expression promotes glutamine metabolism as well as purine and pyrimidine biosynthesis. Furthermore, mutations in genes encoding metabolic enzymes have been identified by cancer genetic association studies (Vander Heiden et al., 2009). However, how specific metabolites contribute to increased proliferation and apoptotic resistance in tumor cells remains a central unanswered question.
The proto-oncogene Bcl-xL has a prominent role in promoting cell survival and cancer development (Boise et al., 1993). It is well-established that Bcl-xL protects against apoptosis by directly binding and inhibiting Bax/Bak-oligomerization mediated mitochondrial permeabilization. However, certain Bcl-xL mutants, such as F131V/D133A and G148E, that are unable to bind to Bax or Bak, nevertheless retain 70–80% anti-apoptotic activity of WT Bcl-xL (Cheng et al., 1996). Curiously, Bcl-xL has also been shown to regulate mitochondrial respiration and metabolism (Gottlieb et al., 2000; Vander Heiden et al., 1999). Whether the metabolic function of Bcl-xL contributes to its role in mediating apoptotic resistance is unclear.
Our unexpected identification of an N-terminal acetyltransferase, Arrest Defective 1 (dArd1) in a genome-wide RNAi screen in Drosophila cells for apoptotic regulators (Yi et al., 2007) prompted us to posit that protein N-alpha-acetylation, a major N-terminal modification, links cell metabolism to apoptotic induction in cancer cells. Since dARD1 is epistatic to Diap1, a direct inhibitor of caspases in Drosophila, and ARD1 is required for caspase activation in mammalian cells (Yi et al., 2007), the role for ARD1 in mediating caspase activation is evolutionarily conserved. How ARD1 regulates caspase activation has not yet been illustrated.
In mammalian cells, protein N-alpha-acetylation is mediated by the highly conserved N-acetyltransferase protein complexes (NatA, NatB, NatC, NatD, and NatE). The NatA complex consists of the catalytic subunit, Arrest Defective 1 (hNaa10p/ARD1), and the auxiliary subunit, N-acetyltransferase 1 (NAT1/hNaa15p/NATH); whereas NatB consists of N-terminal acetyltransferase 3 (hNaa20p/NAT3) and mitochondrial distribution and morphology 20 (hNaa25p/Mdm20). Although the Nat complexes are implicated in regulating cell cycle progression, cell proliferation and tumorigenesis, the mechanisms that connect N-alphaacetylation to the cellular protein apparatus are unknown (Ametzazurra et al., 2008; Polevoda and Sherman, 2003; Starheim et al., 2008; Starheim et al., 2009). Recent N-acetylome studies reveal incomplete acetylation status of proteins (Arnesen et al., 2008; Goetze et al., 2009). Although a commonly accepted view is that partial acetylation results from degenerate nature of protein N-terminal sequences, we considered the possibility that protein N-alpha-acetylation might be regulated, an alternative hypothesis that had not been tested due to technical limitations.
Here we developed a novel biochemical approach to assess the status of endogenous levels of protein N-alpha-acetylation. Using this assay, we show that protein N-alpha-acetylation levels are sensitive to alterations in metabolism and Bcl-xL expression. Bcl-xL overexpression leads to reduced levels of acetyl-CoA and hypoacetylation of protein N-termini through a Bax/Bak independent mechanism. Conversely, Bcl-xL−/− mouse embryonic fibroblasts show increased levels of acetyl-CoA as well as protein N-alpha-acetylation levels. Protein N-alpha-acetylation deficiency in Bcl-xL overexpressing cells contributes to apoptotic resistance since increasing acetyl-CoA production can rescue this deficiency in protein N-alpha-acetylation and sensitize Bcl-xL cells to cell death. Our study suggests that regulation of acetyl-CoA availability and protein N-alpha-acetylation may provide a Bax/Bak independent mechanism for Bcl-xL to regulate apoptotic sensitivity.
Results
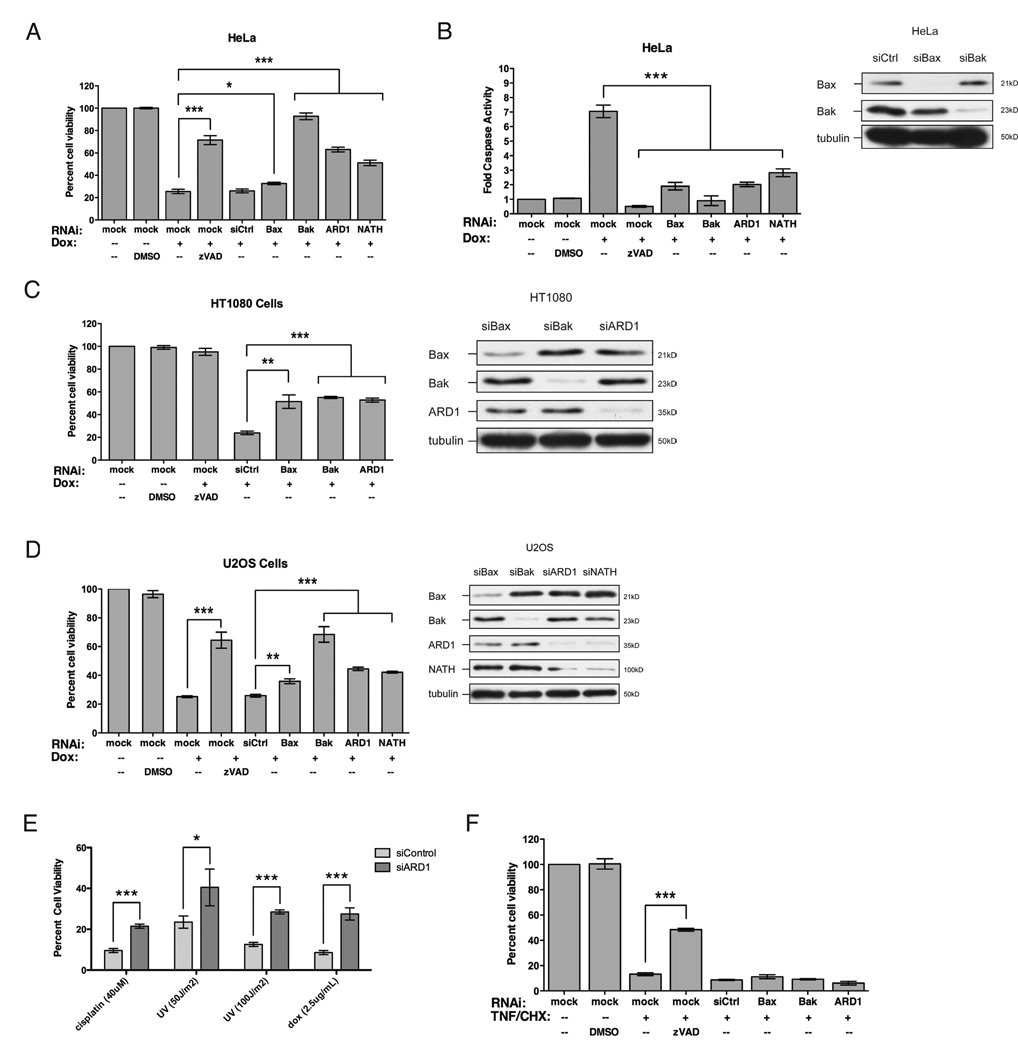
We confirmed that ARD1 is necessary for cell death induced by the DNA damaging agent, doxorubicin, in multiple cell lines of different origins including Drosophila Kc cells (Yi et al., 2007), HeLa, HT1080, and U2OS cells (Figure 1A–D). In addition, HeLa and U2OS cells deficient for NATH were also resistant to doxorubicin treatment, recapitulating the apoptotic resistant phenotype of ARD1 knockdown cells (Figure 1A–D). Thus, the acetylation activity of the NatA complex serves to influence the sensitivity of these cells to apoptosis. Next we tested whether NatA influences apoptotic sensitivity to other DNA damaging agents. We found that ARD1 knockdown cells are also resistant to cisplatin and UV treatment (Figure 1E). However, these cells remained sensitive to tumor necrosis factor (TNFalpha) and cyclohexamide treatment, which specifically activates apoptosis through the death receptor pathway (Figure 1F). Thus, we conclude that protein N-alpha-acetylation regulates apoptotic sensitivity downstream of DNA damage.
Figure 1. NatA knockdown suppresses cell death induced by DNA damage in HeLa, HT1080, and U2OS cells.
