Abstract
A Gram-negative, extremely halophilic, coccoid archaeal strain, CM5T, was isolated from a crude sea-salt sample collected near Qingdao, China. The organism grew optimally at 35–40 °C and pH 6.0 in the presence of 20 % (w/v) NaCl. Its colonies were red in colour and it could use glucose as a sole carbon source for growth. The 16S rRNA gene sequence of CM5T was most closely related to those of Halococcus species. Its pattern of antibiotic susceptibility was similar to those of other described Halococcus species. Biochemical tests revealed no sign of H2S production or gelatin liquefaction. The main polar lipids of strain CM5T were phosphatidylglycerol, phosphatidylglycerol methylphosphate and sulfated diglycosyl diether. No phosphatidylglycerol sulfate was present. The DNA G+C content of strain CM5T was 61.2 mol% and it gave DNA–DNA reassociation values of 33.7, 57.1 and 29.6 %, respectively, with Halococcus salifodinae DSM 8989T, Halococcus dombrowskii DSM 14522T and Halococcus morrhuae ATCC 17082T. Based on its morphological and chemotaxonomic properties and phylogenetic analysis of 16S rRNA gene sequence data, we propose that CM5T should be classified within a novel species, Halococcus qingdaonensis sp. nov., with strain CM5T (=CGMCC 1.4243T=JCM 13587T) as the type strain.
The extremely halophilic bacteria are defined as microorganisms that grow best in media containing 2.5–5.2 M (saturated) NaCl (Kushner & Kamekura, 1988) and include members of both the Archaea and the Bacteria, among them aerobic halophilic archaea that require at least 12 % (2 M) NaCl for growth, which are classified within the family Halobacteriaceae. In recent years, many halobacterial strains have been isolated and described within novel species. The number of genera within this family has increased to 22 (http://www.bacterio.cict.fr/classifgenerafamilies.html#Halobacteriaceae). Halococcus was the second genus after Halobacterium to be classified within the family Halobacteriaceae (Skerman et al., 1980). Currently, there are five recognized species in this genus (Larsen, 1989; Stan-Lotter et al., 2002; Garrity et al., 2004; Goh et al., 2006). Here we report the isolation and taxonomic analysis of a strain representing a novel Halococcus species from a crude sea-salt sample collected near Qingdao in eastern China.
CM medium (Seghal & Gibbons, 1960) with 20 % NaCl at pH 7.0 was used for halobacterial enrichment and growth. The sample was firstly inoculated in liquid medium and cultured on a shaker (120 r.p.m.) at 37 °C in the dark until turbid, and then streaked onto solid medium to produce single colonies. Streaking was repeated several times to obtain pure single colonies. Purified strains were cultured and maintained in liquid or on solid ATCC213 medium with 18 % NaCl at pH 6.0 in the dark.
Seven days after being cultured in liquid medium, living cells were observed under a phase-contrast microscope. Gram staining was performed according to Dussault (1955) and electron microscopy was used to reveal the detailed morphology. Samples were fixed in 5 % (v/v) glutaraldehyde (in 0.2 M phosphate buffer, pH 7.2) for 2 h on ice, washed three times in phosphate buffer and subsequently post-fixed for 1 h in 2 % osmium tetroxide. The pellet was dehydrated with propanol and embedded in resin. Embedding was done according to the protocol of Spurr (1969). Ultrathin sections were prepared with an LKB ultramicrotome and double stained with uranyl acetate and lead citrate. After air-drying, the samples were examined with an electron microscope (JEM-1230). For negative staining, liquid culture of cells at the exponential growth phase was allowed to dry on the grids and stained with 2 % phosphotungstic acid.
Halobacterial growth was determined by measuring optical density at 460 nm at intervals during growth of the liquid culture. The effect of different concentrations of NaCl on growth of strain CM5T was tested in liquid ATCC213 medium. After 7 days of incubation, the optical density at 460 nm was measured. Susceptibility of CM5T to the antibiotics ampicillin, tetracycline, hygromycin, kanamycin, streptomycin, rifampicin, bacitracin, penicillin, chloramphenicol, neomycin and erythromycin (all from Sigma) was tested by placing 6 mm diameter discs containing 20 μg antibiotic on agar plates followed by 7 days incubation at 37 °C. Sensitivity was deemed strong when the diameter of the zone of inhibition was >15 mm (i.e. 4.5 mm beyond the antibiotic disc) and moderate between 6 and 15 mm (1–4.5 mm beyond the antibiotic disc).
