Abstract
The E-box binding zinc finger transcription factors Slug and ZEB1 are important repressors of E-cadherin, contributing to epithelial-mesenchymal transition (EMT) in primary epithelial cancers. Activator or repressor status of EMT transcription factors defines consequences for tumorigenesis. We show that changes in expression levels of Slug in melanoma cell lines lead to concomitant alterations of ZEB1 expression. Electrophoretic mobility shift, luciferase reporter and chromatin immunoprecipitation assays identified Slug as a direct transcriptional activator at E-boxes of the ZEB1 promoter. Transcriptional activation of ZEB1 was demonstrated to be specific for Slug, since EMT regulators Snail and Twist failed to influence ZEB1 expression. Slug and ZEB1 cooperatively repressed E-cadherin expression resulting in decreased adhesion to human keratinocytes, but promoted migration of melanoma cells. Our results show that the transcriptional activity of ZEB1 is increased by Slug, suggesting a hierarchical organized expression of EMT transcription factors through directed activation, triggering an EMT-like process in melanoma.
Introduction
Loss of the epithelial cell adhesion molecule E-cadherin is an initial step of carcinoma cell lines acquiring a more invasive phenotype (Behrens et al, 1989, Frixen et al, 1991). Repression of E-cadherin is mainly mediated by a subset of E-box binding transcription factors referred to as epithelial-mesenchymal transition regulators (EMTRs). Members include the zinc finger transcription factors Snail (Batlle et al, 2000), Slug (Hajra, Chen and Fearon, 2002, Bolos et al, 2003), ZEB1 (Eger et al, 2005) and SIP1 (Comijn et al, 2001), as well as basic helix-loop-helix transcription factors E12/47 (Perez-Moreno et al, 2001) and Twist (Yang et al, 2004). These EMTRs are crucial to the process of epithelial-mesenchymal transition (EMT), a major determinant of metastasis in melanoma (Alonso et al, 2007), in which epithelial cells undergo a conversion into mesenchymal cells. During progression of epithelial tumors, EMTRs promote proliferation, migration and invasion of cancer cells (Thiery, 2003, Kalluri and Weinberg, 2009, Huber, Kraut and Beug, 2005). They also contribute substantially towards resistance to programmed cell death and senescence, chemo- and immunotherapy and escape from immune surveillance (Thiery et al, 2009). In cutaneous melanoma, EMTRs are reported to play an important role in gaining independence from keratinocytes as well as progression towards metastases (Hsu et al, 2000, Poser et al, 2001, Hoek et al, 2004, Gupta et al, 2005).
The zinc finger EMT regulators Slug and ZEB1 are potent repressors of E-cadherin expression and enhancers of migration and invasion in addition to F-actin remodelling (Bolos et al, 2003, Bracken et al, 2008, Drake et al, 2009). Slug regulates integrin expression (Turner et al, 2006), represses Claudin-1 (Martinez-Estrada et al, 2006) and mediates resistance to apoptosis (Kajita, McClinic and Wade, 2004). ZEB1 promotes proliferation in breast cancer (Hu et al, 2010) and represses several master regulators of epithelial cell polarity (Aigner et al, 2007, Spaderna et al, 2008) and basement membrane components (Spaderna et al, 2006). Among others, ZEB1 has been reported to be upregulated by TGF-β (Shirakihara, Saitoh and Miyazono, 2007) and estrogen (Dillner and Sanders, 2002) as well as transcription factors NF-κB (Chua et al, 2007) and HIF-1 (Krishnamachary et al, 2006). Recent reports demonstrated ZEB1 to be subject to negative regulation by the miRNA-200 family and miRNAs-141 and −205 (Gregory et al, 2008, Brabletz and Brabletz, 2010). ZEB1 expression was also found to be promoted by the EMT regulator Snail (Guaita et al, 2002), yet in an indirect way involving not-yet defined pathways or transcription factors. While Slug functions as a repressor of various genes related to EMT, it also serves as an enhancer of gene expression (Moreno-Bueno et al, 2006, Huang et al, 2009).
A hierarchical organization of EMTR expression in the course of tumorigenesis may constitute an appealing concept to better understand the diversity of functions which have been ascribed to these transcription factors. Here we demonstrate the direct, Slug mediated, transcriptional upregulation of ZEB1. Electrophoretic mobility shift, luciferase reporter and chromatin immunoprecipitation assays revealed that Slug is binding to and activating the ZEB1 promoter, leading to a cooperative effect of Slug and ZEB1 in inducing an EMT like phenotype of melanoma cells. The regulation of ZEB1 through Slug is specific, since Snail and Twist are obviously not directly part of alterations in ZEB1 expression.
