Abstract
Currently, little is known about the origin and early evolution of sex chromosomes. This is largely due to the fact that ancient non-recombining sex chromosomes are highly degenerated, and thus provide little information about the early genomic events in their evolution. The Neurospora tetrasperma mating-type (mat) chromosomes contain a young (<6 Mya) and large region (>6.6 Mb) of suppressed recombination, thereby providing a model system to study early stages of sex chromosome evolution. Here, we examined alleles of 207 genes located on the N. tetrasperma mat a and mat A chromosomes to test for signs of genomic alterations at the protein level in the young region of recombination suppression. We report that the N. tetrasperma mat a and mat A chromosomes have each independently accumulated allele-specific non-synonymous codon substitutions in a time-dependent, and gene-specific manner in the recombinationally suppressed region. In addition, examination of the ratio (ω) of non-synonymous substitutions (dN) to synonymous substitutions (dS) using maximum likelihood analyses, indicates that such changes are associated with relaxed purifying selection, a finding consistent with genomic degeneration. We also reveal that sex specific biases in mutation rates or selection pressures are not necessary for genomic alterations in sex chromosomes, and that recombination suppression in itself is sufficient to explain these results. The present findings extend our current understanding of genomic events associated within the young region of recombination suppression in these fungal sex-regulating chromosomes.
Keywords: N. tetrasperma, recombination suppression, mat chromosomes, degeneration, selection
Introduction
The early stages of sex chromosome evolution are currently poorly understood. The reason for this is that most ancient sex chromosomes are highly deteriorated and retain little information about the early stages of genomic differentiation and degeneration (Charlesworth and Charlesworth, 2000). Thus, model systems of young sex chromosomes, that is, systems with newly established regions of recombination suppression, are necessary to study early steps in sex chromosome evolution (Charlesworth et al., 2005). One emerging model system is the Neurospora tetrasperma mating type (mat) chromosomes.
N. tetrasperma is a self-fertile (pseudohomothallic) filamentous ascomycete assumed to have evolved from a self-incompatible (heterothallic) ancestor (Raju and Perkins, 1994; Dettman et al., 2003). As in most filamentous ascomycetes, the N. tetrasperma mat chromosomes contain the mat locus, which is comprised of two highly dissimilar alleles (mat a and mat A idiomorphs) that regulate reproduction and sexual compatibility (Shiu and Glass, 2000; Casselton, 2008). The two mat chromosomes of N. tetrasperma are found in separate haploid nuclei, of which both are present in the sexual ascospores and in vegetative cells, consistent with a predominant heterokaryotic and self-fertile state (Shear and Dodge, 1927; Jacobson, 1995). This feature of N. tetrasperma differs from closely related heterothallic Neurospora species, such as N. crassa, which is haploid and requires two distinct partners containing nuclei of opposite type for mating and sexual reproduction (Shiu and Glass, 2000). The pathway of meiosis and spore development leading to the packaging of nuclei carrying the two opposite mating types into single ascospores of N. tetrasperma is highly novel (Raju and Perkins, 1994). In association with the evolution of a pseudohomothallic mating system, the N. tetrasperma mat chromosomes have evolved the unusual feature that they contain a young and large region of suppressed recombination surrounding the mat locus (<6 Mya and >6.6 Mbp, respectively) (Menkis et al., 2008), which share marked parallels with dimorphic plant and animal sex chromosomes. Specifically, the region of suppressed recombination is located in the central region of the mat chromosomes and surrounds the mat locus (Merino et al., 1996; Menkis et al., 2008). This region is flanked by two normally recombining pseudoautosomal regions, contain genes with markedly diverged alleles, and is comprised of at least two distinct evolutionary strata, that is, recombinationally suppressed regions that have developed successively over time (Fraser and Heitman, 2004, 2005; Menkis et al., 2008). Thus, the N. tetrasperma mat chromosomes provide a model system for the study of genomic changes inherent to young regions of recombination suppression in sex-regulating chromosomes (Menkis et al., 2008).
In ancient sex chromosomes, massive genomic degeneration has occurred on the permanently heterozygous chromosome. This degeneration is believed to be because of a reduced efficiency of natural selection; non-independence of linked loci is expected to reduce the effective population size of the non-recombining chromosome (Hill-Robertson Effect; Hill and Robertson, 1966; Gordo and Charlesworth, 2001), thus decreasing the efficiency of both positive and negative selection, and enhancing the role of genetic drift (Charlesworth and Charlesworth, 2000). The fixation of accumulated mutations on non-recombining regions may be enhanced by background selection (a net effect of accelerating the fixation of mildly deleterious mutations and the loss of positive mutations (Charlesworth and Charlesworth, 2000)) and/or by Muller's Ratchet (wherein mutations accumulate in an asexual manner, with the stochastic losses of chromosome classes carrying the fewest deleterious mutations (Muller, 1964)). It is also plausible that positive selection contributes to degeneration in the recombinationally suppressed regions by dragging deleterious mutations at linked alleles/sites to fixation through selective sweeps (Rice, 1987; Charlesworth and Charlesworth, 2000; Bachtrog, 2004). To date, only limited data has been available regarding evolution of young sex chromosomes, primarily from Silene (Marais et al., 2008) and from the neo-X/Y chromosomes of Drosophila (Bachtrog, 2003, 2004). Each of these studies has shown evidence consistent with genomic degeneration. Further data regarding the onset of genomic changes in young sex regulating chromosomes may be revealed from the study of the recently established region of recombination suppression within the N. tetrasperma mat chromosomes.
In protein coding genes, selective constraint can be measured using the ratio of non-synonymous to synonymous substitution rates over evolutionary time (dN/dS, or ω) (Yang, 2007). Specifically, values of ω<1, ω=1 and ω>1 indicates purifying selection, neutral evolution and positive selection, respectively (Yang and Nielsen, 2002). Methods have been developed based on maximum likelihood analyses that allow the assessment of ω among individual lineages in a phylogeny (Branch Model analyses, Yang, 1998; Yang, 2007). This approach may be particularly useful for the identification of variation in selective pressures inherent to the young region of recombination suppression in the N. tetrasperma mat chromosomes, that is, constraints that have changed after the divergence from a heterothallic ancestor with normal recombination rates.