(A–B) HeLa cells were treated with doxorubicin (1.25µg/mL, 20h for cell viability; 5µg/mL, 8h for caspase activity). (C) HT1080 cells were treated with doxorubicin (1.25µg/mL, 20h). (D) U2OS cells were treated with doxorubicin (1.25µg/mL, 20h). (E) HeLa cells were treated with cisplatin (40µM) or UV (50J/m2 or 100J/m2) for 24h. (F) HeLa cells were treated with TNFalpha (10ng/mL, 24h) and cyclohexamide (1µg/ml, 24h) to induce death receptor mediated cell death. Immunoblots were conducted in parallel to show extent of target knockdown. Data are represented as mean +/− s.d. (n=3). (Student’s T-test; *, p<0.05; **, p<0.01; ***, p<0.001)
Since N-alpha-acetylation has been suggested to affect protein stability (Polevoda and Sherman, 2003), we examined whether protein synthesis and/or protein turnover might be affected by acetylation status. We tested whether ARD1 substrates such as caspase-2 and Chk1 (see results below) are destabilized in ARD1 knockdown cells using cyclohexamide, an inhibitor of protein synthesis. Deficiency in ARD1 did not lead to decreases in the cellular levels of these proteins compared to that of control (Figure S1A). The steady state levels of total cellular proteins in ARD1 knockdown cells were similar to the levels in control cells (Figure S1B). We also tested whether general protein stability is altered in ARD1 or NATH knockdown cells (Figure S1C). By pulse-chase 35S-Met labelling experiments, we observed that neither general protein synthesis nor turnover was affected in ARD1 or NATH knockdown cells. Thus, protein N-alpha-acetylation mediated by NatA complex is not required to maintain protein stability globally. In addition, we verified that cell cycle progression is unaffected in cells deficient for ARD1/NATH (Figure S1D). Taken together, these data suggest that the NatA complex may influence apoptotic sensitivity by mediating protein N-alpha-acetylation of key apoptotic components.
In vitro detection of unmodified protein N-termini
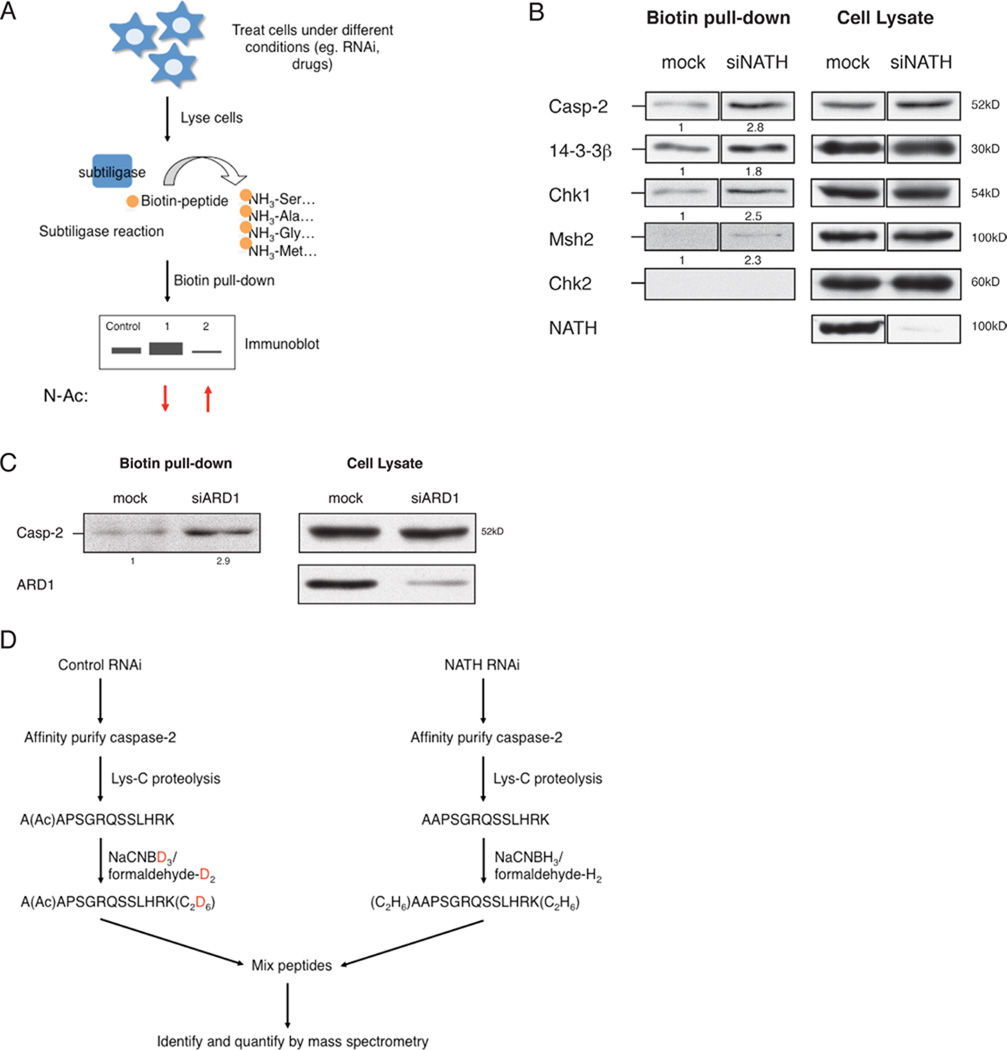
The lack of an immunological method to detect the acetylation status of protein N-termini has limited our understanding of the mechanisms that regulate protein N-alpha-acetylation. To this end, we developed a selective biotin labelling method using an engineered protein ligase, termed subtiligase (Abrahmsen et al., 1991; Tan et al., 2007) that detects non-acetylated N-termini of endogenous proteins. This approach was used to capture unmodified protein N-termini resulting from caspase mediated cleavage during apoptotic cell death (Mahrus et al., 2008). Unblocked N-termini can be labelled using subtiligase, which preferentially biotinylates N-terminal amine groups consistent with the specificity of NatA or NatB (Abrahmsen et al., 1991; Mahrus et al., 2008). As the N-termini of up to 80–90% of cellular proteins may be blocked by a number of different modifications (Martinez et al., 2008), very few proteins will be biotin labelled by subtiligase as previously demonstrated (Mahrus et al., 2008). Thus, any protein that is biotin labelled by subtiligase in our assays most likely results from a specific loss in N-alpha-acetylation.
We utilized subtiligase to biotinylate free N-termini of proteins in whole cell lysates followed by avidin affinity purification and western blot analysis. Decreased levels of protein N-alpha-acetylation are expected to increase subtiligase-mediated protein biotinylation and conversely, increased levels of protein N-alpha-acetylation are expected to decrease subtiligase-mediated protein biotinylation (Figure 2A). First, we asked whether the subtiligase assay could be used to distinguish the N-alpha-acetylation status of protein N-termini when the expression of the NatA complex is diminished by RNAi mediated knockdown. ARD1 acetylates a subclass of proteins with Ser, Ala, or Thr as the newly exposed N-terminal residue after initiator Met (iMet) cleavage (Polevoda and Sherman, 2003). We tested 14-3-3β, which is known to be N-alpha-acetylated (Arnesen et al., 2009; Martin et al., 1993), and proteins that we predict to be N-alpha-acetylated based on their sequences, Chk1 and Msh2. Caspase-2, which is responsive to both DNA damage (Tinel and Tschopp, 2004) and metabolic stress (Nutt et al., 2009; Nutt et al., 2005), is also a good candidate for acetylation by ARD1 as the second amino acid in the caspase-2 polypeptide is Ala. We observed that these proteins as well as caspase-2 were biotinylated to a higher extent by subtiligase in NATH or ARD1 knockdown cells than in control cells (Figure 2B and 2C). These data suggest that subtiligase can distinguish N-alpha-acetylation of multiple proteins that is dependent on NatA expression.
Figure 2. A subtiligase-based assay to label unacetylated protein N-termini.
