Abstract
Post-translational modifications of histones are determining factors in the global and local regulation of genome activity. Phosphorylation of histone H3 is globally associated with mitotic chromatin compaction but occurs in a much more restricted manner during interphase transcriptional regulation of a limited subset of genes. In the course of gene regulation, serine 10 phosphorylation at histone H3 is targeted to a very small fraction of nucleosomes that is highly susceptible to additional acetylation events. Recently, we and others have identified 14-3-3 as a binding protein that recognizes both phosphorylated serine 10 and phosphorylated serine 28 on histone H3. In vitro, the affinity of 14-3-3 for phosphoserine 10 is weak but becomes significantly increased by additional acetylation of either lysine 9 or lysine 14 on the same histone tail. In contrast, the histone H3S28 site matches elements of 14-3-3 high affinity consensus motifs. This region mediates an initial stronger interaction that is less susceptible to modulation by “auxiliary” modifications. Here we discuss the binding of 14-3-3 proteins to histone H3 in detail and putative biological implications of these interactions.
Keywords: histone code, epigenetics, transcription, phosphoacetylation, methylation
Intrinsic Factors Influencing 14-3-3 Histone H3 Interaction
14-3-3 proteins comprise a highly conserved protein family with at least two isoforms expressed in lower eukaryotic organisms and up to 15 in plants. In mammals the 14-3-3 family comprises seven members (β, γ, ε, η, τ/θ, ζ and σ) each encoded by a distinct gene. Despite considerable variability in the coding sequences, 14-3-3 proteins display a high degree of overall conservation in primary and tertiary protein structure. Although most isoform are ubiquitously expressed, 14-3-3σ expression appears restricted to epithelial tissue. Further, spatial and temporal patterns of isoforms expression occur during developmental progression.1-3 Differential post-translational modifications of particular isoforms have also been reported.4-6
Pioneering research revealed the dimeric nature of this protein class7,8 as an important hallmark of 14-3-3 biology.9,10 Moreover, 14-3-3 proteins were identified as the first phosphoserine/threonine dependent adaptor molecules.11 Detailed investigations on substrate preferences demonstrated that 14-3-3 proteins recognize two internal consensus motifs: the sequences RSXS/TphXP (mode 1) and RXXXS/TphXP (mode 2) where S/Tph indicates phosphorylated serine or threonine and X any amino acid except cysteine with position dependent preferences.11-13 Further, a carboxy-terminal consensus referred to as mode 3 has been identified.14-16 Although one of these consensus sequences is frequently found within 14-3-3 associated proteins, several interaction partners contain variations of this motif or do not require phosphorylation for binding at all.17-22
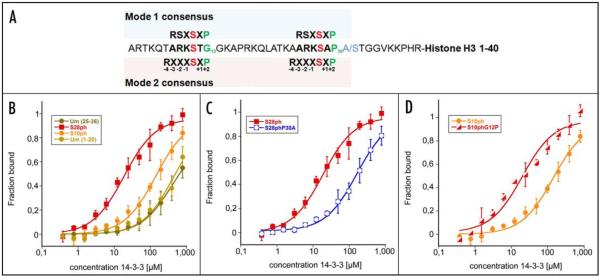
Recently, 14-3-3 proteins were reported to interact with phosphorylated histone H3.23-25 These studies indicated a function of this association in transcriptional activation.25 The two phosphorylated serines within histone H3, S10 and S28 that were shown to mediate interaction with 14-3-3, do not perfectly match one of the two consensus motifs (Fig. 1A). In vitro, H3S28ph however mediates a significantly stronger interaction with 14-3-3 than H3S10ph (Fig. 1B).23,25
Figure 1.
Intrinsic factors influencing the interaction of 14-3-3 with histone H3. (A) Sequence alignment of high affinity 14-3-3 consensus motifs of mode 1 (upper box) and mode 2 (lower box) with histone H3. The critical position at P + 2 (green residues) from the phosphorylated serines (red residues) is formed by glycine 12 for serine 10 and proline 30 for serine 28, respectively. Histone H3.3 contains a serine at position 31 (blue residue) whereas histones H3.1 and H3.2 contain an alanine at this site. (B) Serine 28 phosphorylated histone H3 is bound by 14-3-3ζ with higher affinity than H3 phosphorylated at serine 10. Binding curves determined by fluorescence polarization measurement are shown.60 Binding assays were performed for H3S28ph peptide, H3S10ph peptide and the respective unmodified controls. Data points of at least three independent measurements were averaged. Binding curves were fitted using least square algorithm. Dissociation constants (Kd) values are summarized in Table 1. (C) Proline 30 constitutes an important factor for the higher affinity of the H3S28ph peptide. Proline 30 was mutated to alanine (H3S28phP30A) and affinity for 14-3-3 binding was determined. (D) Proline at position 12 enhances binding to the H3S10ph peptide. The P + 2 position was changed from glycine to proline (H3S10phG12P) and binding assays were performed as described for (B). (D) Proline 30 constitutes an important factor for the higher affinity of the H3S28ph peptide. Proline 30 was mutated to alanine (H3S28phP30A) and affinity for 14-3-3 binding was determined.