In this study, we investigated DNA sequence divergence in the young recombinationally suppressed region of the N. tetrasperma mat chromosomes. We examined 207 genes located on the mat chromosomes of N. tetrasperma and its close normally recombining relatives N. crassa and N. discreta. The main objective was to reveal the frequency of allele-specific non-synonymous and synonymous changes in the recombinationally suppressed region as compared with the normally recombining pseudoautosomal regions. In addition, we conducted branch-model analysis of ω to reveal the variation in evolutionary constraints within and among N. tetrasperma mat chromosomes.
Materials and methods
Several traits of N. tetrasperma mat chromosomes have been previously established, and thus, were assumed in our present analysis. Most importantly, restricted recombination in the central segment of the N. tetrasperma mat chromosomes has been repeatedly demonstrated in scientific analyses, and was assumed for our study. Specifically, recombination suppression has been demonstrated by cytological inspection of the mat chromosomes during meiosis (Gallegos et al., 2000), by analysis of genetic marker segregation in N. tetrasperma wild crosses (Gallegos et al., 2000), and by mat chromosome introgressions between N. crassa and N. tetrasperma in the laboratory (Howe and Haysman, 1966; Jacobson, 2005). Furthermore, population level analyses have revealed that gene alleles located in the recombinationally suppressed region are linked to each other and to the mat a or the mat A chromosome in N. tetrasperma (Merino et al., 1996; Gallegos et al., 2000; Jacobson, 2005). Finally, a key indicator of recombination suppression is that alleles in this region have undergone substantial sequence divergence (Lahn and Page, 1999; Handley et al., 2004; Sandstedt and Tucker, 2004), which is not observed in the normally recombining flanking pseudoautosomal regions, where alleles are nearly completely homogenous (Merino et al., 1996; Menkis et al., 2008, 2009).
We also assumed a conserved gene order between the studied species. It has been reported that the gene order in N. tetrasperma and N. crassa is highly conserved and collinear, including the mat chromosomes (Howe and Haysman, 1966; Perkins, 1985; Jacobson, 2005). Thus, the gene order for N. tetrasperma mat chromosomes examined herein was based on the known order for N. crassa, as has proven effective in previous studies (for example, Gallegos et al., 2000). The gene order for N. crassa is available from MapView (National Center for Biotechnology Information, NCBI, http://www.ncbi.nlm.nih.gov/mapview/).
For our investigation, we obtained sequences for N. crassa and N. discreta available from the N. crassa database (FGSC 2489, Annotation Version 3 http://www.broad.mit.edu/annotation/ genome/neurospora/; the mRNA was also accessed using the RefSeq database at NCBI, http://www.ncbi.nlm.nih.gov/RefSeq/) and from the Joint Genome Initiative (http://www.jgi.doe.gov/), respectively. The data for N. tetrasperma was generated using Solexa technology and were assembled as described below.
Solexa data assembly of N. tetrasperma strain P4492
For our study, we gathered genomic sequence data from each of the two separate homokaryotic single mating-type component strains (that is, mat a and mat A) of the N. tetrasperma heterokaryon P4492. The strain P4492 originates from the Perkins collection of Neurospora from nature, curated by the Fungal Genomics Stock Center (FGSC), University of Missouri (FGSC ID for the mat a component strain is FGSC 9034, and for mat A is FGSC 9033) and has previously been shown to be contained within phylogenetic lineage 1 of the N. tetrasperma species complex (Menkis et al., 2009). The N. tetrasperma mat a and mat A strains/chromosomes are referred to in the present study as NTa and NTA, respectively. Genomic sequences were obtained separately for the NTa and the NTA strain by Solexa technology by Geneservice, Source BioScience plc group (http://www.geneservice.co.uk/home/). The 55 bp paired-end DNA library was constructed with insert size ∼170 bp. The sequencing process followed the protocol instruction for Genome Analyzer II, and the image from the Genome Analyzer II were processed by GA pipeline software in the purpose of base calling. The 55 bp paired-end sequence reads associated with the mat chromosomes were identified and assembled through sequence similarity searches relative to the Linkage Group I (mat chromosomes) for N. tetrasperma FGSC 2508 (available from the Joint Genome Institute, http://www.jgi.doe.gov) using the MAQ (Mapping and Assembly with Qualities) software (http://maq.sourceforge.net/maq-man.shtml). We set the maximum number of mismatches to three in the process of mapping raw reads to the reference N. tetrasperma genome. Consensus sequences were extracted with the minimum read depth of three. The average depth (coverage) across all non-gap regions calculated by MAQ mapcheck module were 12.54 (NTA) and 11.73 (NTa), which represents standard levels for a genomic survey analysis. The average base error probability was lower than 0.24% (NTA) and 0.15% (NTa), thus demonstrating very high quality sequencing and base calling (calculated based on the Illumina quality). The Solexa DNA sequence covered more than 69 and 70% of the ∼10 Mbp mat chromosomes for NTa and NTA, respectively; the fraction not covered (⩽31%) may reflect bias in library construction or sequence amplification. Nonetheless, the fraction of sequences obtained was highly accurate (⩾99.85% accuracy). The sequences include genomic regions identified as coding DNA, intergeneric regions and introns.
Identification of genes
N. crassa has historically served as a highly studied model system for fungi, and genes for this species have been annotated (FGSC 2489 http://www.broad.mit.edu/annotation/genome/neurospora/). Accordingly, the gene sequences and annotation for this species were used as a reference for gene identification in N. tetrasperma and N. discreta, whose genomes have not previously been fully annotated. For gene identification, we compared genes (mRNA) from the N. crassa sequence database to the DNA sequences for linkage group I of N. tetrasperma and to the putative mRNA dataset of N. discreta.
For N. tetrasperma, the available sequence for NTa and NTA linkage group I was compared with the mRNA dataset from N. crassa. We first identified 2309 N. crassa mRNA associated with the mat chromosomes, which have been defined by an NCU number at the Neurospora crassa database. An NCU ID has been assigned for well defined genes, and those encoding hypothetical and predicated proteins (FGSC 2489 http://www.broad.mit.edu/annotation/genome/neurospora). Subsequently, the available sequence from linkage group I sequence for NTa and for NTA were each defined as a database and compared with the N. crassa mRNA gene set using BLASTN (http://www.ncbi.nlm.nih). Note that X's were assigned for regions not covered by the Solexa sequencing. The N. crassa gene with the lowest e-value (with a cutoff of e⩽10−6) for a particular region of NTa and for NTA linkage group I was considered a match. The genomic region contained within and surrounding each match for NTa and for NTA was isolated and manually aligned to the matching N. crassa mRNA using CLUSTALW (Thompson et al., 1994). Introns and/or indel sequences identified within the NTa and NTA sequence from the alignment were removed. Genomic regions from NTa and NTA, which matched an entire mRNA from N. crassa, including the start codon, the stop codon and which did not contain unknown or ambiguous nucleotides were identified for our analysis. Each NTa and NTA gene was translated to ensure the accurate identification of reading frames. For N. discreta, the putative open reading frames from JGI (generated from assembled ESTs and genomic data) were compared against the 2309 genes from N. crassa using TBLASTX (http://www.ncbi.nlm.nih.gov), and the gene with the lowest e-value was used for identification (with a cutoff of e⩽10−6).