(A) A schematic of the subtiligase assay. Briefly, cells were gently lysed in NP-40 lysis buffer on ice. Cell debris was pelleted and resulting supernatant was subjected to a subtiligase reaction (1µM subtiligase, 1mM biotin-peptide, 2mM DTT; 1h at room temperature). Biotinylated proteins were affinity purified using Neutravidin beads (Thermo Scientific) and analyzed by SDS-PAGE. Protein N-alpha-acetylation levels were determined by immunoblot against proteins in which the N-terminal residue corresponds to specificity of a Nat complex. Chk2 was included as a negative control, which should not be recognized by subtiligase based on sequence. (B–C) Subtiligase reaction was conducted on HeLa cells transfected with siRNA against NATH (B) or ARD1 (C) as described in (A). Enrichment of biotin labelled NatA substrates was observed in ARD1 or NATH deficient cells compared to that of control. Enrichment in the amount of pulldown suggests a decrease in protein N-alpha-acetylation levels of NatA substrates in NatA deficient cells. Blot quantification for all subtiligase assays was calculated relative to control and normalized to corresponding lysate sample using ImageJ software. (D) A schematic of detecting protein N-terminal peptide modifications by quantitative mass spectrometry. Briefly, cells were treated with RNAi and lysed in NP-40 buffer. FLAG-tagged caspase-2 (active cysteine mutant C320G) is affinity purified using anti-FLAG agarose and eluted with FLAG peptide. Purified protein was digested by Lys-C, which cleaves after a lysine residue. Dimethyl labelling of peptides was conducted as previously described (Hsu et al., 2003; Khidekel et al., 2007). Briefly, peptides were chemically modified using NaCNBH3 or NaCNBD3 followed by formaldehyde-H2 or formaldehyde-D2 treatment. Heavy or light modified peptides were mixed and analyzed by mass spectrometry. Ratios for levels of N-terminal acetylation were determined by normalizing to 1:1 ratio of shared internal peptides (See Figure S1 for list of peptides).
To determine the validity of subtiligase assay, we measured the extent of protein N-alpha-acetylation by quantitative mass spectrometry using differential isotope labelling (Figure 2D). First, we tested whether we could detect the basal levels of N-alpha-acetylation of caspase-2 by mass spectrometry. We observed that the mass to charge ratio (m/z) of the N-terminal peptide of caspase-2 is shifted as expected with an acetyl modification (Figure S2A and S2B). Furthermore, we found a 30% reduction in the amount of N-alpha-acetylated caspase-2 in NATH deficient cells relative to control by subtiligase assay as well as mass spectrometry (Figure 2B; Figure S2D). These results support the conclusion that caspase-2 is N-alpha-acetylated by ARD1.
Protein N-alpha-acetylation promotes the assembly of caspase-2 complex
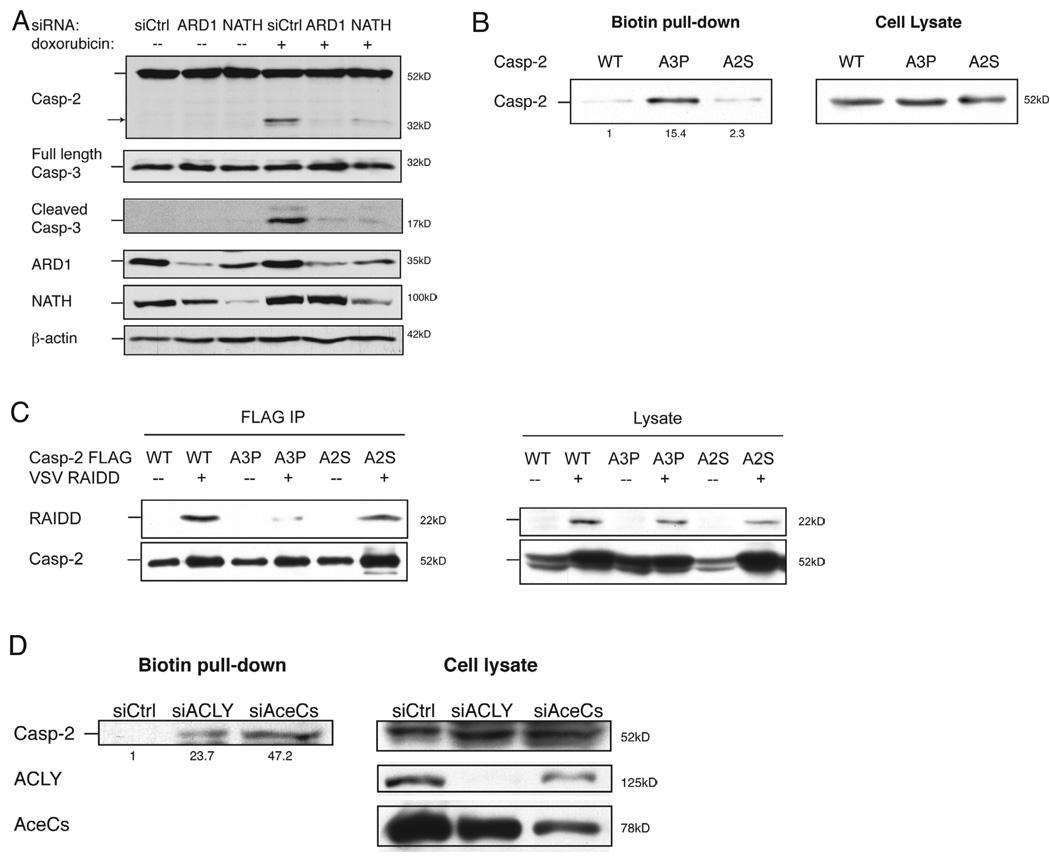
As caspase-2 is a substrate of ARD1 (Figure 2 and S2) and the activation of caspase-2 is inhibited by ARD1 or NATH knockdown (Figure 3A), we asked how N-alpha-acetylation of caspase-2 might influence caspase activation.
Figure 3. N-alpha-acetylation of caspase-2 is required for binding to RAIDD.
(A) HeLa cells were transfected with siRNAs as indicated and treated with doxorubicin (5µg/mL, 8h). Cell lysates were analyzed by SDS-PAGE, and caspase cleavage was assessed by immunoblot. Caspase-2 and caspase-3 activation is suppressed in ARD1 and NATH knockdown cells compared to that of control. Arrow indicates cleaved product of caspase-2. (B) Subtiligase reaction was conducted on HEK 293T cells transfected with FLAG-tagged WT, A3P, and A2S caspase-2 (active cysteine mutant C320G of caspase-2) as described in Figure 1. Enrichment of biotin-labeled A3P caspase-2 was observed compared to WT and A2S caspase-2. This suggests that A3P caspase-2 is hypoacetylated. (C) HEK 293T cells were transfected with FLAG-tagged WT, A3P, and A2S caspase-2 as well as with VSV-RAIDD as indicated. Caspase-2 was affinity purified with anti-FLAG agarose and eluted with FLAG peptide. Resulting eluants were subjected to SDS-PAGE for immunblot analysis. RAIDD efficiently co-immunoprecipitates with WT and A2S but not with A3P caspase-2. Blots are representative of at least three independent experiments. (D) Subtiligase reaction was conducted on lysates from HeLa cells transfected with siRNAs against acetyl-CoA synthetase or ATP citrate lyase as indicated. Increased biotin labelling of caspase-2 was observed in knockdown cells compared to control cells.
First we conducted mutagenesis analysis of caspase-2 to disrupt protein N-alpha-acetylation. We replaced the third residue of caspase-2 with Pro (A3P) as the presence of Pro in this position inhibits protein N-alpha-acetylation. The 3P mutation has been previously demonstrated to inhibit N-alpha-acetylation of other substrates, known as the XPX rule (Goetze et al., 2009). We also replaced the second Ala for Ser as a control to maintain N-alpha-acetylation (A2S) as well as iMet removal (Arnesen et al., 2009; Goetze et al., 2009). Generation of these targeted substitutions allows us to definitively test whether subtiligase can differentiate between acetylated and unacetylated forms of caspase-2. An increase in subtiligase-mediated biotinylation of A3P was detected, while very little A2S or wild-type caspase-2 was detected after biotin pull down, consistent with acetylation as the explanation for the lower biotinylation levels (Figure 3B). A defect in N-alpha-acetylation of A3P caspase-2, but not WT and A2S caspase-2 was confirmed by mass spectrometry (Figure S2A–C). Thus, subtiligase is an effective tool for detecting unmodified protein N-termini.
The caspase-2 scaffolding complex, which promotes caspase-2 activation, includes the adaptor protein, receptor-interacting protein (RIP)-associated ICH-1/CED-3 homologous protein with a death domain (RAIDD) (Duan and Dixit, 1997; Tinel and Tschopp, 2004). The ability of the N-terminal caspase-2 mutants to interact with RAIDD was assessed by co-immunoprecipitation. We found that RAIDD efficiently co-immunoprecipitated with WT and A2S but not with A3P caspase-2 (Figure 3C). This suggests that N-alpha-acetylation of caspase-2 facilitates its interaction with RAIDD.