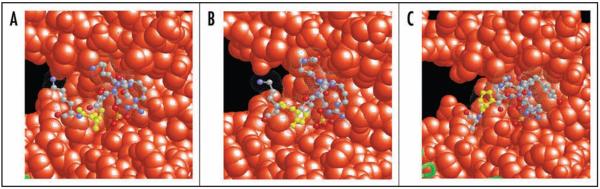
Both mode 1 and mode 2 consensus motifs contain proline at position P + 2 which adopts either cis conformation in mode 1 or trans conformation in mode 2.12 In general there is a strong selection for turn-forming residues at this position.12,13 Histone H3S10 and H3S28 are preceded by the same amino acid motif ARK. The carboxy-terminal sequence however differs considerably between the two sites (Fig. 1A). H3S10 is followed by an additional phosphorylatable threonine at P + 1. Tandem glycine residues follow at P + 2 and P + 3. In the crystal structure of 14-3-3ζ bound to the phosphorylated H3 tail these residues allow the H3 peptide to exit the binding cleft (Fig. 3).23 In contrast, H3S28 is followed by an alanine and contains proline at position 30 (H3P30) matching the strongly preferred proline at position P + 2 contained within the two 14-3-3 consensus motifs (Fig. 1A).12,13 The presence of proline at P + 2 appears to be favorable over tandem glycines as indicated by significantly stronger interaction of 14-3-3 with the H3S28 site compared to the H3S10 site (Fig. 1B).23,25 Further, mutation of H3P30 to alanine (H3P30A) significantly decreased the affinity of 14-3-3ζ for the H3 tail (Fig. 1C). Conversely, exchange of glycine at position 12 by proline (H3G12P) resulted in enhanced 14-3-3ζ binding to the H3S10ph peptide (Fig. 1D). Therefore and in agreement with profound structural data,12,13,23 H3P30 appears to be a crucial residue in mediating the high affinity of 14-3-3 towards the S28 phosphorylated H3 tail.
Figure 3.
Critical amino acids at position P + 2 mediate the exit of the peptide from the 14-3-3 binding cleft. (A) H3S10ph histone H3 peptide (ball and stick view with dotted van der Waals radii) located within the 14-3-3 binding cleft (spacefill view, orange atoms) (PDB entry 2C1N).23 The tandem glycine residues that mediate exit of the peptide from the binding cleft are highlighted in yellow. (B) Representative view of the H3K9acS10phK14ac histone H3 peptide (PDB entry 2C1J) arranged as described for panel A.23 Representative view of the mode 2 binding peptide (PDB entry 1QJA) the proline at position P + 2 that mediates the exit from the binding cleft adopts trans conformation and is highlighted in yellow.12 Figures were rendered using RasMol software on the designated PDB-data files.
Another important parameter of 14-3-3 interaction with histone H3 peptides is a conformational stabilization of the peptide by several intramolecular interactions.23 The phosphate oxyanion forms interactions with the H3G12 backbone amide. In addition, an intramolecular salt bridge is formed between arginine 8 (P − 2) and the phosphate oxyanion of serine 10. This is analogous to the interaction of 14-3-3 with the mode 2 consensus peptide where the guanidine group of the P − 4 arginine forms a salt bridge with the phosphate oxyanion. This conformation is not observed for the P − 3 arginine in mode 1 binding.12,23 Therefore, the interaction between H3S10ph and 14-3-3 exhibits structural features of mode 2 binding. However, in this case the exit of the peptide from the binding cleft is not mediated by the P + 2 proline but via the tandem glycine residues at P + 2 and P + 3. To this point, there are no structural data on the interaction between H3S28ph and 14-3-3. Given the identical amino acid composition amino-terminal of H3S10 and H3S28 it is likely that arginine 26 (P − 2) adopts a similar conformation as arginine 8 (P − 2) and forms a salt bridge with the phosphate oxyanion. This mode of interaction would imply the P + 2 proline (H3P30) adopting trans conformation thereby allowing the peptide to exit the binding cleft.12
Does isomerisation of prolines in the histone H3 tail therefore participate in the regulation of 14-3-3 binding to the histone H3 tail? Proline isomerisation constitutes an important factor in regulation of protein folding. For 14-3-3 interaction with target proteins, proline is strongly preferred at position P + 2 and adopts either cis conformation in mode 1 and trans conformation in mode 2 binding.12 In solution the cis conformation is relatively abundant (5–10% of peptidyl-prolyl bonds) compared to other non-prolyl peptide bonds and proline isomerisation events constitute an important factor for secondary structure formation.26 Peptidyl-prolyl isomerisation has been reported for histone H3P30 and H3P38 via the FKBP proline isomerase family member Fpr4.27 In this study histone H3P30 isomerisation was demonstrated to directly impact Set2 mediated K36 methylation. Therefore, it will be interesting to investigate whether H3P30 isomerisation could impact 14-3-3 binding to phosphorylated histone H3.