Results
Slug regulates ZEB1 expression at the transcriptional level
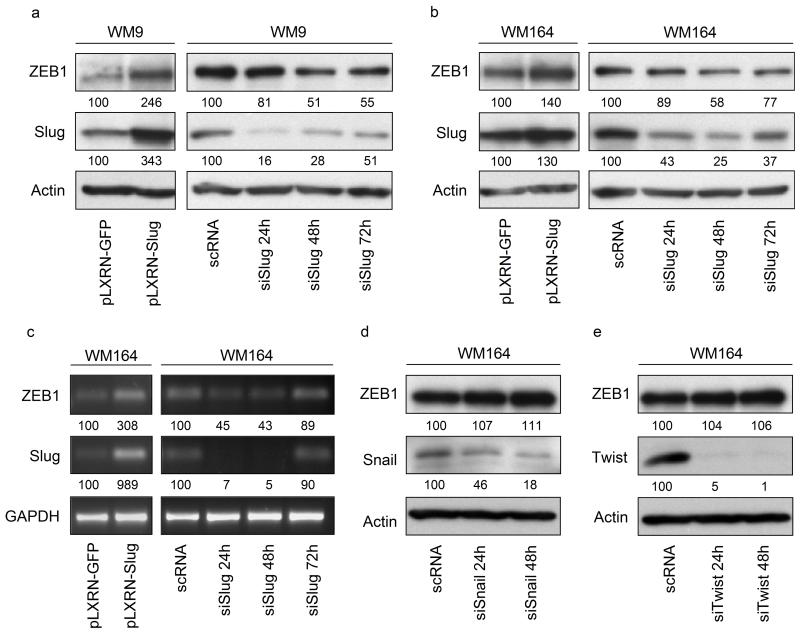
ZEB1 is regulated by the TGF-β signalling pathway (Shirakihara, Saitoh and Miyazono, 2007), NF-κB (Chua et al, 2007), HIF-1 (Krishnamachary et al, 2006), or the miR-200 family and miR-205 (Gregory et al, 2008). We investigated a direct transcriptional activation of ZEB1 through Slug. Analyses of ZEB1 levels by immunoblotting revealed increased ZEB1 expression in melanoma cell lines WM164 and WM9 transduced with retroviral Slug expression vector pLXRN-Slug compared to GFP control (Figure 1a, b, left panels). This effect was reversed by silencing of Slug. ZEB1 was downregulated in both cell lines following Slug silencing (Figure 1a, b, right panels). ZEB1 mRNA was significantly increased in Slug overexpressing cells (Figure 1c, left panel), whereas Slug silencing led to a decrease of ZEB1 transcripts (right panel). These results suggest that Slug regulates ZEB1 at the transcriptional level. To prove the specificity of these findings, we determined whether two related E-box binding EMTRs, Snail and Twist, affected ZEB1 protein levels in WM164 (Figure 1d, e). This was not the case, indicating that ZEB1 regulation is accomplished through Slug.
Figure 1.
Slug regulates ZEB1 expression. Immunoblots of whole cell lysates (15μg/lane) of transduced melanoma cell lines WM164 (a) and WM9 (b) overexpressing Slug (pLXRN-Slug) or GFP control (pLXRN-GFP). Slug knockdown was achieved by transfection with siRNA against Slug mRNA (siSlug) over a time period of 24 – 72h. Results were compared to unspecific scrambled RNA (scRNA). Samples were separated on a 10% SDS polyacrylamide gel. After transfer to a PVDF membrane, blots were probed with antibodies against Slug and ZEB1. β-Actin was used as a loading control. (c) Analysis of total mRNA extracts of either Slug overexpressing or Slug silenced WM164. Slug, ZEB1 and GAPDH mRNA expression was determined by PCR. E-box binding EMT regulators Snail (d) and Twist (e) were silenced for 24 and 48h using specific siRNAs (siSnail, siTwist). Effects of Snail and Twist silencing on ZEB1 expression were determined by immunoblotting using indicated antibodies. Numbers represent changes in % of controls and normalized to β-Actin or GAPDH.
Slug binds to E-boxes in the ZEB1 promoter
Slug binds to E-boxes, acting either as a repressor (Hajra, Chen and Fearon, 2002, Bolos et al, 2003, Martinez-Estrada et al, 2006) or activator (Moreno-Bueno et al, 2006, Huang et al, 2009) of transcription. We found four E-boxes at positions −858, −239, −82 and +36 of the ZEB1 promoter. To investigate the binding capacity of Slug to these E-boxes in vitro, electrophoretic mobility shift assays (EMSA) were performed, incubating nuclear extracts of WM164 (Figure 2, left panel) or WM9 (Figure 2, right panel) with Cy3 labeled 20nt fragments each containing one of the four wild type or mutated E-boxes. Sequences of the fragments used for EMSA are provided in Supplementary Table S1. After detection of shifted Cy3 labeled DNA fragments (Figure 2, top panels), DNA-protein complexes were transferred to PVDF membranes and subjected to immunoblotting. In WM164, binding of Slug to all four wild type E-box-fragments was observed. With respect to the fragments of E-box 1, 2 and 3, a single band representing Slug was detected in each case when protein extracts were incubated with either Cy3 labelled or additional 20fold excess of unlabeled wild type E-box fragments but not with mutated E-boxes. The same was observed for E-box 4 with the exception that a second band of higher molecular weight was detected when a 20fold excess of wild type cold probe was provided, indicating that Slug might be part of a larger complex binding to the E-box. Since Snail has been reported to affect ZEB1 expression (Guaita et al, 2002), we also determined its binding capacity to ZEB1 promoter fragments. Snail binding to fragments containing E-boxes 2 and 4 was detected only when a 20fold excess cold probe was provided additionally to Cy3 labelled probe. It remains unclear whether the detected bands are non-specific or if Snail is binding to E-boxes 2 and 4 specifically but to a lesser extent than Slug. Twist, an E-box binding bHLH transcription factor, is presumably binding specifically to E-boxes 3 and 4, since two bands could be detected for the wild type but not for mutated probes. Interestingly, bands of the same size were found binding to E-boxes 1 and 2 but only if a 20fold excess of cold probe was added (E-box2) or if binding to the mutated sequence (E-box1). Similar results were obtained when using nuclear extracts of WM9. Taken together, these results suggest a preferential binding of Slug to all four wild type E-boxes compared to Snail and Twist.