Alignments were conducted between the gene sequences for NTa, NTA, N. crassa and N. discreta, with N. crassa used as the template for the identification of all open reading frames. Only genomic regions in NTa, NTA, N. crassa and N. discreta fully corresponding to regions in N. crassa were included in analysis. Thus, the genes identified herein are not always the complete mRNA regions (for example, there may be an insertion in NTA, NTa or N. discreta that is not inherent to N. crassa), and do not include premature stop codons, but nonetheless we refer to them using the term ‘genes' herein. Altogether, we identified 207 genes in NTa, NTA and N. discreta that fully match the complete mRNA sequence from N. crassa, which were used in our investigation. The DNA sequences for the N. tetraperma mat a and mat A linked alleles for each of the 207 genes identified herein are available in FASTA format at Dryad (http://dx.doi.org/10.5061/dryad.8429; note that the sequence names that end in P4492_a and P4492A in this file are from the mat a and mat A chromosomes, respectively). The reads data are available from the Sequence Reads Archive at NCBI (http://www.ncbi.nlm.nih.gov/sra/) under the accession numbers SRR099222 (P4492_a) and SRR099270 (P4492A).
The location of each of the 207 genes on the N. tetrasperma mat chromosomes was determined using the gene order for N. crassa. Genes were subsequently categorized as being located in stratum 1, stratum 2, pseudoautosomal region 1 or pseudoautosomal region 2. These categorizations were based on demarcation points within the mat chromosomes previously reported by Menkis et al. (2008) (see Supplementary Table S1). These previous findings, dividing the regions of the chromosome into stratum 1, stratum 2 and the PA regions, were based on the fact that gene alleles are identical or nearly identical in normally recombining genomic regions in a highly inbred taxon such as N. tetrasperma (Menkis et al., 2008) and that that genes located in the recombinationally suppressed regions have greater synonymous divergence among their alleles over time (Lahn and Page, 1999; Handley et al., 2004; Sandstedt and Tucker, 2004; Menkis et al., 2008).
Synonymous and non-synonymous nucleotide substitution rates
For each of the 207 genes identified above, we determined the synonymous (dS or Ks) and non-synonymous (dN or Ka) substitution rates among the mat a and mat A chromosomes in N. tetrasperma. The values were determined for genes located in stratum 1, stratum 2, and for each of the two pseudoautosomal regions. Substitution rates were determined using DNASP (Librado and Rozas, 2009).
Identification of allele-specific codon substitutions
To more fully understand the dynamics of the mat chromosome divergence, we examined each of the 82 793 codon positions spanning all 207 genes to identify allele-specific codon substitutions located on the N. tetrasperma mat a and mat A chromosomes. Allele-specific codon substitutions were utilized to reveal whether there has been independent accumulation of genomic changes on the mat a and mat A chromosomes. For this analysis, we identified each codon position wherein N. crassa and one N. tetrasperma mat linked allele contained the same codon (ancestral codon), and the other N. tetrasperma mat linked allele contained a different codon (note that N. crassa is more closely related to N. tetrasperma than N. discreta (Dettman et al., 2003), therefore N. crassa was used for this purpose). Allele-specific codon substitutions that were associated with the same amino acid on both N. tetrasperma mat chromosomes were identified as allele-specific synonymous codon substitutions. Allele-specific codon substitutions that were associated with a change in the amino acid on one N. tetrasperma mat chromosome and not the other were identified as allele-specific non-synonymous codon substitutions. For comparisons among genes and genomic regions, the number of allele-specific codon substitutions was standardized per 1000 sites as follows: number of allele-specific codon substitutions/total number of codons examined × 1000.
In addition to the analysis of allele-specific codon substitutions, we also assessed all possible categories of codon substitutions among the N. tetrasperma mat chromosomes. These include codon positions showing no evidence of substitutions, those with substitutions that were identical among chromosomes, and those positions having different codons on both mat chromosomes.
Branch model analyses
Differences in ω between the species over evolutionary time were investigated by using the codeml program of phylogenetic analysis by maximum likelihood (PAML; Yang, 2007). For this, the branch-model mode (model=2, NS=0) was utilized to test for differences in ω in the NTa and NTA branches (as a group) relative to the N. crassa and N. discreta branches, the latter two of which exhibit free recombination along the mat chromosomes. The tree files used for this comparison were ((NTa no. 1, NTA no. 1), N. crassa no. 1, N. discreta no. 1) versus ((NTa no. 1, NTA no. 1), N. crassa no. 2, N. discreta no. 2), wherein different numbers within a single tree file represent lineages permitted to have different ω values. Likelihood-ratio tests were conducted among the two models (df=1). An elevated ω for the branches for NTa and NTA was tested both for concatenated data of the region of suppressed recombination and for the pseudoautosomal regions; the latter case was used to reveal whether the pseudohomothallic N. tetrasperma mating system/life history in itself is associated with an elevated ω as compared with the heterothallic species N. crassa and N. discreta, or whether differences in ω are only associated with the N. tetrasperma recombinationally suppressed regions.
Branch-model analysis was also used to reveal differences in ω among N. tetrasperma mat a and mat A linked alleles for each of the individual 207 genes examined in this study. For this, two models were compared; one where two distinct ω were permitted for the NTa and NTA branches, and another with a single ω for the NTa and NTA branches. The tree files utilized in phylogenetic analysis by maximum likelihood for these analyses were ((NTa no. 1, NTA no. 1), N. crassa, N. discreta) versus ((Nta no. 1, NTA no. 2), N. crassa, N. discreta).