Extrinsic Factors Influencing 14-3-3 Histone H3 Interaction
Histone proteins are subject of an extensive and steadily expanding list of post-translational modifications (PTMs).28-30 Several reports indicate interphase H3S10 phosphorylation frequently coinciding with adjacent acetylation events (phosphoacetylation). This process has been investigated in particular for H3K9acS10ph and H3S10phK14ac phosphoacetylation.25,30-35 Studies using antibodies directed against double modified H3 species and mass spectrometry based approaches also demonstrated H3S10 phosphorylation co-existing with neighboring lysine methylation (mono-, di- and trimethylation) on H3K9. In addition, triple modified forms (H3K9meS10phK14ac) were identified.25,36,37
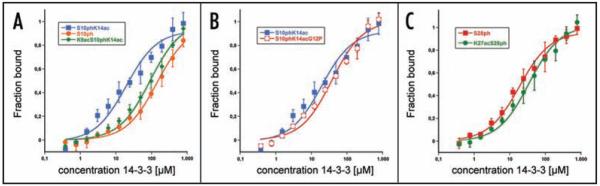
Recent evidence indicates that additional histone modifications modulate interaction between S10 phosphorylated histone H3 and 14-3-3. Additional acetylation on either H3K9 or H3K14 significantly increases the affinity for the S10 phosphorylated H3 tail.24,25 Importantly, this enhancing effect is only observed for single acetylation events, as an H3K9acS10phK14ac peptide showed interaction parameters similar to the single phosphorylated histone H3 tail (Fig. 2A and ref. 23). This demonstrates that discrete degrees of histone H3 acetylation result in different affinities for the interaction with 14-3-3.
Figure 2.
Extrinsic factors influencing the interaction of 14-3-3 with histone H3. (A) Discrete degrees of acetylation result in different effects on the interaction with 14-3-3ζ. Binding curves for the phosphorylated histone H3 peptide (H3S10ph), the single phosphoacetylated peptide (H3S10phK14ac) and the double phosphoacetylated peptide (H3K9acS10phK14ac) were determined as described for Figure 1. Single acetylation of H3K9 or H3K14 results in increased affinity of 14-3-3 for the H3S10ph peptide24,25 whereas for the double phosphoacetylated peptide (H3K9acS10phK14ac) this effect is abolished. (B) Acetylation cannot further increase the affinity for the H3S10phK14ac when proline is at position P + 2 from serine 10. (C) Acetylation of H3K27 does not increase binding to the H3S28ph peptide, which already contains a proline at P + 2.
Since the crystal structure of 14-3-3ζ in complex with the histone H3 tail has been determined with the phosphorylated and double phosphoacetylated form, it is unclear how single acetylation may increase the affinity of 14-3-3. The H3K14 side chain is directed outward of the 14-3-3 binding cleft, but the acetyl moiety is not visible in the crystal structure suggesting a flexible state of this group.23 In contrast, the acetyl group at H3K9 folds back and forms a hydrogen bond with the backbone amide of alanine 7, causing some minor reorganization of the peptide backbone. The selectivity of the 14-3-3 consensus motifs is restricted to the sequence from position P − 4 to P + 2.13 Thus, it is likely that acetylation of H3K14 causes increased affinity via a peptide intramolecular interaction rather than by direct interaction with 14-3-3. The exit of the histone H3S10ph peptide from the binding cleft is mediated by the tandem glycines at P + 2 and P + 3. This structural feature appears to be not optimal for the interaction with 14-3-3 and proline at P + 2 would be clearly favorable (Figs. 1C, D and 3).13,23 Therefore, we speculate that one mode how lysine acetylation could cause increased affinity may be via the stabilization of a kinked structure that might improve the exit of the peptide from the binding cleft directing it outwards analogously to the P + 2 proline in mode 2 consensus motif. Putatively, the peptide may only assume this conformation upon fitting into the cleft, which requires an extended conformation.23 This hypothesis is supported by the observation that mutation of H3G12 to proline results in increased affinity of 14-3-3 for the peptide (Fig. 1D) that is not further increased by additional acetylation of H3K14 (Fig. 2B). Similarly, the high binding affinity of 14-3-3 for H3S28ph is also not affected by additional acetylation of H3K27 (Fig. 2C).