Figure 2.
Differential binding of Slug, Snail and Twist to E-boxes of the ZEB1 promoter. Four Cy3- labeled 20nt wild type (wt) fragments of the ZEB1 promoter (wt-Cy3), each containing one out of four potential Slug binding sites (E-boxes) found in the ZEB1 promoter in a region from −3000 to +200 relative to the transcription start were incubated with 10μg of nuclear extracts of WM164 (a) or WM9 (b) in the absence or presence of a 20-fold molar excess cold probe (20 x wt unlabeled) for 30min at 37°C in binding buffer. DNA-protein complexes were separated by non denaturating PAGE (6%) for 45min, 120V and detected by Bio-Rad Molecular Imager FX (EMSA-Gel). After detection, complexes where transferred to PVDF membrane and subjected to immunoblotting with antibodies against Slug, Snail and Twist. To prove the specificity of the binding of EMT regulators to the contributing E-boxes, wild type sequences CASSTG were mutated to ATSSTA (mut-Cy3).
Slug activates the ZEB1 promoter
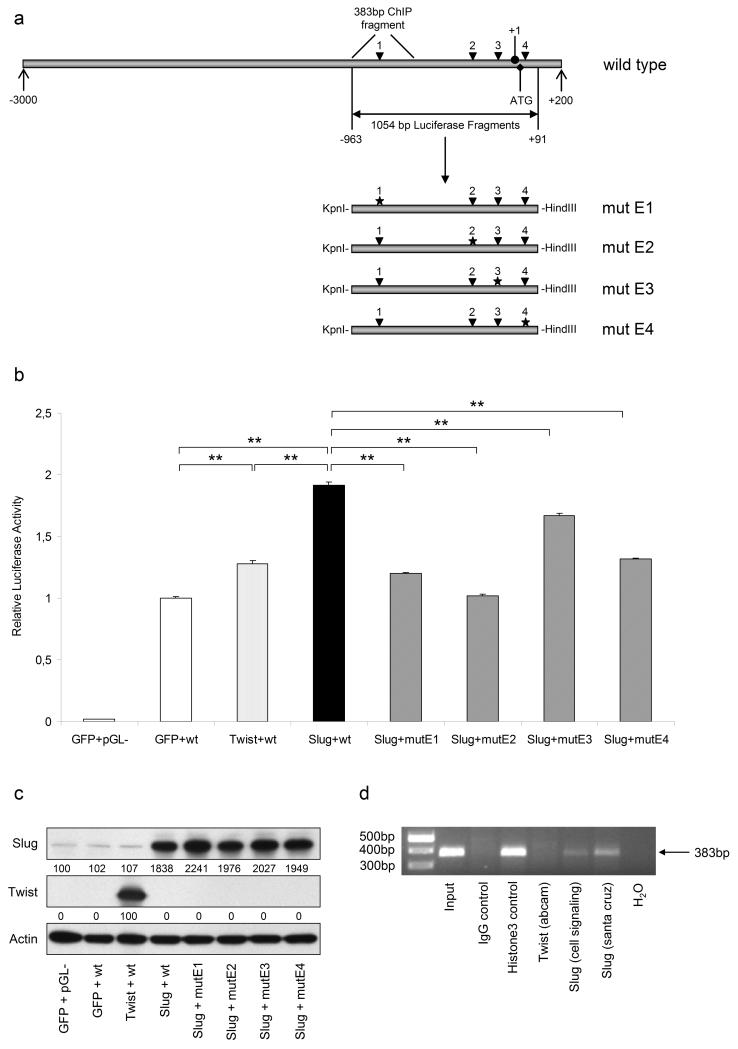
To confirm the results obtained by mobility shift assays, we investigated the effect of Slug on activating the ZEB1 promoter. To this end we cloned a 1054bp fragment covering the region −963 to +91 relative to the transcription start (+1) of the human ZEB1 promoter into pGL3-basic luciferase reporter plasmid (pGL3-promZEB1wt). This fragment contains all four E-box binding sites, each mutated separately by site directed mutagenesis PCR. A scheme of the human ZEB1 promoter as well as wild type and mutated luciferase fragments is provided in Figure 3a. COS-7 cells were co-transfected with reporter plasmids and pcDNA expression vectors containing either GFP, Slug or Twist cDNAs. Figure 3b demonstrates activation of the ZEB1 promoter through Slug as luciferase activity was increased two-fold compared to GFP control. Examination of luciferase expression driven by ZEB1 promoter fragments with single mutated E-boxes revealed the highest reduction when E-box2 (mutE2) was mutated, followed by E-boxes 1, 4 and 3. Since mobility shift assays suggested Twist also binding to ZEB1 promoter E-boxes, we additionally investigated the effect of Twist overexpression on luciferase expression. Only a 28% increase in luciferase activity was observed compared to GFP control, indicating that Slug rather than Twist has an impact on ZEB1 promoter regulation. Overexpression of Slug and Twist in COS-7 cells was confirmed by immunoblotting as shown in Figure 3c. To prove these results and demonstrate binding of Slug to the ZEB1 promoter in WM164 cells in vivo, we performed chromatin immunoprecipitation (ChIP) assays, using two different Slug antibodies and one Twist antibody as well as Histone H3 as a positive and unspecific IgG as a negative control. Figure 3d indicates that Slug indeed binds to the ZEB1 promoter, since a specific 383bp fragment covering E-box1 was amplified by PCR out of chromatin precipitated using both Slug and Histone H3 but not Twist or IgG control antibodies.