Results
Identification and characterization of chromosomal regions
A total of 207 genes were identified for our study. On the basis of the demarcations within the N. tetrasperma mat chromosomes reported by Menkis et al. (2008), we found that 168 genes are located in the recombinationally suppressed region of the N. tetrasperma mat chromosomes. Among these, 27 genes are located in the younger stratum 2 (11 631 codon positions) and 141 genes in the older stratum 1 (54 506 codon positions). A total of 39 genes are located in the pseudoautosomal regions, with 18 genes in pseudoautosomal region 1 (5618 codons) and 21 genes in pseudoautosomal region 2 (11 038 codons) (Table 1 and Supplementary Table S1). We found that the mean dS values among the N. tetrasperma mat a and mat A-linked alleles were markedly higher for genes located in stratum 1 (mean (std error (s.e.))Stratum 1=0.0190 (0.0015)) than for genes located in stratum 2 (mean (s.e.)Stratum 2=0.0046 (0.0018)), and was lowest in the pseudoautosomal regions (equal or near equal to zero (mean (s.e.)PA1=0.0003 (0.0003), mean (s.e.)PA2=0 (0)) (Table 1); this pattern is consistent with previous findings by Menkis et al. (2008) of a greater allelic sequence divergence with extended time periods of recombination suppression. The genes from the pseudoautosomal regions, and from the recombinationally suppressed stratum 1 and stratum 2 regions are listed in sequential order that they occur on the mat chromosomes in Supplementary Table S1.
Table 1. The number of genes located on the mat chromosomes of Neurospora crassa, N. discreta and N. tetrasperma that were identified and examined in the present study.
| Mat chromosomal region | Number of genes identified | Total number of codon positions | Mean dS NTa vs NTA per gene (std error) | Mean dN NTa vs NTA per gene (std error) |
|---|---|---|---|---|
| PA1 | 18 | 5618 | 0.0003 (0.0003) | 0 (0) |
| Stratum 2 | 27 | 11 631 | 0.0046 (0.0018) | 0.0009 (0.0004) |
| Stratum 1 | 141 | 54 506 | 0.0190 (0.0015) | 0.0019 (0.0002) |
| PA2 | 21 | 11 038 | 0 (0) | 0 (0) |
| Total | 207 | 82 793 |
Abbreviations: Nta, N. tetrasperma mat a; NTA, N. tetrasperma mat A; PA1, pseudoautosomal region 1; PA2, pseudoautosomal region 2.
The dN and dS values among the N. tetrasperma mat a and mat A-linked alleles are provided. A total of 207 genes were examined.
The mean dN value among the N. tetrasperma mat a and mat A-linked gene alleles was markedly lower than dS values for genes in stratum 1 (mean (s.e.)Stratum 1=0.0019 (0.0002), representing about 10% of dS. The effect is magnified in stratum 2, where dN was only 2% of dS; in addition, the mean values of dN are markedly lower (mean (s.e.)Stratum 2=0.0009 (0.0004)) and the variation is much higher in stratum 2 than stratum 1 (s.e. is 44% of the mean in stratum 2). No non-synonymous substitutions were detected in the normally recombining pseudoautosomal regions, consistent with normal recombination and selection pressures. In totality, these data suggest that marked time-dependent protein sequence divergence has occurred among alleles of genes located in stratum 1 and stratum 2 (that is, higher divergence in stratum 1 than stratum 2), while no such differences are inherent to the pseudoautosomal regions (Table 1).
Mat allele-specific protein evolution
To further reveal the dynamics of the mat chromosome divergence (Table 1), we examined each of the 82 793 codon positions spanning all 207 genes to identify codon substitutions located on the N. tetrasperma mat a and mat A chromosomes (relative to N. crassa). The data show that the vast majority of codon positions in stratum 1, stratum 2 and the pseudoautosomal regions show no evidence of codon changes (>932 codon positions per 1000; see category 1 in Table 2). This is not surprising given that these species are closely related, with <6 Mya as divergence from a common ancestor (believed to approximately correspond to the formation of stratum 1, Menkis et al., 2008). We also identified numerous codon substitutions that were shared among the N. tetrasperma mat chromosomes relative to N. crassa (Categories 2 and 3 in Table 2). These types of homoallelic changes could result from a codon switch in either the N. tetrasperma or the N. crassa lineages. We noted that the level of homoallelic synonymous and non-synonymous substitutions each decreased in the following manner: PA >stratum 2 >stratum 1 (Table 2). This trend is coincident with an uninterrupted history of normal recombination in the pseudoautosomal regions and a longer history of normal recombination in stratum 2 than in stratum 1 of the N. tetrasperma mat chromosomes. Thus, many of these homoallelic substitutions in stratum 1 and in stratum 2 likely originated in the N. tetrasperma lineage and were fixed in the time period before recombination suppression. Accordingly, these data are consistent with a greater frequency of homoallelic substitutions on the N. tetrasperma mat chromosomes with extended periods of normal recombination.
Table 2. The different categories (Cat) of synonymous and non-synonymous codon substitutions among the N. tetrasperma mat a and mat A chromosomes.
| Cat | mat Allele-specific substitution | Type of change | NC | NTa | NTA |
Pseudoautosomal region 1 |
Stratum 2 |
Stratum 1 |
Pseudoautosomal region 2 |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. codons | No. per 1000 codons | No. codons | No. per 1000 codons | No. codons | No. per 1000 codons | No. codons | No. per 1000 codons | ||||||
| 1 | No | None | Codon1 (AA1) | Codon1 (AA1) | Codon1 (AA1) | 5238 | 932.36 | 10 990 | 944.80 | 51 513 | 945.08 | 10 422 | 944.19 |
| 2 | No | Shared Synonymous | Codon1 (AA1) | Codon2 (AA1) | Codon2 (AA1) | 333 | 59.27 | 522 | 44.88 | 1712 | 31.41 | 515 | 46.66 |
| 3 | No | Shared Non-synonymous | Codon1 (AA1) | Codon2 (AA2) | Codon2 (AA2) | 45 | 8.01 | 81 | 6.96 | 363 | 6.66 | 101 | 9.15 |
| 4 | Yes, mat a | Non-synonymous | Codon1 (AA1) | Codon2 (AA2) | Codon1 (AA1) | 0 | 0 | 6 | 0.52 | 116 | 2.13 | 0 | 0 |
| 5 | Yes, mat A | Non-synonymous | Codon1 (AA1) | Codon1 (AA1) | Codon2 (AA2) | 0 | 0 | 13 | 1.12 | 117 | 2.15 | 0 | 0 |
| 6 | Yes, mat a | Synonymous | Codon1 (AA1) | Codon2 (AA1) | Codon1 (AA1) | 0 | 0 | 11 | 0.95 | 320 | 5.87 | 0 | 0 |
| 7 | Yes, mat A | Synonymous | Codon1 (AA1) | Codon1 (AA1) | Codon2 (AA1) | 2 | 0.35 | 8 | 0.69 | 350 | 6.42 | 0 | 0 |
| 8 | Yes (excluded)a | Non-synonymous Both | Codon1 (AA1) | Codon2 (AA2) | Codon3 (AA3) | 0 | 0 | 0 | 0 | 5 | 0.09 | 0 | 0 |
| 9 | Yes (excluded)a | Synonymous Both | Codon1 (AA1) | Codon 2 (AA1) | Codon3 (AA1) | 0 | 0 | 0 | 0 | 10 | 0.18 | 0 | 0 |
| Total number of codons | 5618 | 11 631 | 54 506 | 11 038 | |||||||||
Abbreviations: AA, Amino acid; Nta, N. tetrasperma mat a; NTA, N. tetrasperma mat A.