Physiological and biochemical tests were performed according to Gibbons (1974) and Tian et al. (1997). Anaerobic growth of CM5T with nitrate as the electron acceptor was tested as described by Mancinelli & Hochstein (1986) and growth with DMSO and fermentation of l-arginine as the electron acceptor were tested as described by Oren et al. (1997) and Oren & Trüper (1990), in closed tubes fully filled with the growth medium and held in the dark for more than 1 month which were then compared with growth on media without the test compounds.
Cells for pigment determination were collected by centrifugation and washed twice with 25 % NaCl. They were extracted with a 1 : 1 (v/v) mixture of acetone and methanol for 1 h. After centrifugation, the absorption spectrum of the supernatant was determined. Polar lipids were extracted from 300 mg freeze-dried cells using the method described by Tindall (1990). They were further purified by extraction with chloroform/methanol/0.3 % NaCl (1 : 2 : 0.8, by vol.) and separated by two-dimensional silica-gel TLC (Ross et al., 1985). Polar-lipid extracts were spotted onto the corner of a 10×10 cm thin-layer silica-gel plate (60F254; Merck). The first direction was developed in chloroform/methanol/water (65 : 25 : 4, by vol.) and the second in chloroform/methanol/acetic acid/water (80 : 12 : 15 : 4, by vol.). Total lipids and specific functional groups were detected using phosphomolybdic acid, molybdenum blue, α-naphthol and Bial’s reagent (orcinol ferric chloride spray reagent) (Stan-Lotter et al., 1999, 2002). The equivalence of spots was determined by co-chromatography of extracts of known haloarchaea in two dimensions and by comparison with published data.
The DNA G+C content of strain CM5T was determined using the thermal denaturation method of Marmur & Doty (1962) with Escherichia coli JM105 as a control. We used the optical renaturation method (De Ley et al., 1970; Huß et al., 1983; Jahnke, 1992) to perform DNA–DNA hybridization experiments. The three most closely related strains, Halococcus dombrowskii DSM 14522T, Halococcus morrhuae ATCC 17082T and Halococcus salifodinae DSM 8989T, were used as reference strains.
PCR amplification of 16S rRNA genes was performed according to Wang et al. (2000) after total DNA extraction by using primers 5′-ATTCCGGTTGATCCTGCCGGA-3′ (primer 1; positions 6–25, according to E. coli numbering) and 5′-AGGAGGTGATCCAAGCCGCAG-3′ (primer 2; positions 1540–1521) with 36 cycles of denaturing (94 °C, 1 min), annealing (52 °C, 1 min) and extension (72 °C, 3 min). The PCR product was ligated to the T-vector and transformed into E. coli DH10 for purification and sequencing. Sequences used for comparison with the 16S rRNA gene sequence from strain CM5T were obtained from GenBank by using the blastn program and the sequence was aligned with closely related 16S rRNA gene sequences with clustal x software program version 1.83 (Thompson et al., 1997). The phylogenetic tree was constructed by the neighbour-joining method (Saitou & Nei, 1987) in mega program version 3.1 (Kumar et al., 2001, 2004). Confidence values of branches of the phylogenetic tree were determined using bootstrap analyses (Felsenstein, 1985) based on 1000 resamplings.
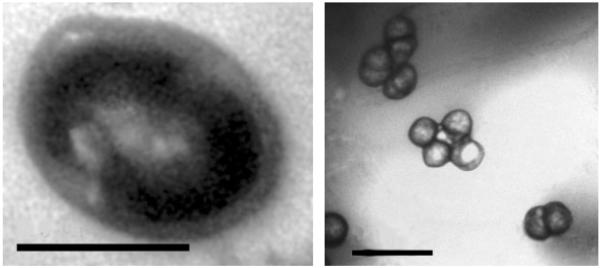
Cells of strain CM5T were coccoids, 0.6–1.5 μm in diameter (Fig. 1). They were non-motile and often arranged in doublets or tetrads; they stained Gram-negative and did not lyse in distilled water. The colonies were red in colour, wet and smooth-surfaced with clear edges and reached 0.5 mm in size after 7 days of culture at 37 °C on ATCC213 medium. Strain CM5T could grow at pH 4.0–9.0, with optimal growth at pH 6.0. It required at least 10 % NaCl for growth and 18 % was the optimum. The optimal Mg2+ concentration was 40 mM. The permissive temperature for growth was between 26 and 45 °C, with optimum growth between 35 and 40 °C.