Figure 3.
Slug enhances the activity of the ZEB1 promoter in COS-7 transfectants. (a) Scheme of the ZEB1 promoter region from −3000 to +200 relative to the transcription start (+1). Four potential Slug binding sites (E-boxes) were found and referred to as E-boxes 1-4 (arrowheads). A 1054bp fragment harbouring all four E-boxes was amplified out of genomic DNA and ligated into pGL3-basic (Promega) luciferase reporter vector (pGL3-promZEB1). Each of the E-boxes (mut E1-4) was mutated separately as indicated by asterisks. (b) COS-7 cells were co-transfected with either pcDNA-GFP, pcDNA-Twist or pcDNA-Slug together with empty vector control (pGL-) or pGL3-promZEB1 wild type (wt) or mutated E-boxes (mutE1-4). Cells were subjected to luciferase expression analysis 48h after transfection. Luciferase activity is given as mean + SD relative to activity in GFP control cells. **, p<0.01 highly significant. (c) Slug and Twist overexpression in pcDNA-Slug or -Twist transfected cells was confirmed by immunoblotting. Numbers represent changes in % of controls and normalized to β-Actin. (d) In vivo binding of Slug but not Twist to the ZEB1 promoter. ChIP was performed with WM164 cells using two different Slug antibodies (Cell Signaling; Santa Cruz)for IP. ZEB1 promoter-specific primers amplified a 383bp fragment.
Slug and ZEB1 cooperatively regulate E-cadherin expression
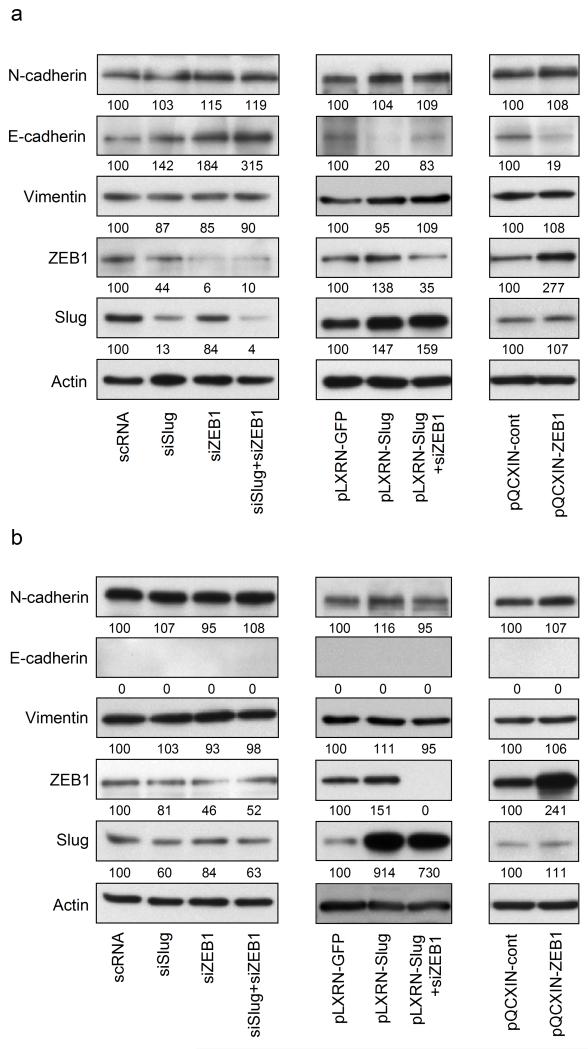
Slug and ZEB1 have been reported to be repressors of epithelial (Hajra, Chen and Fearon, 2002, Eger et al, 2005, Aigner et al, 2007) and activators of mesenchymal (Moreno-Bueno et al, 2006, Graham et al, 2008) markers. To investigate the effects of Slug and ZEB1 on the expression of E- and N-cadherin as well as Vimentin in melanoma, we performed immunoblotting with lysates of cell lines WM164 and WM9 either overexpressing Slug or ZEB1, or transfected with siRNA against both transcription factors. To investigate on the reversibility of Slug mediated effects, ZEB1 was silenced in Slug overexpressing cell lines. In WM164, a melanoma cell line which expresses both E- and N-cadherin, both silencing of Slug and ZEB1 led to an upregulation of E-cadherin, with the most significant effect obtained when both EMTRs were downregulated simultaneously (Figure 4a, left panel). Slug overexpression led to a downregulation of E-cadherin. This effect was reversed by silencing of ZEB1, indicating ZEB1 to be a potent repressor of E-cadherin in melanoma (Figure 4a, middle panel). Overexpression of ZEB1 alone leads to E-cadherin downregulation compared to empty vector control (Figure 4a, right panel). In contrast, N-cadherin and Vimentin expression levels were not significantly altered by any of the chosen conditions in either WM164 (Figure 4a) or WM9 (Figure 4b).
Figure 4.
The effect of Slug and ZEB1 on E-cadherin expression is additive. Regulation of epithelial and mesenchymal markers through Slug and ZEB1 in melanoma cell lines WM164 (a) and WM9 (b) was determined by immunoblotting. Cells were transfected with either scrambled RNA (scRNA), siRNA against Slug (siSlug), ZEB1 (siZEB1) or both Slug and ZEB1 (siSlug+siZEB1) for 48h (left panels). Stable expression of GFP or Slug (pLXRN-GFP, pLXRN-Slug, middle panels) or ZEB1 (pQCXIN-ZEB1 and empty control vector pQCXIN-cont, right panels) was achieved by retroviral transduction of WM164 and WM9. Effects of Slug overexpression through coactivation of ZEB1 were reversed using siRNA against ZEB1 (middle panels). Samples were separated by 10% SDS polyacrylamide gels, transferred to PVDF membrane and probed by indicated antibodies. E-cadherin (expressed only in WM164) was chosen as epithelial marker, N-cadherin and Vimentin as mesenchymal markers. β-Actin was used as a loading control. Numbers represent changes in % of controls and normalized to β-Actin.