The type of substitution was determined based on comparison of codons and amino acids (AA) to the closely related and normally recombining mat chromosomes from N crassa (NC). Data is based on concatenated sequences for each genomic region. Allele-specific substitutions are in bold. Codons and amino acids followed by different numbers within a single category are indicative of a substitution.
Codon positions that have switched to different codons on both the mat a and mat A chromosomes were not included in allele-specific counts and were categorized separately.
The primary focus of our examination of concatenated sequences across all 82 793 codon positions was to identify allele-specific codon substitutions on the N. tetrasperma mat a and mat A chromosomes. These include codon positions wherein N. crassa and one N. tetrasperma mat-linked allele contained the same codon (ancestral codon), and the other N. tetrasperma mat-linked allele contained a different codon (Categories 4, 5, 6 and 7 in Table 2). Our results reveal evidence of the independent accumulation of protein level changes in the recombinationally suppressed region of the mat a and mat A chromosomes. For example, we found ∼2.13 allele-specific non-synonymous substitutions per 1000 codons in stratum 1 of the mat a chromosome (Table 2). A similar value of 2.15 was detected within the mat A chromosome (Table 2); these findings were attributable to different (allele-specific) substitutions on each chromosome. Similar findings were detected for stratum 2, wherein 0.52 and 1.12 allele-specific non-synonymous codon substitutions were identified per 1000 codons on the mat a and mat A chromosomes, respectively. This is consistent with independent accumulation of genomic changes in this chromosomal region. The level of allele-specific non-synonymous and allele-specific synonymous codon substitutions were each found to be markedly higher in stratum 1 than in stratum 2 (Table 2 Supplementary Figure S1), consistent with a longer time period of recombination suppression in stratum 1. Overall, these data further support the conclusion that the mat chromosomes have independently accumulated substitutions at the protein level in a time-dependent manner within the region of suppressed recombination.
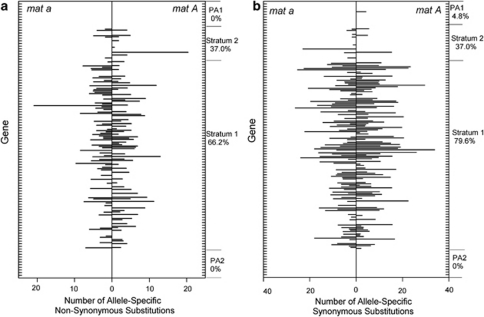
As a further analysis, we determined the frequency of allele-specific non-synonymous codon substitutions on the N. tetrasperma mat chromosomes for each of the 207 genes examined herein. These results reveal that 37% of genes in stratum 2 have at least one allele-specific non-synonymous codon substitution, similar to the fraction of genes with synonymous codon substitutions (not necessarily the same genes, Figure 1). In contrast, in stratum 1, 66.2% of genes contain at least one allele-specific non-synonymous codon substitution, which is marginally lower than for synonymous substitution (79%). No allele-specific non-synonymous substitutions have occurred in genes located in the pseudoautosomal regions (Figure 1). In sum, these data show that there has been time-dependent and independent accumulation of allele-specific non-synonymous codon substitutions on the mat a and on the mat A-linked alleles for the majority of genes located in the recombinationally suppressed region (Figure 1).
Figure 1.
The number of allele-specific codon substitutions per gene (per 1000 codons) on the N. tetrasperma mat a chromosome and on the mat A chromosome (based on comparison to N. crassa). (a) Number of allele-specific non-synonymous codon substitutions. (b) Number of allele-specific synonymous codon substitutions. The percentage of genes in each genomic region having at least one allele-specific substitution (on either of the mat chromosomes) is provided. Genes are listed on the Y axis in the order they occur on the mat chromosomes. The various segments of the mat chromosomes, including pseudoautosomal region 1 (PA1), stratum2, stratum 1 and pseudoautosomal region 2 (PA2) are indicated on the Y axis.
Correlation coefficients among allele-specific substitutions per gene in stratum 2 revealed a strong-positive correlation between non-synonymous and synonymous changes on the mat A chromosome (R=0.71, P=2.2 × 10−5), suggesting that many codon sites may be evolving neutrally in this region. No such correlation was found for the mat a chromosome in stratum 2 (R=−0.007 P=0.972), indicating that there may be marked differences in how the two mat chromosomes evolve in this genomic segment. Weaker, but statistically significant correlations were detected in stratum 1 (mat a R=0.236 P=0.004; mat A R=0.174 P=0.040), suggesting that there is a less direct and more complex relationship between synonymous and non-synonymous allele-specific codon substitutions in this genomic region, possibly reflecting variation in selection pressures.
Branch-model analysis
Phylogenetic analysis by maximum likelihood (PAML) branch-model analysis was utilized to assess whether the switch in mating-system and/or the onset of recombination suppression in the N. tetrasperma mat chromosomes (NTa and NTA as a group) are associated with changes in ω as compared with its close relatives N. crassa and N. discreta. The results show no differences in ω among N. tetrasperma (mat a and mat A lineages) and N. crassa and N. discreta for the concatenated gene sequences from the two normally recombining pseudoautosomal regions (Table 3). One would expect differences in the branch-model analysis in the pseudoautosomal region if mating system in itself (pseudohomothallism) alters ω in N. tetrasperma; thus, this finding suggests that mating system does not substantially alter ω. In contrast, however, we found that the concatenated gene sequences of the N. tetrasperma mat chromosomes have a statistically significantly higher ω value in the recombinationally suppressed regions, stratum 1 and stratum 2, than the homologous genes in the heterothallic and normally recombining species N. crassa and N. discreta (Table 3). Thus, the totality of these findings reveals that there is an elevated ω in the recombinationally suppressed regions of N. tetrasperma mat chromosomes as compared with the homologous normally recombining regions in N. crassa and N. discreta, and that this result is directly associated with recombination suppression in N. tetrasperma and not the pseudohomothallic mating system.