Fig. 1.
Transmission electron micrographs of strain CM5T. Left, ultrathin section (bar, 0.5 μm); right, negative staining showing doublet/tetrad arrangements (bar, 2 μm).
Pigment determination revealed three absorbance spectrum peaks at 389, 495 and 527 nm for CM5T, which are characteristic of carotenoids in extremely halophilic archaea. Two-dimensional TLC analysis of the polar lipids in strain CM5T (Fig. 2) as well as the three reference strains revealed the presence of C20C20 and C20C25 archaeal core lipids, which were detected by double spots. Glycerol diethers of phosphatidylglycerol and phosphatidylglycerol methylphosphate and sulfated diglycosyl diether were present in strain CM5T; no phosphatidylglycerol sulfate was present. These patterns were similar to those of Hcc. morrhuae DSM 1307T, Hcc. salifodinae DSM 8989T and Hcc. dombrowskii DSM 14522T. Two unknown phospholipids and one unknown glycolipid were present in strain CM5T. The overall phospholipid pattern of strain CM5T was characteristic of members of the genus Halococcus (Ross et al., 1985; Wainø et al., 2000).
Fig. 2.
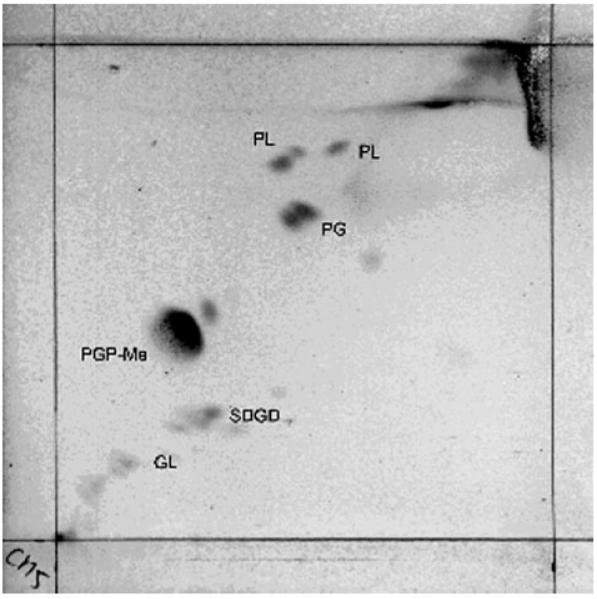
Polar lipid pattern of strain CM5T. PG, Glycerol diether of phosphatidylglycerol; PGP-Me, glycerol diether of phosphatidylglycerol methylphosphate; SDGD, sulfated diglycosyl diether; PL, unidentified phospholipid; GL, unidentified glycolipid.
Growth of strain CM5T was strongly inhibited by the antibiotics rifampicin and bacitracin and moderately inhibited by neomycin and chloramphenicol. No inhibition was observed when the strain was grown in the presence of erythromycin, ampicillin, kanamycin, streptomycin, tetracycline, hygromycin or penicillin. The susceptibility pattern of strain CM5T was similar to those of Hcc. dombrowskii DSM 14522T and Hcc. morrhuae DSM 1307T (determined in our experiments), except that the latter strains were moderately susceptible to erythromycin, similar to the result described by Stan-Lotter et al. (2002).
Strain CM5T was distinct from other Halococcus species in a number of biochemical properties (Table 1). Unlike Hcc. morrhuae and Halococcus saccharolyticus, it did not produce H2S and was negative for gelatin hydrolysis. Nitrate reduction was not detected with CM5T. Indole but not organic acids could be produced from sugars. It had catalase, but no oxidase, arginine dihydrolase or urease activity. It could use glucose, galactose, sucrose, inositol, fructose and rhamnose, but not sorbitol, cellobiose, mannitol, dextran or lactose, as sole carbon sources. CM5T could not grow anaerobically in the presence of nitrate or DMSO or by fermenting l-arginine.