The effect of Slug and ZEB1 on cell-cell adhesion and cell migration is additive
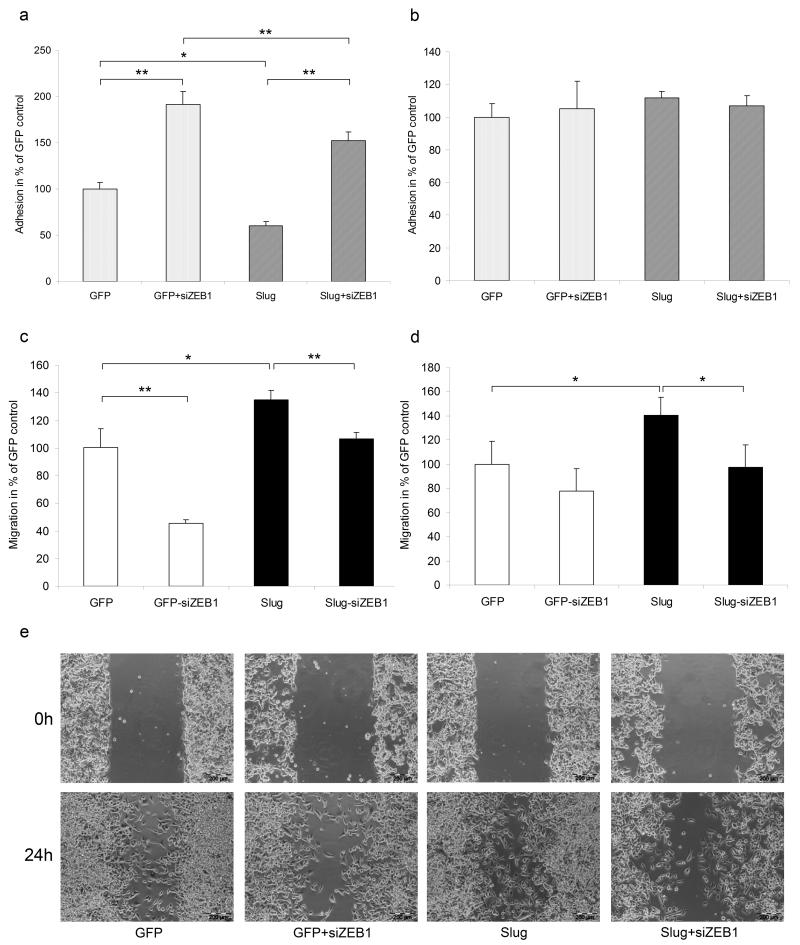
Both Slug and ZEB1 have been shown to change functional properties of cancer cells including adhesion and migration (Hajra, Chen and Fearon, 2002, Eger et al, 2005), accompanied by F-Actin remodelling (Bolos et al, 2003, Bracken et al, 2008). Overexpression of Slug in WM164 cells led to a repression of E-cadherin which in turn was followed by a significant decrease of adhesion to keratinocytes (Figure 5a). Silencing of ZEB1 resulted in an almost two-fold increased adhesion to keratinocytes in GFP transduced WM164 and also compensated for the effect obtained by Slug overexpression. In fact, silencing of ZEB1 in Slug overexpressing cells even increased adhesion to keratinocytes compared to GFP control cells. However, decreased binding to keratinocytes was observed compared to siZEB1 alone, indicating a net effect of Slug due to ZEB1 activation (Figure 5a). Adhesion to fibroblasts remained unaltered under the same conditions (Figure 5b) in line with the results shown in Figure 4, indicating that Slug and ZEB1 do not influence N-cadherin expression. Silencing of ZEB1 in WM164 and WM9 not only resulted in a decreased directed migration of pLXRN-GFP transduced cells but also compensated for the gain in migration obtained by Slug overexpression in both cell lines (Figure 5c,d and S1a,b). These results were confirmed by wound healing assays representing random migration (Figure 5e and S1c). Additionally, we determined the effect of Slug and ZEB1 on F-Actin remodelling in WM164. Results showed that there was no significant difference between untransduced and Slug transduced cells, since both revealed filamentous structures of stress fibres, however these structures were lost in ZEB1 silenced cells (Supplementary Figure S1d). Cell proliferation of WM164 and WM9 as determined by MTT assays was not altered in siSlug, siZEB1 or double silenced cells (data not shown).
Figure 5.
Slug and ZEB1 cooperate in regulating cell-cell-adhesion and migration. To test for Slug and ZEB1 dependent alterations in adhesion to layers of keratinocytes (a) and fibroblasts (b), ZEB1 was silenced in GFP or Slug overexpressing WM164. After incubation for 30min, non adherent cells were removed and adherent cells were counted. The effect of Slug and ZEB1 on directed migration was investigated in GFP or Slug transduced WM164 (c) and WM9 (d), transfected with scRNA or siZEB1. DiI stained cells were seeded in the upper compartment of 8μm pore size cell culture inserts. Migration towards the lower compartment was monitored by fluorescent microscope. All results are given as mean + SD in % compared to GFP control. *, p<0.05 significant; **, p<0.01 highly significant. (e) Wound healing assay (24h) of GFP or Slug transduced WM164 transfected with scRNA or siZEB1. Bar = 200μm.