Table 3. Branch model analysis of ω for N. tetrasperma mat a and mat A (Nta, NTA) versus N. crassa and N. discreta (NC, ND).
| Chromosomal region | Direction of difference in ω | P-value |
|---|---|---|
| PA1 | NA | 0.354 |
| Stratum 1 | (NTa, NTA)>(NC, ND) | <10−4 |
| Stratum 2 | (NTa, NTA)>(NC, ND) | <10−4 |
| PA2 | NA | 1 |
Abbreviations: NA, not applicable; Nta, N. tetrasperma mat a; NTA, N. tetrasperma mat A; PA1, pseudoautosomal region 1; PA2, pseudoautosomal region 2.
P-values of likelihood ratio tests (LRTs) are provided.
Bold values are statistically significant.
We subsequently used branch-model analysis to assess differences in ω between N. tetrasperma mat a and mat A-linked alleles for all genes examined in this study. For the analysis, we focused on the older region of suppressed recombination, stratum 1 (N=141 genes), as the younger stratum 2 is insufficiently diverged among mat chromosomes to give reliable estimates among these chromosomes (Table 1; Yang, 2007). The results from the branch-model analysis of concatenated N. tetrasperma sequences for the mat a and mat A chromosomes for stratum 1 show that ω does not differ among the mat chromosomes (constrained versus unconstrained models P=0.3482, Table 4; note that only the N. tetrasperma branch lengths, which are being tested, are shown). This finding, of similar ω for both mat lineages does not suggest that there are not major differences among the mat a and mat A chromosomes, but rather suggests that ω has shifted independently and to similar levels on both chromosomes (for mat a and mat A, ω=0.1321 and 0.1132, respectively). That is, this can be inferred given that allele-specific non-synonymous substitutions are prevalent on both the mat a and on the mat A chromosomes in stratum 1 (Tables 1 and 2, Figure 1, Supplementary Figure S1). We also found that the ω values are markedly <1 in stratum 1 for each of the mat chromosomes, which is consistent with a history of purifying selection. Nonetheless, given that the values of ω in stratum 1 are markedly higher (ω>0.1121) than for the pseudoautosomal regions (wherein dN and thus ω=0, Tables 1 and 4), the results suggest that there has been relaxed selection and/or positive selection in the recombinationally suppressed region.
Table 4. Genes in stratum 1 with markedly different (P<0.1) dN/dS (ω) ratios among the Neurospora tetrasperma mat a and mat A-linked alleles based on the branch model analysis in PAML.
| Gene | Gene descriptiona | ωmat ab | ωmat Ab | Direction of difference | LRT P-value |
|---|---|---|---|---|---|
| Concatenated sequences of all stratum 1 genes | 0.1321 | 0.1142 | ωmat a>ωmat A | 0.3482 | |
| Specific genes with differences | |||||
| NCU02502 | Hypothetical protein similar to S-adenosyl-methionine-sterol-C | >0.0001 | 0.0001 | ωmat a>ωmat A | 0.0514 |
| NCU02510 | Clathrin heavy chain | 0.0001 | 0.3233 | ωmat A>ωmat a | 0.0427 |
| NCU02633 | Hypothetical protein similar to nuclear mRNA splicing factor | >0.0001 | 0.0001 | ωmat a>ωmat A | 0.0010 |
| NCU02675 | Hypothetical protein similar to RNA-3′-phosphate cyclase family protein | 0.0001 | 0.13 | ωmat A>ωmat a | 0.0558 |
| NCU02761 | Hypothetical protein | >0.0001 | 0.0001 | ωmat a>ωmat A | 0.0534 |
| NCU09382 | Predicted protein | 0.0001 | 0.6752 | ωmat A>ωmat a | 0.0496 |
| NCU09317 | Predicted protein | >0.0001 | 0.0001 | ωmat a>ωmat A | 0.067 |
| NCU09310 | Conserved hypothetical protein | >0.1311 | 0.1311 | ωmat a>ωmat A | 0.0254 |
| NCU03152 | Conserved hypothetical protein | 0.3087 | 0.0001 | ωmat a>ωmat A | 0.0978 |
| NCU09142 | Hypothetical protein similar to cytoplasmic dynein intermediate chain | 0.1328 | 0.0001 | ωmat a>ωmat A | 0.0496 |
| NCU08348 | Conserved hypothetical protein | 0.0001 | 0.5494 | ωmat A>ωmat a | 0.0338 |
| NCU07362 | Hypothetical protein similar to mitochondrial cytochrome b2 | 0.3567 | 0.0001 | ωmat a>ωmat A | 0.0025 |
| NCU00921 | Conserved hypothetical protein | 0.4264 | >0.4264 | ωmat A>ωmat a | 0.0936 |
| NCU00771 | Conserved hypothetical protein | 0.0001 | 0.3085 | ωmatA>ωmat a | 0.0111 |
| NCU00499 | All development altered-1 | >0.0001 | 0.0001 | ωmat a>ωmat A | 0.0964 |
| NCU03013 | Anchored cell wall protein-10 | 0.0001 | >0.0001 | ωmatA>ωmat a | 0.0592 |
Abbreviations: LRT, likelihood ratio test; PAML, phylogenetic analysis by maximum likelihood.
P-values of likelihood ratio tests (LRT) are provided.
Hypothetical proteins are predicted from genomic data, but have no evidence of expression to date; predicted proteins have evidence of expression from ESTs or other mRNA analysis.
Cases with a value beginning with > indicates that the dS level was very low (<0.0001) or equal to zero while dN was markedly >0, wherein PAML assigns no estimate of ω (that is, a default value of 999), but only indicates whether a statistically significant difference is present and the direction of the difference.