Table 1. Characteristics that differentiate strain CM5T from type strains of other Halococcus species.
Strains: 1, strain CM5T; 2, Hcc. morrhuae ATCC 17082T; 3, Hcc. dombrowskii H4T; 4, Hcc. salifodinae DSM 8989T (data in columns 1–4 determined in this study unless indicated); 5, Hcc. saccharolyticus ATCC 49257T (data from Montero et al., 1989); 6, Hcc. hamelinensis 100A6T (data from Goh et al., 2006). +, Positive reaction or growth; −, no reaction or growth; v, variable; nd, no data available.
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Optimum NaCl concentration (% w/v) | 18 | 20–30a* | 20–30b | 20–30c | 20–30 | 15 |
| pH range for growth | 4.0–9.0 | 5.5–8.0a | 5.2–8.0b | 6.8–9.5c | 6.0–8.0 | 4.0–9.0 |
| Oxidase | − | +a | + | + | + | − |
| Nitrate reduction | − | + | + | + | + | + |
| Starch hydrolysis | + | − | − | − | − | + |
| Gelatin liquefaction | − | +a | + | + | v | − |
| Tween 80 hydrolysis | − | +a | − | + | − | nd |
| Use of carbohydrates: | ||||||
| Glucose | + | − | − | + | nd | + |
| Lactose | − | − | − | + | + | − |
Data taken from following references:
The G+C content of strain CM5T was 61.2 mol%, similar to the values determined for Hcc. morrhuae [62.3 mol% (this study); 61–66 mol% (Larsen, 1989)], Hcc. dombrowskii (61.3 mol%; Stan-Lotter et al., 2002) and Hcc. salifodinae [62.7 mol% (this study); 62±1 mol% (Denner et al., 1994)]. DNA–DNA reassociation values between strain CM5T and related type strains were 33.65 % with Hcc. salifodinae DSM 8989T, 57.1 % with Hcc. dombrowskii DSM 14522T and 29.6 % with Hcc. morrhuae ATCC 17082T (each repeated three times). Levels of 70 % or more relatedness can be considered indicative of species levels of similarity (Gutierrez et al., 1989, 1990); hence, strain CM5T represented a novel species of the genus Halococcus.
The full sequence (1476 bases) of the 16S rRNA gene of strain CM5T was determined. Phylogenetic analysis of strain CM5T using 16S rRNA gene sequences showed high similarity to Hcc. morrhuae ATCC 17082T (99.3 %) and Hcc. dombrowskii H4T (99.2 %), but less to Hcc. salifodinae DSM 8989T (94 %), Hcc. saccharolyticus ATCC 49257T (93.8 %) and Halococcus hamelinensis 100A6T (93.2 %). Phylogenetic analysis based on neighbour-joining showed that strain CM5T forms a branch within the Hcc. morrhuae lineage (Fig. 3). The genus Halococcus appears to contain at least two lineages on the basis of 16S rRNA gene sequence data: one contains Hcc. salifodinae and Hcc. saccharolyticus, while the other consists of Hcc. morrhuae and other coccoid strains (Stan-Lotter et al., 2002).
Fig. 3.

Phylogenetic relationships of strain CM5T and other related taxa, constructed using neighbour-joining method and based on 16S rRNA gene sequences. Halalkalicoccus tibetensis DS12T was used as an outgroup. Bootstrap values are shown as percentages of 1000 replicates. Bar, 0.01 changes per nucleotide position.
On the basis of its 16S rRNA gene sequence, G+C content, polar-lipid content, antibiotic sensitivity and other characteristics, the coccoid strain CM5T was identified as being a halophilic archaeon of the genus Halococcus. Unlike Hcc. morrhuae and Hcc. dombrowskii, CM5T produced no H2S, was not sensitive to tetracycline and could use glucose as a sole carbon source. Hcc. dombrowskii requires at least 15 % NaCl for growth, and the optimal concentration is 25–30 % (Stan-Lotter et al., 2002), whereas the minimal and optimal NaCl concentrations for growth of strain CM5T were 10 and 18–20 %, respectively. Strain CM5T differed from Hcc. dombrowskii and Hcc. morrhuae with respect to its cellular morphology and arrangement, DNA–DNA reassociation value, susceptibility to some antibiotics and usage of carbohydrates. The values of less than 70 % DNA–DNA reassociation between CM5T and Hcc. morrhuae ATCC 17082T (29.6 %) and Hcc. dombrowskii DSM 14522T (57.1 %) also support the suggestion to designate CM5T as representing a novel species. In conclusion, we describe CM5T as the type strain of a novel species, for which we propose the name Halococcus qingdaonensis sp. nov.