Discussion
The E-box binding zinc finger transcription factor ZEB1 is as a repressor of E-cadherin and other epithelial markers as well as promoter of migration and invasion in various cancer types ( reviewed in Vandewalle, Van Roy and Berx, 2009, Brabletz and Brabletz, 2010,). Of the E-box binding, EMT regulating transcription factors, Snail has been reported to be an enhancer of ZEB1 expression in different cell types including HT-29 M6 colon cancer cells (Guaita et al, 2002), however the authors did not consider Snail to directly regulate ZEB1 at the transcriptional level. A delay of four days between the induction of Snail expression and ZEB1 upregulation was observed and the time required to activate the ZEB1 promoter in luciferase reporter assays was longer than the time taken to repress the promoter of E-cadherin (Guaita et al, 2002). In accordance, Taube et al. (Taube et al, 2010) showed elevated ZEB1 mRNA levels in Snail and Twist overexpressing HMLE cells. These data suggest that EMTRs might be involved in ZEB1 regulation. This study now provides evidence for the specific, transcriptional regulation of ZEB1 through Slug, but not Snail or Twist in melanoma cell lines, consistent with results of Huang et al. (Huang et al, 2009), who reported Slug, but not Snail or Twist to induce expression of Membrane-type 4 matrix metalloproteinase. The proof of a direct, specific regulation of one EMT transcription factor by another constitutes an appealing concept of a potential hierarchy of EMTR expression in the course of melanoma progression.
Intact E-box sequences have been described as a requirement for Slug mediated regulation of E-cadherin (Hajra, Chen and Fearon, 2002, Bolos et al, 2003), Claudin-1 (Martinez-Estrada et al, 2006), integrins (Turner et al, 2006), occludin (Wang et al, 2007) and membrane-type 4 matrix metalloproteinase (Huang et al, 2009). Performing electrophoretic mobility shift assays, we revealed Slug binding to wild type but not mutated E-box sequences which were identified in the ZEB1 promoter in a range from −3000 to +200 relative to the transcription start. Transcriptional activation at these sites was confirmed by luciferase reporter assays employing a 1.054kb ZEB1 promoter fragment containing all four E-boxes. Introduction of mutations in single E-boxes revealed preferable sites of activation. Slug binds to distinct E-boxes of the E-cadherin (Hajra, Chen and Fearon, 2002) and Claudin (Martinez-Estrada et al, 2006) promoters with different affinities. Binding the mouse E-cadherin E-pal sequence was suggested to even occur in a di- or multimeric form (Bolos et al, 2003). Similarly, our results indicate that the binding to and activation of the ZEB1 promoter E-boxes by Slug occurs with different affinities, in part generating different sized bands on mobility shift assays. Whether these differences in size depict multimeres of Slug alone or complexes of Slug together with other proteins is yet to be determined. Since Slug has been described to mediate its repressing function at least partially by recruiting C-terminal-binding protein-1 (CtBP-1) and histone deacetylase-1 (HDAC-1) (Tripathi et al, 2005), it is tempting to speculate that other transcription factors or co-factors might be involved in Slug dependent ZEB1 activation as well. The ZEB1 promoter is induced approximately 2.5-fold by NF-κB subunit p65 compared to empty vector control (Chua et al, 2007) and 2-fold by Slug, but only marginally by Twist (this study). Thus, the affinity of Twist to the ZEB1 promoter may not be strong enough to result in significant changes of ZEB1 protein levels, indicating Slug preferentially regulating ZEB1 expression in melanoma compared to other EMTRs. These findings are supported by the results of chromatin immunoprecipitations, indicating Slug but not Twist binding to the ZEB1 promoter in vivo. We assume that Twist may be able to bind to the ZEB1 promoter fragments if provided in in vitro (EMSA) affinity studies but not in the cellular context, thus giving evidence for the specificity of Slug as a ZEB1 regulating transcription factor.
Activation of ZEB1 transcription by Slug might have consequences for functionality. To this end the impact of Slug and ZEB1 on melanoma cell adhesion and migration as prototypic features of EMT was determined. Specifically, we focused on the cooperation of the two transcription factors and the ability of ZEB1 knockdown to at least partially abrogate the effects obtained by Slug overexpression. Regarding the regulation of epithelial and mesenchymal markers, Slug and ZEB1 have been consistently described as repressors of E-cadherin in various cell lines (Hajra, Chen and Fearon, 2002, Bolos et al, 2003, Eger et al, 2005, Shirakihara, Saitoh and Miyazono, 2007). In prostate cancer cells, N-cadherin has been reported to be upregulated by ZEB1 by Graham et al. (Graham et al, 2008), but not by Drake et al. (Drake et al, 2009). N-cadherin is slightly upregulated in Slug transduced MCDK cells (Moreno-Bueno et al, 2006) whereas it is downregulated in ZEB1 silenced human esophageal epithelial cells (Ohashi et al, 2010). Shirakihara et al. (Shirakihara, Saitoh and Miyazono, 2007) reported ZEB1 to have no influence on the expression of mesenchymal markers N-cadherin, Vimentin and Fibronectin. In contrast, Vimentin is upregulated by ZEB1 in PC-3 cells (Drake et al, 2009), and upregulated by Slug in DLD1 colon cancer (Medici, Hay and Olsen, 2008) and OE33 esophageal adenocarcinoma cell lines (Jethwa et al, 2008). These reports strongly suggest that the effect of Slug and ZEB1 on N-cadherin and Vimentin expression may depend on the cellular background and the microenvironment. In this study, we demonstrate Slug and ZEB1 to be repressors of E-cadherin in melanoma, observing an additive effect of both transcription factors. Slug mediated downregulation of E-cadherin can be reversed by ZEB1 silencing, indicating a cooperative repressor function of both EMTRs. N-cadherin and Vimentin levels, however, are not affected by overexpression or silencing of either transcription factor. Respective changes in E- and N-cadherin expression are reflected by adhesion assays of melanoma cells to keratinocytes and fibroblasts.