Structural analysis shows the H3K9 acetyl group forming a hydrogen bond with the backbone amide of H3A7. This residue points outward the binding cleft and is not involved in intramolecular interactions in the non-acetylated peptide.23 Further, the acetyl group of H3K14 appears not to participate in any interactions in the double acetylated peptide and remains flexible.23 Interestingly, double H3K9/K14 acetylation does not significantly increase the interaction with the H3S10ph peptide (Fig. 2A and ref. 23). Likewise, dimethylation of H3K9 has no significant effect on the affinity of 14-3-3 for the peptide.25 These observations imply that charge neutralization via acetylation of one lysine (H3K9 or H3K14) is favorable for the interaction with 14-3-3, whereas charge neutralization of both lysines (H3K9 and H3K14) abrogates the enhancing effect. Based on these observations it is tempting to speculate that acetylation of one lysine may result in the formation of either an inter- or intramolecular interaction (see above), that supports the organization of the peptide in the 14-3-3 binding cleft. If both lysines are acetylated, this interaction cannot be established. The acetyl group of H3K9 folds back to form the hydrogen bond with the backbone amid of alanine 7, while the acetyl group on H3K14 adopts a flexible conformation.23
Another possible way how PTMs may facilitate the interaction with 14-3-3 comes from structural predictions of the histone H3 tail. The histone H3 amino-terminal region is assumed to be mainly unstructured. Several stretches were recently predicted to have a high probability of adopting α-helical conformation, in particular the stretch from H3T3 to H3S10.38 Computational simulation showed that the stability of this helical population may be significantly reduced upon lysine acetylation,38 whereas serine phosphorylation was predicted to stabilize helical conformations.39 In addition, the computer model predicts that single dimethylation of H3K9 shows no major shift in α-helix population, but in concert with H3K4 dimethylation leads to reduced stability of the α-helical conformation.38 In general, the interaction of proteins with the histone H3 tail requires an extended conformation. The relaxation of putative α-helical structures by PTMs could therefore enhance the contact with binding proteins by facilitating a shift in the equilibrium between the helical and non-helical conformation states. Although, the impact of H3K9/H3K14 double acetylation was not simulated, it appears that these effects are rather cumulative. Such an interpretation is therefore hardly compatible with the observation that double acetylation abolishes enhanced 14-3-3 binding (Fig. 2A).
Besides the discussed examples, the impact of several other possible modifications on the interaction between histone H3 and 14-3-3 has not yet been investigated. For example, methylation of arginine 8 (P − 2) could probably impact the interaction with the phosphate-oxyanion (see above). Also, phosphorylation of threonine 11 might modulate the accessibility of phosphorylated serine 10. However, it is not clear whether such hypothetical modification patterns are indeed established in vivo.
Implications for Combinatorial Modification Patterns
Several examples for modulation of protein binding via combinatorial modification patterns have been described,24,25,36,37,40-43 suggesting that histone modifications are frequently cooperative. A biological effect of a PTM might rather depend on the complete modification “make-up” of the histone tail or even entire nucleosomes than on a singular readout.
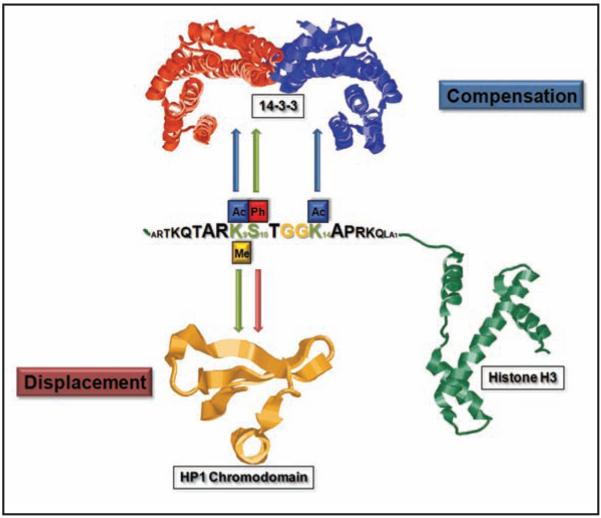
One obvious advantage of modulating the binding of PTM detector proteins to substrates via combinatorial modification patterns is the increased ability to fine tune the interaction and gain additional control levels. In the case of histone H3S10 phosphorylation the interplay with the binding protein of H3K9 methylation, heterochromatin protein 1 (HP1), and 14-3-3 proteins constitutes a reciprocal system. HP1 is displaced by additional S10 phosphorylation36,41 or phosphoacetylation44 while 14-3-3 is recruited at the same time (Fig. 4).25 Additional acetylation of H3K14 increases the affinity of 14-3-3 for H3K9me2S10ph histone H3 thereby supporting recruitment to relevant genomic regions. Hence, the triple modified form allows to efficiently “override” the transcriptional repressive H3K9 methylation not only by HP1 displacement but also via 14-3-3 recruitment.25 Since H3S10ph already leads to displacement of HP1, why should “overriding” of H3K9 methylation be important for transcriptional activation?