Description of Halococcus qingdaonensis sp. nov.
Halococcus qingdaonensis (qing.dao.nen’sis. N.L. masc. adj. qingdaonensis pertaining to Qingdao, from where the type strain was isolated).
Aerobic, Gram-negative, non-motile cocci, 0.6–1.5 μm in diameter, often occurring as doublets or tetrads. Colonies are red in colour, wet and smooth-surfaced with clear edges, reaching 0.5 mm in size after 7 days of cultivation at 37 °C. The optimum temperature for growth is 35–40 °C; the pH range for growth is 4.0–9.0, with optimum growth at pH 6.0. It requires at least 10 % NaCl for growth and 18 % is the optimum. No lysis is observed in distilled water. The optimal Mg2+ concentration is 40 mM. Catalase reaction is positive. It has no oxidase, arginine dihydrolase or urease activity. Gelatin is not liquefied. The reaction for Tween 80 is negative. Glucose can be used as the sole carbon source for growth. The type strain is susceptible to rifampicin and bacitracin, moderately susceptible to neomycin and chloramphenicol and resistant to erythromycin, ampicillin, kanamycin, streptomycin, tetracycline, hygromycin and penicillin. Main polar lipids are phosphatidylglycerol, phosphatidylglycerol methylphosphate, sulfated diglycosyl diether. No phosphatidylglycerol sulfate is present. The G+C content of the type strain is 61.2 mol%.
The type strain, CM5T (=CGMCC 1.4243T=JCM 13587T), was isolated from a crude sea-salt sample collected near Qingdao in eastern China.
Acknowledgements
This work was supported by the National Science Foundation of China (NSFC, 3027) and the Austrian Science Foundation (FWF), P18256. We thank Mr Bing Pi for his help in the phylogenetic analysis. We are indebted to Dr Wen-jun Li, Yunan University, China, and Professor Pei-jin Zhou, Institute of Microbiology, CAS, for their excellent technical assistance in determination of G+C content and DNA–DNA hybridization experiments.
Footnotes
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain CM5T is AY243109.
References
- De Ley L, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Denner EBM, McGenity TJ, Busse H-J, Grant WD, Wanner G, Stan-Lotter H. Halococcus salifodinae sp. nov., an archaeal isolate from an Austrian salt mine. Int J Syst Bacteriol. 1994;44:774–780. [Google Scholar]
- Dussault HP. An improved technique for staining red halophilic bacteria. J Bacteriol. 1955;70:484–485. doi: 10.1128/jb.70.4.484-485.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn TG. Bergey’s Manual of Systematic Bacteriology. 2nd edn, release 5.0. Springer; New York: 2004. Taxonomic outline of the prokaryotes. http://141.150.157.80/bergeysoutline/main.htm. [Google Scholar]
- Gibbons NE. Family V. Halobacteriaceae fam. nov. In: Buchanan RG, Gibbons NE, editors. Bergey’s Manual of Determinative Bacteriology. 8th edn Williams & Wilkins; Baltimore: 1974. pp. 269–273. [Google Scholar]
- Goh F, Leuko S, Allen MA, Bowman JP, Kamekura M, Neilan BA, Burns BP. Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int J Syst Evol Microbiol. 2006;56:1323–1329. doi: 10.1099/ijs.0.64180-0. [DOI] [PubMed] [Google Scholar]
- Grant WD, Larsen H. Group III. Extremely halophilic archaeobacteria. Order Halobacteriales ord. nov. In: Staley JT, Bryant MP, Pfennig N, Holt JG, editors. Bergey’s Manual of Systematic Bacteriology. vol. 3. Williams & Wilkins; Baltimore: 1989. pp. 2216–2219. [Google Scholar]
- Gutierrez MC, Ventosa A, Ruiz-Berraquero F. DNA-DNA homology studies among strains of Haloferax and other halobacteria. Curr Microbiol. 1989;18:253–256. [Google Scholar]
- Gutierrez MC, Ventosa A, Ruiz-Berraquero F. Deoxyribonucleic acid relatedness among species of Haloarcula and other halobacteria. Biochem Cell Biol. 1990;68:106–110. [Google Scholar]