Since acetyl-CoA is a key co-factor in N-alpha acetylation, we speculated that the levels of N-alpha-acetylated caspase-2 might be dependent on expression of key metabolic enzymes that are responsible for production of cytoplasmic acetyl-CoA. To explore this question, we tested whether knockdown of ATP citrate lyase or acetyl-CoA synthetase, the two metabolic enzymes that utilize citrate and acetate, respectively, to generate acetyl-CoA, results in decreased levels of N-alpha-acetylated caspase-2. Indeed, we observed increased biotin labelling of caspase-2 in knockdown cells compared to control cells following subtiligase assay (Figure 3D). This suggests that caspase-2 is hypoacetylated when acetyl-CoA generation is reduced and therefore, protein N-alpha-acetylation is subject to metabolic regulation.
Regulation of protein N-alpha-acetylation by Bcl-xL
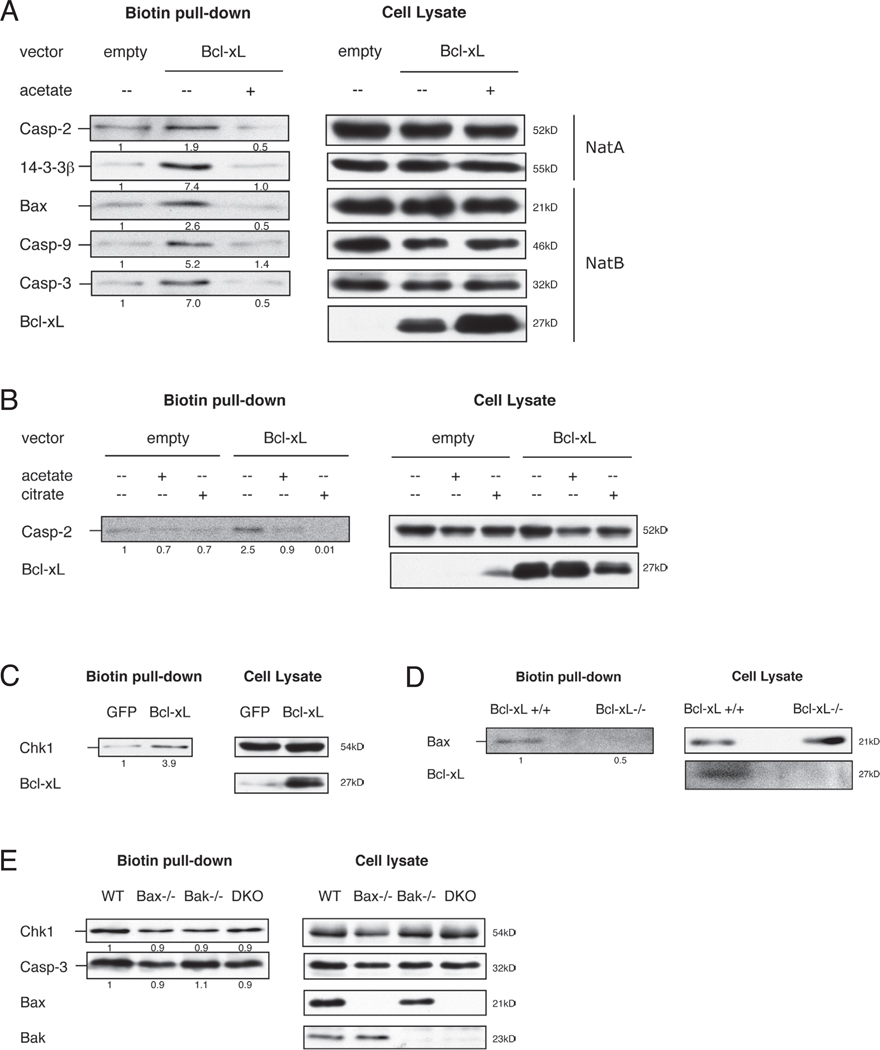
Since decreased levels of protein N-alpha-acetylation leads to apoptotic deficiency, we reasoned that regulation of protein N-alpha-acetylation might provide a mechanism for certain apoptotic regulators to control apoptotic sensitivity. Bcl-xL, an anti-apoptotic Bcl-2 family member, is known to have an effect on metabolism (Shimizu et al., 1999; Vander Heiden et al., 2001). We asked whether protein N-alpha-acetylation levels are sensitive to Bcl-xL expression using subtiligase assay. An increase in biotin labelling of caspase-2, -9, -3, and Bax was observed by Bcl-xL expression in 293T, HeLa, and Jurkat cells compared to that of control (Figure 4A, 4B, and 4C). Conversely, a decrease in biotin labelling was apparent in Bcl-xL−/− MEFs compared to that of Bcl-xL+/+ MEFs (Figure 4D). Since Bcl-xL is known for maintaining mitochondrial integrity by blocking oligomerization of Bax/Bak, we measured the levels of protein N-alpha-acetylation in Bax/Bak deficient cells. Surprisingly, the levels of protein N-alpha-acetylation were similar in Bax−/−, Bak−/− or Bax/Bak−/− DKO MEFs compared to that of WT MEFs by subtiligase assay (Figure 4E). This suggests that Bcl-xL mediated regulation of protein N-alpha-acetylation is independent of Bax/Bak.
Figure 4. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL.
(A–C) HEK 293T, HeLa, or Jurkat cells were generated to express empty GFP vector or Bcl-xL. Cells were treated with acetate (50mM, 24h) or citrate (10mM, 24h) as indicated. Subtiligase reaction was conducted as described in Figure 1. Enrichment of biotin-labelled NatA and NatB substrates is observed in Bcl-xL cells compared to control cells. Biotin labelling is reduced to control levels by acetate or citrate treatment in Bcl-xL cells. (D) Lysates generated from Bcl-xL +/+ or Bcl-xL −/− MEFs were subjected to subtiligase reaction as described in Figure 1. Bax is hypoacetylated in Bcl-xL +/+ MEFs compared to Bcl-xL −/− MEFs. (E) Lysates generated from wild type, Bax −/−, Bak −/−, or Bax/Bak −/− (DKO) MEFs were subjected to subtiligase assay as described in Figure 1. Blots are representative of at least three independent experiments.
Recent studies show that histone lysine acetylation is dependent on acetyl-CoA production in yeast and mammalian cells (Takahashi et al., 2006; Wellen et al., 2009). However, we found that lysine acetylation of histone H3 and H4 were unaffected in Bcl-xL cells compared to control (Figure S3). This suggests that histone lysine acetylation is not sensitive to the changes in acetyl-CoA levels associated with Bcl-xL expression.
We next tested whether protein N-alpha-acetylation levels in Bcl-xL cells are affected by changes in acetyl-CoA metabolism. Addition of acetate or citrate stimulates cytosolic acetyl-CoA production by acetyl-CoA synthetase or ATP citrate lyase respectively (Sullivan et al., 1974a; Sullivan et al., 1974b). We verified that these metabolites increase acetyl-CoA levels in mammalian cells (data not shown). Under metabolite treatment, protein N-alpha-acetylation levels were restored in Bcl-xL expressing cells to that of control levels (Figure 4A and 4B). Thus, a reduction in acetyl-CoA production in Bcl-xL cells may be responsible for the observed hypoacetylation.
Metabolic alterations caused by Bcl-xL expression
The expression of Bcl-xL is often elevated in tumors (Adams and Cory, 2007). To explore the effect of Bcl-xL on tumor metabolism, we conducted a systematic search using a combination of 2D-NMR and mass spectrometry to identify metabolic changes associated with increased Bcl-xL expression. Principal component analysis (PCA) of one-dimensional (1D) proton spectra shows that the metabolome of Bcl-xL expressing cells was significantly different from the metabolome of control cells (Figure S4).