Figure 4.
Modulation of detector protein binding by combinatorial modification patterns. The panel depicts two distinct modes of action for combinatorial modification patterns. In the case of the 14-3-3 histone H3 interaction phosphorylation of serine 10 (green residue) is the initial trigger for binding (green arrow upwards), which is of low affinity. Additional acetylation of lysine 9 or 14 compensates for the non-optimal interaction platform provided by the P + 2/3 tandem glycine backbone (orange residues) and supports the interaction between histone H3 and 14-3-3 (blue arrows up). The binding of the HP1 chromodomain exemplifies another mode how complex modification patterns can impact the interaction of modification dependent binding proteins. The initial trigger for the interaction with the chromodomain is di- or trimethylation of H3K9 (green arrow down), which in general generates transcriptional repressive chromatin. The additional phosphorylation of serine10 results in displacement of HP1 and allows for subsequent interaction with 14-3-3 proteins (upper part).
Knockdown of particular 14-3-3 isoforms resulted in reduced transcriptional induction of genes regulated by histone H3 phosphoacetylation.25 Therefore, it appears that establishment of H3 phosphoacetylation and displacement of HP1 is not sufficient for gene activation. An H3S10ph binding protein with activator function seems to be required.
Shut-down of transcription in this system might involve the activity of histone deacetylases (HDACs) as well as serine/threonine phosphatases. These enzymes re-establish the repressive H3K9me2 signature without the requirement of histone methyltransferases and allow for re-association of HP1, provided that the promoter-associated nucleosomes are not exchanged during transcription.
Active deprivation of H3K9 methylation was demonstrated for other systems such as androgen receptor mediated transcriptional activation.45-47 In the case of some phosphoacetylation target genes H3K9 methylation is rather transformed into complex modification forms like H3K9meS10ph or H3K9meS10phK14ac.25,48 Why then is H3K9 methylation not always removed? The repressive effect of H3K9 methylation is in these modification states “ignored” since both activating modifications are efficiently bound by 14-3-3.25 Obviously, cellular regulation relies on memory systems and certain histone methylation signals must be retained. In these cases, the readout is rather regulated than the mark itself. Removal of phosphorylation and acetylation signals suffices for re-establishing a transcriptional repressive environment.
Two enzymes are able to remove methylation of H3K9. In the context of androgen receptor mediated transcriptional induction LSD1 resolves the dimethylated and monomethylated states. JmjC-domain containing demethylases like JMJD2C are active on trimethylated H3K9.45,47,49 Interestingly, both enzymes appear to be excluded from their substrates when H3S10 is phosphorylated. Also, the activity of LSD1 on hyperacetylated nucleosomal substrates is reduced.50,51 Obviously, phosphorylation and acetylation signals therefore can protect H3K9me against demodification. Interestingly, phosphorylation of threonine 11 was recently demonstrated to stimulate H3K9 demethylation and to facilitate androgen receptor mediated transcription46 suggesting that histone phosphorylation can also propagate demethylation depending on the particular modified residue.
Implications for Intrinsic Factors
All different enzymes adding or removing diverse but spatial closely located modifications on histones have to recognize and act on the same amino acid “platform” (apart from previously positioned modifications). Therefore, evolutionary constraints imposed on the amino-terminal tails of histones may have favored the generation of multi-modifiable patches, accessible to a vast variety of different enzymatic machineries, probably at the cost of substrate efficiency.
The same restrictions might limit the interaction with PTM-dependent binding proteins. One concrete example, the interaction between 14-3-3 and phosphorylated histone H3 was discussed in this article. H3S10 phosphorylation mediates only weak interaction with 14-3-3 proteins and one particular factor for this low affinity is the lack of the P + 2 proline, which is functionally replaced by tandem glycine residues (Fig. 1A). Substitution of the P + 2 position by proline would be clearly favorable for 14-3-3 binding (Fig. 1D), but evolutionary constraint retained tandem glycines, indicating that these residues may be important for other interactions and therefore indispensable.51 The insufficiency of the amino acid patch surrounding H3S10, to mediate strong initial interaction with 14-3-3 can be compensated by additional acetylation of either H3K9 or H3K14.24,25 One particular function of combinatorial modification patterns for this interaction may therefore be compensation of non-optimal interaction platforms (Fig. 4). Such non-optimal motifs may have originated from evolutionary constraint amino-acid mutability required to maintain modification versatility. However, acetylation of H3K9 or H3K14 has additional effects than modulating 14-3-3 binding. Also this modification is much more abundant than interphase histone phosphorylation. Therefore direct coevolution of both events appears unlikely.