There is unanimous agreement that both Slug (Bolos et al, 2003, del Barrio and Nieto, 2002, reviewed in Barrallo-Gimeno and Nieto, 2005) and ZEB1 (Bracken et al, 2008, Drake et al, 2009, Aigner et al, 2007, Das et al, 2009) are inducers of cell migration in various cell types. We were able to confirm these results in melanoma, again suggesting a cooperative regulation of Slug and ZEB1, since enhanced migration after Slug overexpression was reversed by ZEB1 silencing. Additionally, it has been shown that F-actin remodelling from a cortical to a stress-fiber pattern, representing a more motile phenotype, occurs as a consequence of Slug (Bolos et al, 2003) and ZEB1 (Bracken et al, 2008) overexpression. In this study, silencing of ZEB1 in wild type and Slug overexpressing metastatic melanoma cell line WM164 led to an actin rearrangement from a filamentous to a cortical structure.
Overall, these data demonstrate the direct and specific upregulation of ZEB1 by Slug, increasing the cooperative effect of these two transcription factors on epithelial-mesenchymal transition. According to Bolos et al. (Bolos et al, 2003), who claimed the priority of a rapid and efficient repression of E-cadherin at initial stages of invasion, we speculate that a directed regulation of EMT transcription factors may promptly execute the repression of epithelial markers, enabling detachment and migration during early stages of melanomagenesis.
Materials and Methods
Cell Culture
Human melanoma cell lines, fibroblasts and keratinocytes were kindly provided by Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA). The human metastatic melanoma cell lines WM9 and WM164 were cultured in RPMI 1640 (Sigma Aldrich, St. Louis, MO) supplemented with 2% FCS and 2% L-Glutamine (PAA, Pasching, Austria). The HEK-293 derived cell line GP-293 (Clontech, Mountain View, CA), stably transfected with gag/pol genes and used for the production of retroviruses and human fibroblasts FF2462 were maintained in DMEM (Sigma) containing 10% FCS and 2% L-Glutamine. Monkey COS-7 cells were cultured in DMEM Nutrient Mixture F-12 HAM (Sigma), supplemented with 4% FCS and 2% L-Glutamine. Primary human keratinocytes FK-181 were kept in Keratinocyte SFM (Invitrogen, Carlsbad, CA). All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
RNA inhibition experiments
siRNAs targeting mRNAs of Slug, ZEB1 and Twist were purchased from Santa Cruz, siRNA against Snail was obtained from Dharmacon Inc. (Lafayette, CO). Control siRNA was purchased from Qiagen. 150 pmol siRNA were transfected into cells seeded in 6-well-plates using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocol. Cells were processed for immunoblot analyses or functional assays 24, 48 or 72h after transfection.
Vectors and Vector Production
GFP and Slug cDNAs were subcloned into the modified retroviral vector pLXRN-CMV1, kindly provided by Dr. Chengjiang Li (Department of Medicine, The First Affiliated Hospital, College of Medicine, Zhejiang University, PR China). pcDNA3.hSlug was a gift of Dr. Pierre Savagner (Batiment de Recherche en Cancerologie, Montpellier, France). Retroviral pQCXIN-ZEB1 and pQCXIN-cont vectors were kindly provided by Dr. Harikrishna Nakshatri (Department of Surgery, Indiana University School of Medicine, Indianapolis, IN, USA). pcDNA4-hTwist was a kind gift of Dr. Carlotta Glackin (Department of Neurosciences, Beckman Research Institute of City of Hope, Duarte, CA, USA). For luciferase assays, a 1.054kb fragment of the ZEB1 promoter covering the region −963 to +91 relative to the transcription start was amplified out of genomic DNA using Phusion High-Fidelity DNA Polymerase (Finnzymes, Espoo, Finland) and cloned into pGL3-basic vector (Promega, Madison, WI) using restriction sites KpnI and HindIII (pGL3-promZEB1wt).
Electrophoretic Mobility Shift Assay
EMSA was performed according to standard protocols, using 20 nucleotide fragments of the ZEB1 promoter containing either wild type (CASSTG) or mutated (ATSSTA) E-boxes (Table S1). Nuclear extracts (10μg) of WM9 or WM164 were incubated with Cy3-labeled double strand oligonucleotides for 30 min at 37°C in binding buffer [10mM Tris (pH 7.5), 50mM NaCl2, 1mM DTT, 0,1mM EDTA, 5% glycerol] with 1μg of poly(dI-dC). After incubation the reaction batches were separated by non-denaturing PAGE (6%). DNA-protein complexes were detected by Bio-Rad Molecular Imager FX, transferred to PVDF membrane and probed with Slug, Snail or Twist antibodies by immunoblotting.