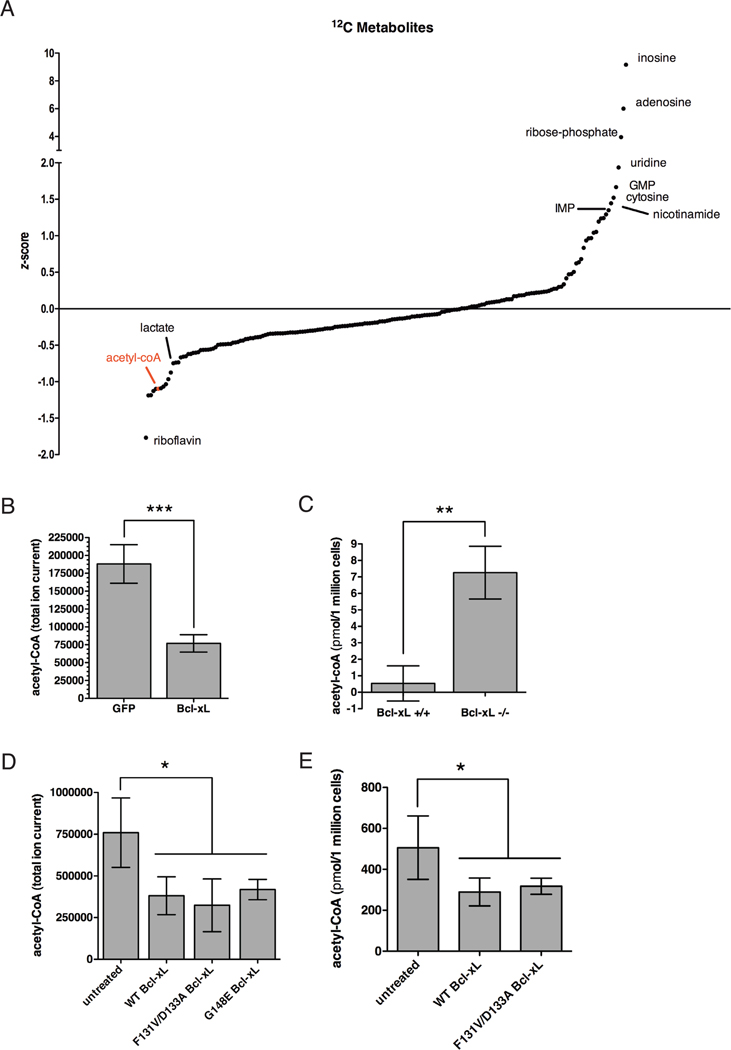
We then used triple-quadruple mass spectrometry via selected reaction monitoring (SRM) to identify metabolite changes in Bcl-xL cells relative to GFP control cells as mass spectrometry is a more sensitive approach (Bajad et al., 2006; Lu et al., 2008). This is particularly relevant for intermediates of glucose metabolism as these metabolites are difficult to decipher by NMR due to their similar proton content. Thus, both NMR and mass spectrometry provide complementary approaches for a comprehensive understanding of the metabolite changes resulting from a specific perturbation. Indeed we found that acetyl-CoA levels were decreased by two-fold in Bcl-xL expressing cells relative to GFP expressing cells by mass spectrometry as well as an enzyme-based assay (Figure 5A–B). Conversely, acetyl-CoA levels were substantially increased in Bcl-xL−/− mouse embryonic fibroblasts (MEFs) compared to Bcl-xL+/+ MEFs (Figure 5C). These data provide strong evidence that Bcl-xL expression reduces the levels of acetyl-CoA, suggesting that reduced levels of acetyl-CoA in Bcl-xL overexpressing cells leads to hypoacetylation.
Figure 5. Bcl-xL expression reduces acetyl-CoA production.
(A) Identification of metabolites using an AB/Sciex 5500 QTRAP mass spectrometer separated with normal phase HILIC NH2 chromatography using positive/negative ion switching in Jurkat cells stably expressing GFP empty vector or Bcl-xL. (B) Acetyl-CoA levels were measured in Jurkat cells stably expressing GFP empty vector or Bcl-xL by mass spectrometry. Peak areas of total ion current (TIC) were processed using MultiQuant 1.1 software. Average TIC for acetyl-CoA analyte is shown. Acetyl-CoA levels are lower in Bcl-xL cells relative to GFP cells. (C) Acetyl-CoA levels were measured in Bcl-xL +/+ or BclxL −/− MEFs using an enzyme-based assay (Abcam). Metabolite concentration was determined by generating a standard curve. Bcl-xL deficiency results in higher acetyl-CoA levels compared to control. (D) 293T cells were transiently transfected with empty vector, WT Bcl-xL, or specific Bcl-xL mutants (F131V/D133A or G148E). Acetyl-CoA levels were measured by mass spectrometry. WT or mutant Bcl-xL cells show a decrease in acetyl-CoA levels compared to empty vector cells. (E) Acetyl-CoA levels were measured in 293T cells transiently expressing WT or mutant Bcl-xL as described in (C). Data are represented as mean +/− s.d. (n=3). (Student’s T-test; *, p<0.05; **, p<0.01; ***, p<0.001).
Since Bax/Bak DKO cells are not defective in protein N-alpha-acetylation, we reasoned that Bcl-xL might be able to negatively regulate the levels of acetyl-CoA independent of Bax/Bak binding. Cheng et al. reported that certain Bcl-xL mutants, such as F131V/D133A and G148E, are unable to bind to Bax or Bak but nevertheless retain 70–80% anti-apoptotic activity of WT Bcl-xL (Cheng et al., 1996). We measured acetyl-CoA levels in cells expressing WT Bcl-xL or these specific Bcl-xL mutants. A similar reduction in acetyl-coA levels was observed in cells expressing these Bcl-xL mutants and in cells expressing WT Bcl-xL (Figure 5D and 5E). Thus, Bcl-xL’s metabolic function in regulating the levels of acetyl-CoA does not depend on its interaction with Bax/Bak.
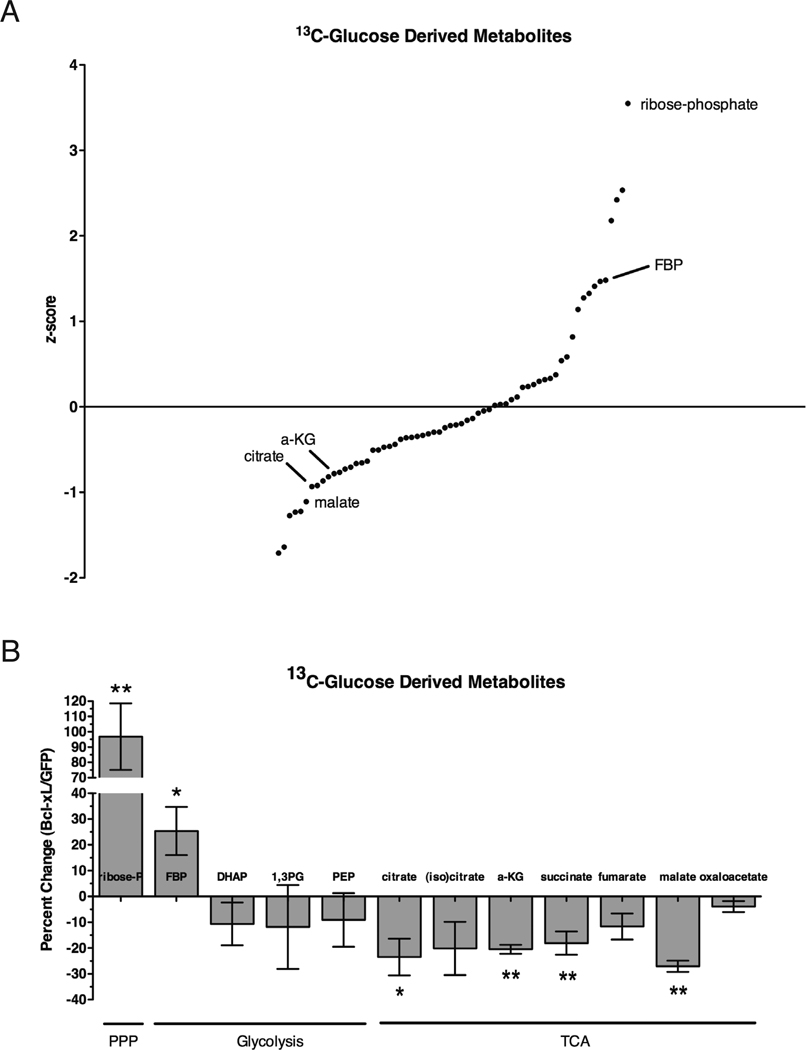
As the majority of cellular acetyl-group in acetyl-CoA is produced from glucose (DeBerardinis et al., 2007), we asked whether glucose metabolism might be altered in Bcl-xL expressing cells. We fed Bcl-xL cells uniformly labelled 13C-glucose to differentiate glucose-derived metabolites from those derived from other carbon sources (Figure 6A; Table S1). We found that the levels of glucose-derived citrate were decreased by approximately 25% in Bcl-xL expressing cells relative to control (Student’s t-test, p-value<0.05; Figure 6B; Table S1). As citrate is the direct precursor of cytoplasmic pools of acetyl-CoA, the lower levels of glucose-derived citrate might explain the decrease in acetyl-CoA levels observed in Bcl-xL expressing cells. Consistent with this hypothesis, levels of alpha-ketoglutarate, which is also derived from citrate, were lower in Bcl-xL expressing cells relative to control (Figure 6B; Table S1).