Histone H3S10 phosphorylation can be mediated via mitogen activated protein (MAP) kinase pathways and some immediate early genes (IE) are rapidly and transiently induced by stress stimuli.33 It however appears desirable to restrict the plethora of potentially activated genes in a manner adequate to the precise stimulus. One possibility for such tight control is to limit the kinase substrate interaction. Indeed it was demonstrated that overexpression of the histone H3S10 kinase MSK1 does not change either distribution or overall amounts of histone H3S10 or H3S28 phosphorylation, despite full activation of the exogenous kinase.52 Obviously additional factors are critical to allow for the placement of either histone H3S10 or H3S28 phosphorylation. Not all genes targeted by histone H3S10 phosphorylation are activated upon transient MAP kinase stimulation but rather require complex modification patterns.25,34 The requirement for a dual modification also allows for a more refined binding regulation and transcriptional activation. Expression of these genes is more tightly regulated as two distinct pathways are required for full transcriptional activation.
Another example on the function of combinatorial modification patterns was provided by studies on the displacement of HP1 proteins bound to H3K9me2/3 via H3S10 phosphorylation (Fig. 4).36,41 In this context the combinatorial modification obviously does not compensate for non-optimal binding conditions but rather provides a rapid displacement of the binding protein without need to “erase” the epigenetic information of H3K9 methylation.53 Obviously this epigenetic “memory maintenance” system is extremely valuable during mitotic progression.
These examples demonstrate possible functional outcomes of combinatorial modification patterns: positive compensation for non-optimal binding motifs and increased regulatory control in the case of the 14-3-3 and H3S10ph interaction, or generation of unfavorable binding platforms epitomized by H3K9me2/3S10ph in the case of HP1 proteins (Fig. 4). It is important to mention that in vivo the complexity of these events may be significantly expanded by the putative contribution of additional factors and also the more limited access to nucleosomal histones.
Consideration of Histone Variants
Posttranslational modification of histone amino-terminal tails constitutes an important mechanism for the regulation of genome accessibility.54 Within the nucleosomal core histones specialized variants have evolved. These take over particular functions in genome organization like centromere maintenance or constitutive heterochromatin formation.30,54 Besides the centromere specific isoform, Centromeric protein A (CenpA), three additional histone H3 isoforms are expressed in mammals, designated as H3.1, H3.2 and H3.3.
Concerning the transcription-associated interaction between histone H3 and 14-3-3 proteins the latter isoform H3.3 is particular interesting. Histone H3.3 can be incorporated into chromatin outside of S-phase in a replication independent manner (RI), which is important for nucleosome exchange during transcription.55 Phosphorylation of H3S10 and H3S28 may be spatially separated52,56 and asymmetrically targeted to specific isoforms, as H3.3 was found to be the main species phosphorylated at serine 28 in chicken erythrocytes.57
Because of the spatial separation in interphase cells, H3S10ph and H3S28ph may correlate with transcriptional activation of distinct target genes. As interaction between 14-3-3 and H3S28ph is significantly stronger,23,25 target genes for H3S28 phosphorylation may be less dependent on additional histone acetylation to enable 14-3-3 binding. For genes targeted by serine 10 phosphorylation, additional lysine acetylation, besides other functions, is important to stabilize the interaction with 14-3-3.25
Although H3S28 phosphorylation may not require additional acetylation for 14-3-3 binding, the modification co-exists with additional histone H3 acetylation in vivo and is even stimulated by preceding HDAC inhibition.52 This indicates that histone acetylation may be nevertheless important for transcriptional activation of H3S28ph targets but in a different context than reinforcement of 14-3-3 binding. Based on the in vitro interaction studies the reason for this may differ between both systems. In the case of H3S10 phosphorylation, binding of 14-3-3 is stabilized by additional acetylation (H3K9 or H3K14). The interaction with the H3S28 phosphorylated histone H3 tail is not modulated (Fig. 2C). However, increased acetylation supports phosphorylation of H3S2852 and thereby creation of an efficiently bound 14-3-3 substrate. Therefore, the recruitment 14-3-3 to H3S10ph and H3S28ph, may be modulated by additional acetylation. H3S28 phosphorylation is more abundant in the context of hyperacetylated histone H3 and this may directly correlate with increased recruitment of 14-3-3 proteins and transcriptional activation.
It will be important to determine whether histone H3 acetylation directly mediates increased S28 phosphorylation, or if HDAC inhibition modulates the activity of H3S28 kinase activity.