Examination of each of the 141 individual genes in stratum 1 reveals that ω is substantially higher on one of the mat-linked alleles than the other (that is, mat a versus mat A) for 16 genes (P<0.1 Table 4, Supplementary Table S2). We used P-values of <0.1 for discerning genes with substantial differences in ω because lower values are overly conservative for our purposes to detect lineage-level differences in ω (as ω represents a global value per branch). These 16 genes span a range of functions, including genes encoding hypothetical proteins (predicted from genomic data, but not supported by expression analyses), predicted proteins (evident from genomic and expression data) and genes involved in mitochondrial processes (NCU02502, NCU07362) and RNA splicing and/or synthesis (NCU02633, NCU02675) (http://www.broadinstitute.org/annotation). We found that the allele with the higher values of ω occurred on the mat a chromosome for some of the genes and on the mat A chromosome for others, and this trait was gene-specific (Table 4). We also found that ω values are <1 for mat a and mat A lineages for most of these genes, suggesting that both gene alleles have evolved under purifying selection (Table 4). The findings of higher ω on one allele in each of these genes strongly suggest that there has been relaxed purifying selection, which gives rise to the enhanced fixation of slightly deleterious mutations on one of the copies. Another potential factor that could also contribute to different ω values among gene alleles is that one allele has been subjected to positive selection (Gerrard and Filatov, 2005).
Discussion
Previous reports have shown that alleles of ancient non-recombining sex chromosomes have elevated non-synonymous substitution rates as compared with the gametologs found on their homologous recombining chromosome (that is, Y- versus X-linked alleles in humans and W- versus Z-linked alleles in birds (Fridolfsson and Ellegren, 2000; Wyckoff et al., 2002)). The few available studies in young or neo-sex chromosomes have also revealed signs of genomic degeneration in regions of suppressed recombination, including accelerated protein evolution (Bachtrog, 2003; Marais et al., 2008). A key indicator of reduced molecular level adaptation in recombinationally suppressed genes is elevated non-synonymous substitutions (Bachtrog, 2003; Marais et al., 2008). Accordingly, we conclude that our findings showing the accumulation of allele-specific non-synonymous substitutions in the region of suppressed recombination N. tetrasperma mat chromosomes (Figure 1, Table 2) are best explained by genomic degeneration in this young system.
The marked accumulation of allele-specific substitutions reported herein (Figure 1 and Table 2) combined with the variation in ω among the N. tetrasperma mat chromosomes, suggest that relaxed purifying selection is inherent to the region of suppressed recombination. Hill-Robertson effects, background selection and/or Muller's Ratchet may have a role driving these genomic changes in the region of suppressed recombination (Hill and Robertson, 1966; Charlesworth and Charlesworth, 2000; Gordo and Charlesworth, 2001). Rare events of positive selection could also substantially contribute to these genomic alterations (Bachtrog, 2004). In particular, positive selection (that is, selective sweeps) for strongly beneficial mutations might lead to the fixation of linked deleterious mutations in recombinationally suppressed regions (Charlesworth and Charlesworth, 2000; Bachtrog, 2004). Although we did not find evidence of positive selection herein, adaptive evolution likely affects only a few sites and is often not detectable from branch-model analysis (Yang et al., 2000; Yang and Nielsen, 2002; Yang, 2007). Thus, as more genomic data becomes available across a broader range of species, future studies may reveal whether site-specific positive events are associated with the region of suppressed recombination (Yang and Nielsen, 2002; Yang, 2007). Altogether, it is evident that both relaxed purifying selection (and/or background selection, Muller's Ratchet) and positive selection models could each have a role in shaping the genomic alterations in the region of suppressed recombination in N. tetrasperma mat chromosomes.
Pattern of genomic changes
In contrast to ancient dimorphic X/Y and W/Z sex chromosome systems, wherein only one chromosome is recombinationally suppressed and degenerates, our results reveal that each of the N. tetrasperma mat chromosomes has independently undergone changes consistent with degeneration. This is evidenced by the prevalence of allele-specific non-synonymous substitutions on each of the two mat chromosomes, and by the fact that the allele with higher ω value could occur on either the mat a or mat A chromosome (Figure 1 Table 4). The most readily evident explanation for our observations of genomic alterations on each of the mat chromosomes is the fact that both chromosomes are recombinationally suppressed in N. tetrasperma, which differs from ancient dimorphic sex chromosomes. Nonetheless, another possible contributing factor for the genomic changes on both mat chromosomes is the novel mating system inherent to N. tetrasperma. In N. tetrasperma, both of the mat chromosomes (mat a and mat A) are normally present in the sexual spores and in vegetative cells, consistent with a predominant heterokaryotic state (Jacobson, 1995). Thus, the evolution of the mat chromosomes in N. tetrasperma may parallel that expected for organisms with haploid mating systems, wherein the sexes are haploid and the zygote is diploid. In such systems, genomic degeneration is expected to occur at similar, and low, levels among sex chromosomes, because of extensive haploid selection and the lack of sheltering of mutations (Bull, 1983). Our data are consistent with degeneration on both mat chromosomes, but not necessarily at the very low levels predicted for haploid systems (Figure 1 and Table 2). Accordingly, it may be speculated that the recombinationally suppressed regions of the N. tetrasperma mat chromosomes might evolve in a hybrid manner, sharing traits associated with ancient dimorphic sex chromosomes, such as a high level of degeneration, as well as those inherent to haploid systems, for example, degeneration on both chromosomes. Thus, even though the data presented here suggest that mating system does not alter evolution in the normally recombining regions of the N. tetrasperma mat chromosomes (as shown in Table 3), it could substantially contribute to the pattern of divergence among alleles in the recombinationally suppressed region. Further data from a range of very young sex chromosomes will be needed to ascertain whether a hybrid system of evolution is inherent to a range of organisms.
The role of sex-specific biases
It is worthwhile to note that most studies of sex chromosome evolution to date have been conducted in diecious species, wherein male and female gametes are confined to different individuals. In many of these organisms, sex-specific biases in mutation rates and sex-specific selection pressures have been widely reported for example, humans, mice, birds and plants (Filatov and Charlesworth, 2002; Bachtrog, 2004; Ellegren, 2007). These biases are believed to contribute to the degeneration of the permanently heterozygous chromosome, especially in systems with male heterogamety. For example, in Drosophila, male-driven mutation rates, resulting from greater number of cell divisions in male than female germlines, are believed to contribute to Y chromosome sequence divergence at synonymous sites, while positive selection events for male fitness in this taxon are believed to be at least partially responsible for degeneration at non-synonymous sites (Bachtrog, 2004, 2005, 2008). In contrast to these systems, however, sex-specific mutation and selection biases cannot contribute to our findings in N. tetrasperma, because each individual in this organism is able to produce both male and female reproductive structures, and the sexual spores contain both the mat a and mat A chromosomes (Raju and Perkins, 1994; Jacobson, 1995). Accordingly, our findings of changes consistent with degeneration on both mat chromosomes in N. tetrasperma suggest that sex-specific mutation rates and/or sex-specific selection pressures are not required for genomic alterations in sex-regulating chromosomes. Recombination suppression alone is sufficient to explain the observed genomic changes.