- Huß VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Jahnke K-D. BASIC computer program for evaluation of spectroscopic DNA renaturation data from Gilford System 2600 spectrophotometer on a PC/XT/AT type personal computer. J Microbiol Methods. 1992;15:621–627. [Google Scholar]
- Kumar S, Tamura K, Jakobson I-B, Nei M. mega2: molecular evolution analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobson I-B, Nei M. mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kushner DJ, Kamekura M. Physiology of halophilic eubacteria. In: Rodriguez-Valera F, editor. Halobacteria. vol. 1. CRC Press; Boca Raton, FL: 1988. pp. 109–140. [Google Scholar]
- Larsen H. Genus IV. Halococcus Schoop 1935, 817AL. In: Staley JT, Bryant MP, Pfennig N, Holt JG, editors. Bergey’s Manual of Systematic Bacteriology. vol. 3. Williams & Wilkins; Baltimore: 1989. pp. 2228–2230. [Google Scholar]
- Mancinelli RL, Hochstein LI. The occurrence of denitrification in extremely halophilic bacteria. FEMS Microbiol Lett. 1986;35:55–58. doi: 10.1111/j.1574-6968.1986.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its denaturation temperature. J Mol Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Montero CG, Ventosa A, Rodriguez-Valera F, Kates M, Moldoveanu N, Ruiz-Berraquero F. Halococcus saccharolyticus sp. nov., a new species of extremely halophilic non-alkaliphilic cocci. Syst Appl Microbiol. 1989;12:167–171. [Google Scholar]
- Oren A, Trüper HG. Anaerobic growth of halophilic archaeobacteria by reduction of dimethylsulfoxide and trimethylamine N-oxide. FEMS Microbiol Lett. 1990;70:33–36. [Google Scholar]
- Oren A, Ventosa A, Grant WD. Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol. 1997;47:233–238. [Google Scholar]
- Ross HM, Grant WD, Harris JE. Lipids in archaeabacterial taxonomy. In: Goodfellow M, Minnikin DE, editors. Chemical Methods in Bacterial Systematics. Academic Press; London: 1985. pp. 289–299. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seghal SN, Gibbons NE. Effect of some metal ions on the growth of Halobacterium cutirubrum. Can J Microbiol. 1960;6:165–169. doi: 10.1139/m60-018. [DOI] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA, editors. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stan-Lotter H, McGenity TJ, Legat A, Denner EBM, Glaser K, Stetter KO, Wanner G. Very similar strains of Halococcus salifodinae are found in geographically separated Permo-Triassic salt deposits. Microbiology. 1999;145:3565–3574. doi: 10.1099/00221287-145-12-3565. [DOI] [PubMed] [Google Scholar]
- Stan-Lotter H, Pfaffenhuemer M, Legat A, Busse H-J, Radax C, Gruber C. Halococcus dombrowskii sp. nov., an archaeal isolate from a Permian alpine salt deposit. Int J Syst Evol Microbiol. 2002;52:1807–1814. doi: 10.1099/00207713-52-5-1807. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak K, Jeanmougin F, Higgins DG. The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XY, Xu Y, Liu HC. A new species of Natronobacterium. Wei Sheng Wu Xue Bao. 1997;37:1–6. (in Chinese) [PubMed] [Google Scholar]
- Tindall BJ. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol. 1990;13:128–130. [Google Scholar]
- Wainø M, Tindall BJ, Ingvorsen K. Halorhabdus utahensis gen. nov., sp. nov., an aerobic, extremely halophilic member of the Archaea from Great Salt Lake, Utah. Int J Syst Evol Microbiol. 2000;50:183–190. doi: 10.1099/00207713-50-1-183. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xu Y, Zhou P. Taxonomy of a new species of haloalkalophilic archaeon. Wei Sheng Wu Xue Bao. 2000;40:115–120. (in Chinese) [PubMed] [Google Scholar]