Luciferase Assay
Slug dependent ZEB1 promoter activity was determined by co-transfection of COS-7 cells with either pcDNA-GFP, pcDNA-Slug or pcDNA-Twist (1.6μg), together with empty pGL3-basic, pGL3-promZEB1wt or pGL3-promZEB1 containing mutations in one of four E-boxes (1.6μg) using Lipofectamine 2000 following the manufacturer’s protocol for transfection in 12-well-plates. Luciferase activity was measured 48h post transfection using the Luciferase Assay System from Promega and normalized to 1μg of protein. Detection was done using LUMIstar Omega luminometer (BMG Labtech, Offenburg, Germany). Experiments were performed in triplicate.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling), following the manufacturer’s protocol. In brief, 4×107 WM164 cells were crosslinked with 1% formaldehyde for 30 min. After isolation of nuclei, chromatin was digested by micrococcal nuclease (15000 gel units) for 20 min at 37°C. Subsequently, the nuclear membrane was disrupted by an UP50H Ultrasonic Processor (Dr. Hielscher GmbH, Tetlow, Germany), duty cycle 1, 100% amplitude, 4 × 20 sec. For immunoprecipitation, 100 μl (~15μg of chromatin DNA) of the cross-linked chromatin preparation were incubated over night at 4°C with the following antibodies: Twist [2C1a] (Abcam, dilution 1:50), Slug [L40C6] (Cell Signaling, dilution 1:50), Slug [G-18] (Santa Cruz, dilution 1:50). Histone H3 [D2B12] and normal rabbit IgG, both provided by the ChIP kit, were used as a positive or negative control respectively. After incubation with 30μl Protein G Agarose Beads (2hrs, 4°C, rotation), antibody-DNA complexes were washed, eluted from the beads and Proteinase K digested for 2h at 65°C. Following spin column based DNA purification, PCR was performed using ZEB1 promoter specific primers 5′ TCATGGCCTGTGGATACCTTAGC 3′ (forward) and 5′ TTTGGGGACGGCGAGGA 3′ (reverse), producing a 383bp fragment.
Supplementary Material
Acknowledgements
We would like to thank the staff of the Center for Medical Research (ZMF), Graz, for technical assistance. We are grateful to Terri Campbell from the Dermatology Research Center, University of Queensland (Brisbane, Australia) for editing the manuscript. This work was supported by the Austrian Science Fund (grants No. P18630-B05 to PK and P21156-B18 to CW), S. Joshi was funded by the PhD program Molecular Medicine of the Medical University of Graz, Austria, by the Jubiläumsfond der Österreichischen Nationalbank (12552) and by the Austrian Science Fund (grant Nr. P21156-B18).
Footnotes
Note: Further information is given in the Supplementary Methods section.
Conflict of interest
The authors state no conflict of interest.
References
- Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The snail genes as inducers of cell movement and survival: Implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Behrens J, Mareel MM, Van Roy FM, Birchmeier W. Dissecting tumor cell invasion: Epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell-cell adhesion. J Cell Biol. 1989;108:2435–2447. doi: 10.1083/jcb.108.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: Potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Das S, Becker BN, Hoffmann FM, Mertz JE. Complete reversal of epithelial to mesenchymal transition requires inhibition of both ZEB expression and the rho pathway. BMC Cell Biol. 2009;10:94. doi: 10.1186/1471-2121-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Barrio MG, Nieto MA. Overexpression of snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- Dillner NB, Sanders MM. The zinc finger/homeodomain protein deltaEF1 mediates estrogen-specific induction of the ovalbumin gene. Mol Cell Endocrinol. 2002;192:85–91. doi: 10.1016/s0303-7207(02)00088-6. [DOI] [PubMed] [Google Scholar]
- Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell. 2009;20:2207–2217. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–2488. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Hsu MY, Meier FE, Nesbit M, Hsu JY, Van Belle P, Elder DE, et al. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am J Pathol. 2000;156:1515–1525. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Wang C, Du J, Sun W, Yan J, Mi D, et al. DeltaEF1 promotes breast cancer cell proliferation through down-regulating p21 expression. Biochim Biophys Acta. 2010;1802:301–312. doi: 10.1016/j.bbadis.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, et al. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11:1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Jethwa P, Naqvi M, Hardy RG, Hotchin NA, Roberts S, Spychal R, et al. Overexpression of slug is associated with malignant progression of esophageal adenocarcinoma. World J Gastroenterol. 2008;14:1044–1052. doi: 10.3748/wjg.14.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von hippel-lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–2731. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Culleres A, Soriano FX, Peinado H, Bolos V, Martinez FO, et al. The transcription factors slug and snail act as repressors of claudin-1 expression in epithelial cells. Biochem J. 2006;394:449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici D, Hay ED, Olsen BR. Snail and slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, Villa S, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for snail, slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 2006;66:9543–9556. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Poser I, Dominguez D, de Herreros AG, Varnai A, Buettner R, Bosserhoff AK. Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor snail. J Biol Chem. 2001;276:24661–24666. doi: 10.1074/jbc.M011224200. [DOI] [PubMed] [Google Scholar]
- Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Tripathi MK, Misra S, Khedkar SV, Hamilton N, Irvin-Wilson C, Sharan C, et al. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J Biol Chem. 2005;280:17163–17171. doi: 10.1074/jbc.M501375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, Hughes S, et al. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wade P, Mandell KJ, Akyildiz A, Parkos CA, Mrsny RJ, et al. Raf 1 represses expression of the tight junction protein occludin via activation of the zinc-finger transcription factor slug. Oncogene. 2007;26:1222–1230. doi: 10.1038/sj.onc.1209902. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.