Figure 6. Metabolite profiling of glucose-derived intermediates in Bcl-xL cells by mass spectrometry.
(A) Jurkat cells stably expressing GFP empty vector or Bcl-xL were fed uniformly labelled 13C-glucose and metabolites were isolated by methanol extraction. Lysates were subjected to mass spectrometry as described in Figure 3A. (B) Percent change in 13C-glucose derived metabolites of the pentose phosphate pathway (PPP), glycolysis, and TCA cycle. Fructose-1,6 bisphosphate (FBP), dihydroxyacetone-phosphate (DHAP), 1,3-bis-phosphoglycerate (1,3PG), phosphoenolpyruvate (PEP), α–ketoglutarate (a-KG). Data are represented as mean +/− s.d. (n=3). (Student’s T-test; *, p<0.05; **, p<0.01; ***, p<0.001).
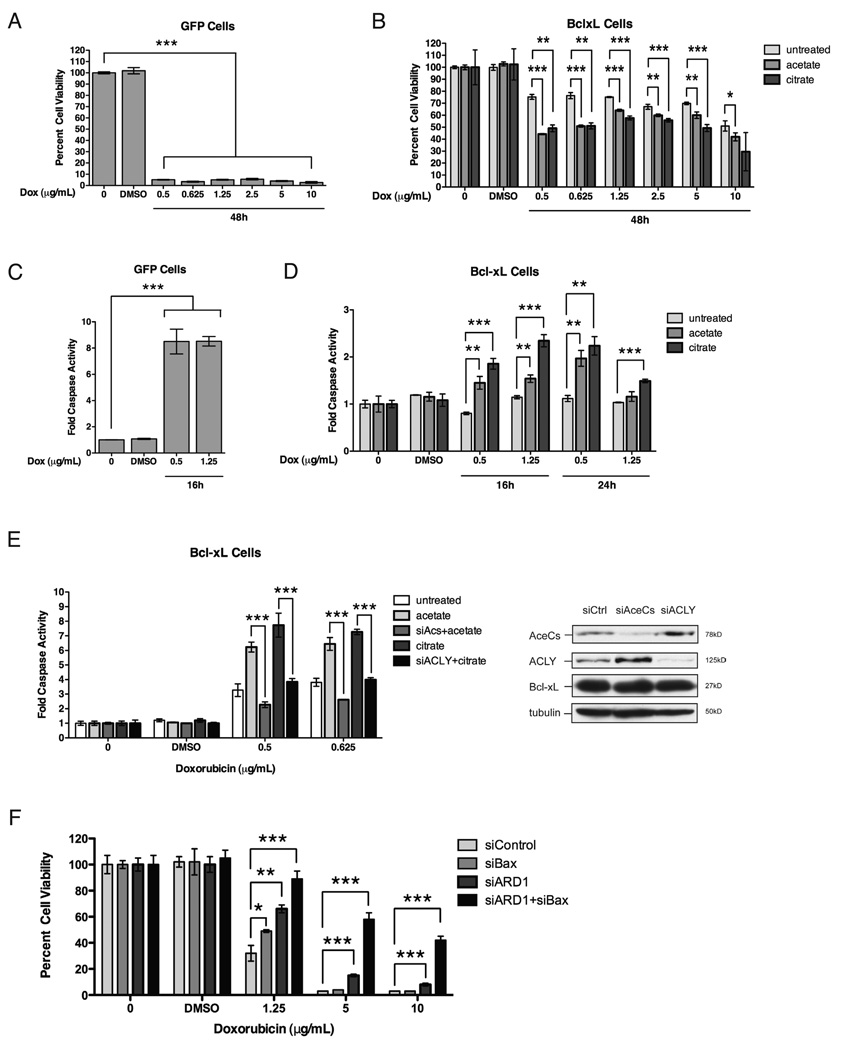
Since metabolite addition rescues the defect on protein N-alpha-acetylation by Bcl-xL (Figure 4A–B), we asked whether these metabolites could alter cell survival that is supported by Bcl-xL expression. Bcl-xL expression effectively protects against a wide range doses of doxorubicin (Figure 7A–D). Remarkably, increasing levels of citrate or acetate sensitized HeLa cells stably expressing Bcl-xL to doxorubicin-induced cell death compared to that of untreated cells (Figure 7B) with a two-fold increase in caspase activity (Figure 7D). Importantly, RNAi against acetyl-CoA synthetase or ATP citrate lyase completely suppressed the sensitization to doxorubicin elicited by addition of acetate or citrate, respectively (Figure 7E). This indicates that metabolite-induced apoptotic sensitization of cells expressing Bcl-xL specifically results from changes in acetyl-CoA production.
Figure 7. Metabolic function of Bcl-xL contributes to cell survival.
(A–D) HeLa cells stably expressing empty GFP vector or Bcl-xL were treated with doxorubicin as indicated. Cell viability was determined by measuring cellular ATP levels using a luminescence-based assay (Promega). Caspase activity was determined by measuring cleavage of a luciferin substrate containing the caspase cleavage site, DEVD (Promega). (A–B) Doxorubicin treatment efficiently kills GFP cells but not Bcl-xL cells. Pretreatment with acetate (50mM) or citrate (10mM) for 24h sensitizes Bcl-xL cells to doxorubicin-induced cell death. (C–D) Doxorubicin treatment induces substantial caspase activity in GFP cells but is completely suppressed in Bcl-xL cells. Pre-treatment of Bcl-xL cells with acetate or citrate enhances caspase activity induced by doxorubicin treatment. (E) Bcl-xL cells were transiently transfected with a pool of siRNAs (50nM, Dharmacon) as indicated followed by pre-treatment with acetate or citrate prior to doxorubicin treatment. Metabolite enhancement of doxorubicin-induced caspase activity in Bcl-xL cells is suppressed to control levels by RNAi against acetyl-CoA synthetase (Acs) or ATP citrate lyase (ACLY), respectively. Immunoblot shows extent of knockdown. (F) HeLa cells were transiently transfected with a pool of siRNAs as indicated. Double knockdown of ARD1 and Bax provides additive protection against doxorubicin-induced cell death compared to single knockdown of these genes. Graphs are representative of three independent experiments. Data are represented as mean +/− s.d. (n=3). (Student’s T-test; *, p<0.05; **, p<0.01; ***, p<0.001).
The above data suggest that Bcl-xL may mediate apoptosis resistance through two parallel pathways by inhibiting Bax/Bak oligomerization and by down-regulating protein N-alpha-acetylation. We therefore directly tested whether the effects of inhibiting Bax and ARD1 are additive in protecting against apoptosis. We found that double knockdown of both ARD1 and Bax indeed provided enhanced protection against apoptosis compared to that of knockdown individually, which was especially significant at higher concentrations of doxorubicin (Figure 7F). This finding supports the notion that Bcl-xL has dual functions in regulating protein N-alpha-acetylation levels and Bax/Bak oligomerization.
Discussion
The ability to rapidly assess protein modifications immunologically has been essential for exploring the significance and regulation of multiple protein posttranslational modifications such as phosphorylation, histone methylation, and acetylation. Since an antibody for protein N-alpha-acetylation does not exist, the ability to assess this modification was severely limited. In this regard, the subtiligase assay as described in the present study provides a powerful tool to allow us to rapidly assess the endogenous levels of protein N-alpha-acetylation. Using this assay, we discovered that protein N-alpha-acetylation status is reduced in cells overexpressing Bcl-xL. Furthermore we show that protein N-alpha-acetylation is sensitive to acute changes in acetyl-CoA availability. Since incomplete N-alpha-acetylation status of proteins has been described by recent proteome studies (Arnesen et al., 2008; Goetze et al., 2009), we propose that protein N-alpha-acetylation might be a regulated process. In the course of our studies, we noted that protein N-alpha-acetylation levels as assessed by subtiligase were sensitive to nutrient levels in the culture medium (Yi CH and Yuan J, unpublished observations). Because ARD1-mediated N-alpha-acetylation requires acetyl-CoA as an essential cofactor, we hypothesized that protein N-alpha-acetylation is influenced by cellular metabolism.