Conclusions
The high degree of conservation and slow evolution of histone molecules emphasizes the evolutionary constraints imposed on these proteins that provide the structural basis for genome organization.54 Besides specialized histone variants, PTMs provide an additional tool for the generation of diversity in a more dynamic manner. Several studies indicate that PTMs can operate as combinatorial rather than single entity. These investigations demonstrated different readouts of combinatorial modifications either by generating positive or repulsive effects. The advantages of combinatorial modification patterns discussed here are multi-layered, which is emphasized by the different effects of known combination systems. These include increase in binding affinity and thereby gain of control options, as demonstrated for 14-3-3, or reduced affinity as demonstrated for HP1.24,25,36,41
A major step to investigate this interplay is the mapping of combinatorial modification patterns in vivo. Mass spectrometry based approaches are promising tools towards a profound understanding in combinatorial modification patterns.25,36,58-63 Such studies may also provide a basis for the generation of antibodies against complex PTM patterns and genome wide mapping approaches.
Complex PTM patterns may constitute biological relevant factors and yield a single readout rather than simple additive effects (e.g., the more acetylation the more binding the more transcription). This is supported by the observation that increased affinity of 14-3-3 for histone H3S10ph is mediated by single acetylation, but abandoned by double acetylation (Fig. 2A). Different “forms” of phosphoacetylation therefore result in different impacts on the interaction with 14-3-3 and not in simple additive effects.
Also the displacement of HP1 proteins from H3K9me2/3 by H3S10 phosphorylation constitutes a specific non-additive event. The obvious advantage of this system is the retention of epigenetic information. This appears to be desirable not only during mitotic progression36,41,44,53 but also during transcriptional activation of particular target genes.25,48 The generation of multiple modified histone forms may comprise an important tool to temporarily switch from an epigenetically determined state to another. The epigenetic information is not removed but temporarily “faded out” by assembly into complex PTM patterns. The generation of complex PTM combinations may therefore provide an elegant system to dynamically regulate the maintenance of cellular memory.
Table 1.
Dissociation constants in μM for differentially modified histone H3 peptides determined by fluorescence polarization measurements
| Peptide | Kd [μM] |
|---|---|
| H3Um (1–20) | 450,86 ± 158,67 |
| H3S10ph (1–20) | 140,68 ± 22,68 |
| H3S10phG12P (1–20) | 19,92 ± 5,36 |
| H3S10phK14ac (1–20) | 19,28 ± 5,6 |
| H3S10phK14acG12P (1–20) | 39,98 ± 7,9 |
| H3K9acS10phK14ac | 108,88 ± 18,6 |
| H3Um (25–38) | 403,55 ± 51,55 |
| H3S28ph (25–38) | 18,65 ± 2,9 |
| H3K27acS28ph (25–38) | 34,86 ± 5,68 |
| H3S28phP30A (25–38) | 162,0 ± 18,56 |
Values are average of at least three independent measurements (Kd [μM]) and Standard deviation is indicated.
Acknowledgements
This work was supported by the Austrian Science Fund (FWF P18746-B12) and the GEN-AU project “Epigenetic Plasticity of the Mammalian Genome” (Austrian Federal Ministry for Education, Science and Culture). Stefan Winter is a fellow of the Vienna Biocenter PhD program (Austrian Science Fund).
References
- 1.Darling DL, Yingling J, Boris A Wynshaw. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- 2.Thomas D, Guthridge M, Woodcock J, Lopez A. 14-3-3 protein signaling in development and growth factor responses. Curr Top Dev Biol. 2005;67:285–303. doi: 10.1016/S0070-2153(05)67009-3. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MB. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–7. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 4.Aitken A, Howell S, Jones D, Madrazo J, Martin H, Patel Y, Robinson K. Post-translationally modified 14-3-3 isoforms and inhibition of protein kinase C. Mol Cell Biochem. 1995;149:41–9. doi: 10.1007/BF01076562. [DOI] [PubMed] [Google Scholar]
- 5.Aitken A, Howell S, Jones D, Madrazo J, Patel Y. 14-3-3 alpha and delta are the phosphorylated forms of raf-activating 14-3-3 beta and zeta. In vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys MOTIF. J Biol Chem. 1995;270:5706–9. doi: 10.1074/jbc.270.11.5706. [DOI] [PubMed] [Google Scholar]
- 6.Woodcock JM, Murphy J, Stomski FC, Berndt MC, Lopez AF. The dimeric versus monomeric status of 14-3-3 zeta is controlled by phosphorylation of Ser58 at the dimer interface. J Biol Chem. 2003;278:36323–7. doi: 10.1074/jbc.M304689200. [DOI] [PubMed] [Google Scholar]
- 7.Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–4. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 8.Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–91. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 9.Aitken A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol. 2002;50:993–1010. doi: 10.1023/a:1021261931561. [DOI] [PubMed] [Google Scholar]
- 10.Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14:4721–33. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–97. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 12.Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–66. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 13.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 14.Coblitz B, Shikano S, Wu M, Gabelli SB, Cockrell LM, Spieker M, Hanyu Y, Fu H, Amzel LM, Li M. C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J Biol Chem. 2005;280:36263–72. doi: 10.1074/jbc.M507559200. [DOI] [PubMed] [Google Scholar]