Conclusions
In totality, the present data provide important new insights into the evolution of the young region of recombination suppression in the N. tetrasperma mat chromosomes. These insights include findings that the region of recombination suppression in the mat chromosomes accumulates non-synonymous substitutions in a time-dependent manner, that each of the mat chromosomes are subjected to genomic changes consistent with degeneration, that the mat chromosomes evolve independently and have marked levels of allele-specific substitutions, and that these processes are likely driven by weakening of purifying selection (possibly because of Hill-Robertson effects, background selection and/or Muller's Ratchet) as well as by rare positive selection events. As more data becomes available, further studies across a greater number of species, and among populations, will be highly valuable in precisely defining the relative roles of relaxed purifying selection and positive selection. In addition, studies of transpositions, insertions/deletions and other degenerative traits in the N. tetrasperma mat chromosomes are warranted to reveal whether these types of degenerative changes also accumulate in a manner similar to non-synonymous mutations.
Acknowledgments
The authors gratefully acknowledge research funding from Royal Swedish Academy of Sciences (Hierta-Retzius Research Grant) (CAW), The Royal Physiographic Society in Lund (CAW), The Lars Hierta Minne Foundation (CAW), the Wenner-Gren Foundation (CAW), The Magn Bergvall Foundation (HJ) and The Swedish Research Council (HJ). We thank Yu Sun for conducting the assembly of N. tetrasperma Solexa data. We would also like to thank Pádraic Corcoran, the Associate Editor and the two anonymous reviewers for their valuable comments on our paper.
Data deposited at Dryad http://dx.doi.org/10.5061/dryad.8429
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bachtrog D. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics. 2003;165:1221–1232. doi: 10.1093/genetics/165.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nature Genet. 2004;36:518–522. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Sex chromosome evolution: molecular aspects of Y chromosome degeneration in Drosophila. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Evidence for male-driven evolution in Drosophila. Mol Biol Evol. 2008;25:617–619. doi: 10.1093/molbev/msn020. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Evolution of Sex Determination Mechanisms. Benjamin Cummings: Menlo Park; 1983. [Google Scholar]
- Casselton LA. Fungal sex genes-searching for the ancestors. Bioessays. 2008;30:711–714. doi: 10.1002/bies.20782. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond Ser B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobon DJ, Taylor JW. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003;57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc R Soc B Biol Sci. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. Substitution rates in the X-linked and Y-linked genes of the plants, Silene latifolia and S.dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Evolution of fungal sex chromosomes. Mol Microbiol. 2004;51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr Opin Genet Dev. 2005;15:645–651. doi: 10.1016/j.gde.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Fridolfsson AK, Ellegren H. Molecular evolution of the avian CHD1 genes on the Z and W sex chromosomes. Genetics. 2000;155:1903–1912. doi: 10.1093/genetics/155.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos A, Jacobson DJ, Raju NB, Skupski MP, Natvig DO. Suppressed recombination and a pairing anomaly on the mating-type chromosome of Neurospora tetrasperma. Genetics. 2000;154:623–633. doi: 10.1093/genetics/154.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard DT, Filatov DA. Positive and negative selection on mammalian Y chromosomes. Mol Biol Evol. 2005;22:1423–1432. doi: 10.1093/molbev/msi128. [DOI] [PubMed] [Google Scholar]
- Gordo I, Charlesworth B. Genetic linkage and molecular evolution. Curr Biol. 2001;11:R684–R686. doi: 10.1016/s0960-9822(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Handley LL, Ceplitis H, Ellegren H. Evolutionary strata on the chicken Z chromosome: Implications for sex chromosome evolution. Genetics. 2004;167:367–376. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Howe HB, Haysman P. Linkage group establishment in Neurospora tetrasperma by interspecific hybridization with N.crassa. Genetics. 1966;54:293–302. doi: 10.1093/genetics/54.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DJ. Sexual dysfunction associated with outcrossing in Neurospora tetrasperma, a pseudohomothallic ascomycete. Mycologia. 1995;87:604–617. [Google Scholar]
- Jacobson DJ. Blocked recombination along the mating-type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics. 2005;171:839–843. doi: 10.1534/genetics.105.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page D. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Marais GA, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, Monéger F, et al. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr Biol. 2008;18:545–549. doi: 10.1016/j.cub.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Menkis A, Bastiaans E, Jacobson DJ, Johannesson H. Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J Evol Biol. 2009;22:1923–1936. doi: 10.1111/j.1420-9101.2009.01801.x. [DOI] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsoon T, Johannesson H. The mating-type chromosome in the filamentous asycomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genetics. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino ST, Nelson MA, Jacobson DJ, Natvig DO. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics. 1996;143:789–799. doi: 10.1093/genetics/143.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Perkins DD.1985The Neurospora genomeIn: WE Timberlake (ed).Molecular Genetics of Filamentous Fungi Alan R. Liss Inc.: New York [Google Scholar]
- Raju NB, Perkins DD. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora and Podospora. Dev Genet. 1994;15:104–118. doi: 10.1002/dvg.1020150111. [DOI] [PubMed] [Google Scholar]
- Rice WR. Genetic hitch-hiking and the evolution of reduced genetic activity of Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstedt SA, Tucker PK. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 2004;14:267–272. doi: 10.1101/gr.1796204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shear CL, Dodge BO. Life histories and heterothallism of the red bread-mold fungi of the Monilia sitophila group. J Agr Res. 1927;34:1019–1041. [Google Scholar]
- Shiu PKT, Glass NL. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr Opin Microbiol. 2000;3:183–188. doi: 10.1016/s1369-5274(00)00073-4. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff G, Li JJ, Wu CI. Molecular evolution of functional genes on the mammalian Y chromosome. Mol Biol Evol. 2002;19:1633–1636. doi: 10.1093/oxfordjournals.molbev.a004226. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites among specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.