Our study directly links a specific metabolite, acetyl-CoA, to apoptotic sensitivity and supports a growing number of studies that describe a role for cell metabolism in controlling apoptosis (Nutt et al., 2009; Nutt et al., 2005; Schafer et al., 2009; Vaughn and Deshmukh, 2008). Interestingly, the cellular levels of acetyl-CoA are sensitive to Bcl-xL status in a Bax/Bak-independent manner because the expression of Bcl-xL mutants that are unable to bind to Bax or Bak (Cheng et al., 1996) can also affect the acetyl-CoA levels to the same extent as that of wild type Bcl-xL. Hardwick and colleagues demonstrated that these Bcl-xL mutants preserve 70–80% anti-apoptotic activity of WT Bcl-xL despite their inability to bind to Bax or Bak (Cheng et al., 1996). Thus, inhibiting acetyl-CoA production might provide an additional mechanism for Bcl-xL to protect against apoptosis in a Bax/Bak-independent manner. Taken together, these data suggest that Bcl-xL might protect against apoptosis through two parallel mechanisms: by directly binding and inhibiting Bax/Bak oligomerization and by regulating mitochondrial metabolism, which leads to reduced levels of acetyl-coA and protein N-alpha-acetylation.
We propose that Bcl-xL integrates metabolism to apoptotic resistance by modulating acetyl-CoA levels. Previous studies show that Bcl-xL directly binds to the voltage-dependent anion channel (VDAC), a component of the mitochondrial permeability transition pore, which controls mitochondrial metabolite exchange (Shimizu et al., 1999; Vander Heiden et al., 2001). It is possible that Bcl-xL expression may alter levels of acetyl-coA by regulating mitochondrial membrane permeability. Citrate carrier (CiC), a nuclear-encoded protein located in the mitochondrial inner membrane and a member of the mitochondrial carrier family, is responsible for the efflux of acetyl-CoA from the mitochondria to the cytosol in the form of citrate (Gnoni et al., 2009; Kaplan et al., 1993). We found that the levels of glucose-derived citrate were decreased by approximately 25% in Bcl-xL expressing cells relative to control. This reduction in citrate levels could explain the observed decrease in acetyl-CoA levels in Bcl-xL expressing cells and contribute to anti-apoptotic function of Bcl-xL. Indeed, addition of citrate to Bcl-xL expressing cells leads to increased protein N-alpha-acetylation and sensitization of these cells to apoptosis. Perturbations in acetyl-CoA production may extend to other oncogenic contexts beyond that of Bcl-xL. For example, the levels of acetyl-CoA pools derived from glucose were found to be approximately 20% higher in myc +/+ cells relative to myc −/− cells (Morrish et al., 2009). The ability of myc to increase in acetyl-CoA levels might contribute to enhanced apoptotic sensitivity of cells overexpressing c-Myc (Evan et al., 1992). We propose that the basal levels of acetyl-CoA may influence the apoptotic threshold in multiple oncogenic contexts.
The ability of Bcl-xL to control the levels of acetyl-CoA and protein-N-acetylation provides a clear example by which metabolism is mechanistically linked with apoptotic sensitivity. Loss-of-function ard1 mutant yeast are specifically defective in alpha-factor response but not to a-factor (Whiteway and Szostak, 1985), indicating that protein N-alpha-acetylation status can dictate specific cellular behavior or process. Since protein N-alpha-acetylation affects a large number of cellular proteins, we propose that metabolic regulation of this process exerts its control on cellular processes through regulating a group(s) of proteins rather than individual proteins. Ard1 deficient mammalian cells are defective in the activation of caspase-2, caspase-3 and caspase-9 in response to DNA damage (Yi et al., 2007). Consistently, the N-alpha-acetylation of multiple caspases, including caspase-2, caspase-3 and caspase-9 was reduced in Bcl-xL overexpressing cells. It is possible that defects in N-alpha-acetylation of multiple caspases, which may negatively regulate their activation, contribute collectively to the apoptosis resistance in Ard1 deficient cells and Bcl-xL overexpressing cells. Thus, the levels of acetyl-CoA that control the N-alpha-acetylation of multiple proteins involved in a pathway may collectively determine the specific outcome of the cell fate. In this regard, the role of acetyl-CoA is clearly beyond what a cofactor normally serves: we propose that acetyl-CoA is a signalling molecule that provides an important liaison between metabolism and multiple cellular processes.
Experimental Procedures
Cell Death Assays
For RNAi studies, low passage HeLa cells were transiently transfected (5 × 105/well in 96-well plates) with a pool of four siRNAs (pre-designed ON-TARGETplus siRNAs available from Dharmacon, 50nM) using Oligofectamine transfection reagent (Invitrogen). Following a 48-hour incubation, cells were treated with doxorubicin (concentrations and incubation times noted in figure legends). siRNAs were tested in triplicate for each independent experiment. For detection of caspase cleavage by western blot, HeLa cells were transfected as described above (1.5 × 105/well in 6-well plates) followed by treatment with doxorubicin. Cells were lysed directly in SDS samples buffer and subjected to SDS-PAGE and western blot analysis using standard procedures.
For metabolite sensitization experiments, HeLa cells stably expressing GFP or Bcl-xL (3 × 105/well in 96-well plates) were pre-treated with acetate (50mM) or citrate (10m) for 24 hours followed by treatment with doxorubicin as indicated.
Cell viability was determined by measuring cellular ATP levels (CellTiter-Glo Luminescent Cell Viability Assay, Promega). Caspase-3/7 activity was quantified using a luciferin-labeled DEVD peptide substrate (Caspase-Glo 3/7 Assay, Promega). Luminescence was measured using a Wallac Victor2 plate reader.
Subtiligase Biotinylation Experiments
Synthesis of peptide substrate
The biotinylated peptide substrate for subtiligase with a TEV protease cleavage site, biotin-ahx-ahx-GGTENLYFQSY-glc-Y-NH2, was synthesized manually on a rink amide MBHA resin according to standard 9-Fluorenylmethoxycarbonyl (Fmoc) chemistry-based solid phase peptide synthesis protocols (Mahrus et al., 2008). The crude peptide was purified using a C18 semi-preparative reverse phase column on a Waters HPLC system. The identity of the purified product was confirmed by ESI-MS (isotopic MW calculated 1949.9 Da, m/z [M+H]+ found 1951.0).
The peptide substrate could be made more soluble by incorporating D-arginine residues into the sequence (Yoshihara et al., 2008). So a more soluble form of the substrate, biotin-ahx-ahx-dRdRdR-ahx-ahx-GGTENLYFQSY-glc-Y-NH2, was also synthesized, and its integrity was verified by HPLC and ESI-MS (MW calculated 2644.4 Da, m/z [M+H]+ found 2645.8).
Expression and purification of subtiligase
The expression construct for subtiligase was prepared using the plasmid pBS42 (ATCC 37279) according to published procedures (Wells et al., 1983) except that a His6 tag was added to the C-terminus (Tan et al., 2007). The gene of the subtiligase variant contains point mutations S221C, P225A, M124L and S125A on wild-type subtilisin BPN′. The recombinant protein was expressed in B. subtilis RIK1285, which is deficient in the production of neutral and alkaline proteases. Purification of subtiligase was performed as previously described (Abrahmsen et al., 1991) except that a Co2+ affinity chromatography step was used instead of ion-exchange chromatography (Tan et al., 2007). The affinity-purified subtiligase was desalted using a PD-10 column (Amersham) with deionized H2O. Aliquots of the desalted enzyme solution were flash-frozen, lyophilized and stored at −80°C until utilized. The prepared subtiligase was analyzed by SDS-PAGE gel and MALDI-TOF MS, which confirmed its purity and identity. The ligase activity of the enzyme preparation was confirmed in model ligation reactions using known peptide substrates for subtiligase.
Subtiligase ligation reaction
Cells used for subtiligase assay were plated at 50–60% confluency to support exponential growth followed by pre-treatment with acetate (50mM) or citrate (10mM) for 24 hours as indicated. Cells were lysed in 0.2% Tween 20 and 0.2% Triton X-100 buffer, and resulting lysates were used for subtiligase reaction that includes 1mM purified subtiligase, 1mM purified biotin-peptide, and 2mM DTT as previously described (Mahrus et al., 2008). Reactions were allowed to proceed for 1 hour at room temperature. Biotinylated proteins were affinity purified with Neutravidin agarose at 4° overnight (Thermo Scientific). The following day, agarose was pelleted and washed three times in lysis buffer. Purified proteins were eluted directly in 2X SDS sample buffer and eluants were analyzed by SDS PAGE.