- 15.Coblitz B, Wu M, Shikano S, Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580:1531–5. doi: 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci USA. 2005;102:1222–7. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, Peden A, Zemlickova E. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans. 2002;30:351–60. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- 18.Andrews RK, Harris SJ, McNally T, Berndt MC. Binding of purified 14-3-3 zeta signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry. 1998;37:638–47. doi: 10.1021/bi970893g. [DOI] [PubMed] [Google Scholar]
- 19.Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. Interaction of 14-3-3 with a non-phosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:5216–21. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- 20.Vincenz C, Dixit VM. 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J Biol Chem. 1996;271:20029–34. doi: 10.1074/jbc.271.33.20029. [DOI] [PubMed] [Google Scholar]
- 21.Wang B, Yang H, Liu YC, Jelinek T, Zhang L, Ruoslahti E, Fu H. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- 22.Zhang SH, Kobayashi R, Graves PR, Piwnica-Worms H, Tonks NK. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3 beta protein. J Biol Chem. 1997;272:27281–7. doi: 10.1074/jbc.272.43.27281. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 24.Walter W, Clynes D, Tang Y, Marmostein R, Mellor J, Berger SL. 14-3-3 interaction with histone H3 involves dual modification pattern of phosphoacetylation. Mol Cell Biol. 2008 doi: 10.1128/MCB.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, Ammerer G, Seiser C. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. Embo J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J Mol Biol. 1998;279:449–60. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 27.Nelson CJ, Rosa H Santos, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–16. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–36. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Winter S, Simboeck E, Seiser Christian. Open Chromatin. Genes, Genomes and Genomics. 2007;1:209–25. [Google Scholar]
- 31.Barratt MJ, Hazzalin CA, Cano E, Mahadevan LC. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc Natl Acad Sci USA. 1994;91:4781–5. doi: 10.1073/pnas.91.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung P, Tanner KG, Cheung WL, Corsi P Sassone, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 33.Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. Embo J. 2000;19:3714–26. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauser C, Schuettengruber B, Bartl S, Lagger G, Seiser C. Activation of the mouse histone deacetylase 1 gene by cooperative histone phosphorylation and acetylation. Mol Cell Biol. 2002;22:7820–30. doi: 10.1128/MCB.22.22.7820-7830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson S, Clayton AL, Mahadevan LC. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell. 2001;8:1231–41. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- 36.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 37.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–9. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Duan Y. Effects of Post-translation Modifications on the Structure and Dynamics of Histone H3 N-Terminal Peptide. Biophys J. 2008 doi: 10.1529/biophysj.107.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smart JL, McCammon JA. Phosphorylation stabilizes the N-termini of alpha-helices. Biopolymers. 1999;49:225–33. doi: 10.1002/(SICI)1097-0282(199903)49:3<225::AID-BIP4>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 41.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–80. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 42.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 43.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross talk between histone modifications in response to HDAC inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem. 2006 doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 44.Mateescu B, England P, Halgand F, Yaniv M, Muchardt C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 2004;5:490–6. doi: 10.1038/sj.embor.7400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 46.Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, Patnaik D, Higgins JM, Potier N, Scheidtmann KH, Buettner R, Schule R. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol. 2008;10:53–60. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–53. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 48.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–81. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 50.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280:41360–5. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 51.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 52.Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci. 2005;118:2247–59. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- 53.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 2006;5:2842–51. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- 54.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–91. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 56.Dunn KL, Davie JR. Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 2005;24:3492–502. doi: 10.1038/sj.onc.1208521. [DOI] [PubMed] [Google Scholar]
- 57.Sun JM, Chen HY, Espino PS, Davie JR. Phosphorylated serine 28 of histone H3 is associated with destabilized nucleosomes in transcribed chromatin. Nucleic Acids Res. 2007;35:6640–7. doi: 10.1093/nar/gkm737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–8. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2007;11:66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 60.Jacobs SA, Fischle W, Khorasanizadeh S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 2004;376:131–48. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
- 61.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–22. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci USA. 2007;104:2086–91. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K, Siino JS, Jones PR, Yau PM, Bradbury EM. A mass spectrometric “Western blot” to evaluate the correlations between histone methylation and histone acetylation. Proteomics. 2004;4:3765–75. doi: 10.1002/pmic.200400819. [DOI] [PubMed] [Google Scholar]