13C Mass Spectrometry Sample Preparation
3 replicates of Jurkat cells stably expressing GFP or Bcl-xL (5×106 for each replicate) were washed twice in PBS and resuspended in RPMI medium with glutamine, 10% dialyzed NCS, and 10mM uniformly labeled 13C-glucose (Cambridge Isotope Laboratories) followed by incubation for 24 hours. Cells were washed twice, and metabolites were extracted in 3mL 80% ice-cold methanol. Insoluble material in lysates was pelleted at 13,000g for 10 minutes and methanol from resulting supernatant was evaporated. Samples were resuspended using 20µL HPLC grade water for mass spectrometry.
12C and 13C Mass Spectrometry Data Acquisition and Analysis
7µL of 20µL were injected using a 5500 QTRAP mass spectrometer (Applied Biosystems/MDS Sciex) equipped with a Prominence UFLC HPLC system (Shimadzu) via single reaction monitoring (SRM) of a total of 249 endogenous metabolites for 12C analyses of GFP and BcL-xL samples. For analyses of 13C labeled GFP and BcL-xL samples, 153 endogenous metabolites were targeted via SRM. Reliable quantitative data are only acquired from approximately 60% of the targeted metabolites (~160 metabolites from the 12C method and ~55% from the 13C method (~80 metabolites). Some metabolites were targeted in both positive and negative ion mode including acetyl-CoA. ESI voltage was 5000V in positive ion mode and −4500V in negative ion mode. The dwell time was 5ms per SRM transition and the collision energy was optimized for each SRM transition. Total cycle time was 2.09 seconds for the 12C method and 1.24 seconds for the 13C method. Scheduled SRMs were not utilized. Samples were delivered to the MS via normal phase chromatography using a 2.0 mm i.d × 10 cm HILIC Luna NH2 column (Phenomenex) at 250µL/min at basic pH using positive and negative ion switching within the same 30 minute LC/MS/MS analytical run. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0–5 minutes; 42% B to 0% B from 5–16 minutes; 0% B was held from 16–24 minutes; 0% B to 85% B from 24–25 minutes; 85% B was held for 7 minutes to re-equilibrate the column. Buffer A was comprised of 20mM ammonium hydroxide/20mM ammonium acetate in 95/5 water/acetonitrile. All metabolomic measurements were performed in triplicate. Peak areas from the total ion current (TIC) for each metabolite SRM transition were integrated using MultiQuant v1.1 software (Applied Biosystems).
Statistics
Statistical analysis was conducted using unpaired, two-tailed Student’s t-test.
Highlights.
N-alpha-acetylation of caspase-2 is required for binding to RAIDD.
Protein N-alpha-acetylation is sensitive to acetyl-CoA availability.
Acetyl-CoA levels are reduced in Bcl-xL expressing cells.
The ability of Bcl-xL to reduce acetyl-CoA levels contributes to apoptotic resistance.
Supplementary Material
Acknowledgements
We are indebted to Alfred Goldberg, Tom Rapoport, Andrew Steele, Marta Lipinski, and Benedicte Py for critical reading of the manuscript, to Pere Puigserver, Harvey Lodish, Kristin White, and Danny Chou for helpful discussions and to Xuemei Yang for help in acquiring mass spectrometry metabolomics data. We thank Thomas Arnesen and Johan Lillehaug (University of Bergen) for anti-NATH as well as with help in designing caspase-2 N-alpha-acetylation mutants. We also thank Tullia Lindsten and Craig Thompson (U Penn) for providing anti-Bcl-xL as well as Bcl-xL +/+ and Bcl-xL −/− MEFs. VSV-RAIDD and Bcl-xL mutant constructs were generously provided by Jurg Tschopp (University of Lausanne) and Emily Cheng (Wash U), respectively. This work was supported, in part, by an Innovator’s Award from the U.S. Army Breast Cancer Research Program (grant DAMD17-02-1-0403), a grant from the National Institute of Aging (R37-012859) and a NIH Director’s Pioneer Award to J. Yuan, NIH grants (DK070299 and GM47467) to G. Wagner, NIH grant (5P01CA120964-03) to J.M. Asara, and a Kirchstein National Research Service Award from the National Institute of Neurological Disorders and Stroke (grant 1F31NS057872-01) to C.H. Yi.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahmsen L, Tom J, Burnier J, Butcher KA, Kossiakoff A, Wells JA. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ametzazurra A, Larrea E, Civeira MP, Prieto J, Aldabe R. Implication of human N-alpha-acetyltransferase 5 in cellular proliferation and carcinogenesis. Oncogene. 2008;27:7296–7306. doi: 10.1038/onc.2008.332. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Thompson PR, Varhaug JE, Lillehaug JR. The protein acetyltransferase ARD1: a novel cancer drug target? Curr Cancer Drug Targets. 2008;8:545–553. doi: 10.2174/156800908786241113. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM. Bax-independent inhibition of apoptosis by Bcl-XL. Nature. 1996;379:554–556. doi: 10.1038/379554a0. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new 'death' adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Gnoni GV, Priore P, Geelen MJ, Siculella L. The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB Life. 2009;61:987–994. doi: 10.1002/iub.249. [DOI] [PubMed] [Google Scholar]
- Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, et al. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000236. e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Vander Heiden MG, Thompson CB. Bcl-x(L) prevents the initial decrease in mitochondrial membrane potential and subsequent reactive oxygen species production during tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2000;20:5680–5689. doi: 10.1128/mcb.20.15.5680-5689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JL, Huang SY, Chow NH, Chen SH. Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- Kaplan RS, Mayor JA, Wood DO. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J Biol Chem. 1993;268:13682–13690. [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Patel Y, Jones D, Howell S, Robinson K, Aitken A. Antibodies against the major brain isoforms of 14-3-3 protein. An antibody specific for the N-acetylated amino-terminus of a protein. FEBS Lett. 1993;331:296–303. doi: 10.1016/0014-5793(93)80356-y. [DOI] [PubMed] [Google Scholar]
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, Meinnel T. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt LK, Buchakjian MR, Gan E, Darbandi R, Yoon SY, Wu JQ, Miyamoto YJ, Gibbon JA, Andersen JL, Freel CD, et al. Metabolic control of oocyte apoptosis mediated by 14-3-3zeta-regulated dephosphorylation of caspase-2. Dev Cell. 2009;16:856–866. doi: 10.1016/j.devcel.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt LK, Margolis SS, Jensen M, Herman CE, Dunphy WG, Rathmell JC, Kornbluth S. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem Biophys Res Commun. 2003;308:1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- Starheim KK, Arnesen T, Gromyko D, Ryningen A, Varhaug JE, Lillehaug JR. Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem J. 2008;415:325–331. doi: 10.1042/BJ20080658. [DOI] [PubMed] [Google Scholar]
- Starheim KK, Gromyko D, Velde R, Varhaug JE, Arnesen T. Composition and biological significance of the human Nalpha-terminal acetyltransferases. BMC Proc. 2009;3 Suppl 6:S3. doi: 10.1186/1753-6561-3-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AC, Triscari J, Hamilton JG, Miller ON. Effect of (−)-hydroxycitrate upon the accumulation of lipid in the rat. II. Appetite. Lipids. 1974a;9:129–134. doi: 10.1007/BF02532137. [DOI] [PubMed] [Google Scholar]
- Sullivan AC, Triscari J, Hamilton JG, Miller ON, Wheatley VR. Effect of (−)-hydroxycitrate upon the accumulation of lipid in the rat. I. Lipogenesis. Lipids. 1974b;9:121–128. doi: 10.1007/BF02532136. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tan XH, Wirjo A, Liu CF. An enzymatic approach to the synthesis of peptide thioesters: mechanism and scope. Chembiochem. 2007;8:1512–1515. doi: 10.1002/cbic.200700284. [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Schumacker PT, Thompson CB. Bcl-xL prevents cell death following growth factor withdrawal by facilitating mitochondrial ATP/ADP exchange. Mol Cell. 1999;3:159–167. doi: 10.1016/s1097-2765(00)80307-x. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Li XX, Gottleib E, Hill RB, Thompson CB, Colombini M. Bcl-xL promotes the open configuration of the voltage-dependent anion channel and metabolite passage through the outer mitochondrial membrane. J Biol Chem. 2001;276:19414–19419. doi: 10.1074/jbc.M101590200. [DOI] [PubMed] [Google Scholar]
- Vaughn AE, Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat Cell Biol. 2008;10:1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, Ferrari E, Henner DJ, Estell DA, Chen EY. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983;11:7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Szostak JW. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985;43:483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, Yuan J. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J Cell Biol. 2007;179:619–626. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara HA, Mahrus S, Wells JA. Tags for labeling protein N-termini with subtiligase for proteomics. Bioorg Med Chem Lett. 2008;18:6000–6003. doi: 10.1016/j.bmcl.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.