The biological role of 2′-O-methylation of host and viral mRNA has remained elusive. Thiel and co-workers show that this modification modulates the induction of type I interferon and sensitivity to interferon.
Supplementary information
The online version of this article (doi:10.1038/ni.1979) contains supplementary material, which is available to authorized users.
Subject terms: Innate immunity, Virus-host interactions, RNA modification
Abstract
The 5′ cap structures of higher eukaryote mRNAs have ribose 2′-O-methylation. Likewise, many viruses that replicate in the cytoplasm of eukaryotes have evolved 2′-O-methyltransferases to autonomously modify their mRNAs. However, a defined biological role for 2′-O-methylation of mRNA remains elusive. Here we show that 2′-O-methylation of viral mRNA was critically involved in subverting the induction of type I interferon. We demonstrate that human and mouse coronavirus mutants lacking 2′-O-methyltransferase activity induced higher expression of type I interferon and were highly sensitive to type I interferon. Notably, the induction of type I interferon by viruses deficient in 2′-O-methyltransferase was dependent on the cytoplasmic RNA sensor Mda5. This link between Mda5-mediated sensing of viral RNA and 2′-O-methylation of mRNA suggests that RNA modifications such as 2′-O-methylation provide a molecular signature for the discrimination of self and non-self mRNA.
Supplementary information
The online version of this article (doi:10.1038/ni.1979) contains supplementary material, which is available to authorized users.
Main
Innate immune recognition of pathogen-associated molecular patterns facilitates the distinction between immunological self and non-self1. For cytoplasmic viral RNA, this involves detection by cytoplasmic RIG-I-like receptors, such as RIG-I and Mda5, and the initiation of signaling cascades that induce the expression of cytokines, including type I interferon. Interferon-α (IFN-α) and IFN-β are secreted and signal through the type I interferon receptor (IFNAR) to transmit a danger signal to neighboring cells. The activated IFNAR triggers a signaling pathway consisting of the kinase Jak and STAT transcription factors, inducing the expression of a large array of interferon-stimulated genes encoding molecules with antiviral activity, thus establishing the so-called 'host cell antiviral state'2,3,4. These interferon-stimulated genes include those encoding the protein kinase PKR, as well as stress-inducible proteins such as IFIT1 and IFIT2 (also known as ISG56 and ISG54, respectively), which impair the host cell protein synthesis apparatus4,5,6,7.
Although the distinction between self and non-self RNA is believed to rely on the molecular signatures found in pathogen-associated molecular patterns, the exact nature of such signatures remains elusive. Both cytosolic RIG-I-like receptors (RIG-I and Mda5) bind to double-stranded RNA (dsRNA); however, RIG-I seems to 'prefer' short dsRNA, whereas Mda5 can specifically bind long dsRNA8. In addition, RIG-I can specifically recognize 5′-triphosphate groups on single-stranded RNA and (partially) dsRNA9,10,11. In contrast, eukaryotic mRNA, which is not recognized by RIG-I or Mda5, usually has a 5′ cap structure methylated at the N7 position of the capping guanosine residue (cap 0), the ribose-2′-O position of the 5′-penultimate residue (cap 1) and sometimes at adjoining residues (cap 2)12. Two evolutionary forces are thought to be responsible for the presence of 5′ cap structures on eukaryotic mRNA: the appearance of 5′ exonucleases in eukaryotes and the need for a means of directing mRNA to the eukaryotic ribosome13. Thus, eukaryotic mRNA 5′ cap structures are known to increase mRNA stability and translational efficacy. Notably, although N7 methylation is important in many mRNA-related processes, such as transcriptional elongation, polyadenylation, splicing, nuclear export and efficient translation, there is no obvious reason for the evolution of 2′-O-methylation of mRNA ribose in cap 1 and cap 2 structures in higher eukaryotes.
The functional importance of mRNA 5′-structures is inferred from the fact that many viruses that replicate in the cytoplasm (such as picornaviruses, flaviviruses, coronaviruses and poxviruses) have evolved alternative 5′ elements, such as small viral proteins linked to the 5′ end of genomic RNA14, or encode functions associated with the formation of a 5′ cap that are homologous to those found in eukaryotic cells, such as RNA 5′-triphosphatase, RNA guanylyltransferase, RNA guanine-N7-methyltransferase (N7-methyltransferase), and 2′-O-methyltransferase. The fact that RNA 5′-triphosphate groups activate RIG-I (refs. 9,10) suggests that viruses must hide or modify their RNA 5′ structures to evade innate immune recognition. In addition, RIG-I activation is diminished when 5′-triphosphate RNA contains modified nucleotides9. Thus, RNA modifications such as methylation could be critical factors for the activation of RNA-specific pattern-recognition receptors. Notably, this idea of methylation-based distinction of self and non-self nucleic acids is well established for DNA, as the methylation status of CpG dinucleotide motifs in DNA is the structural basis of the activation of Toll-like receptor 9 (TLR9)15. Moreover, DNA methylation represents the basis for the ancient bacterial restriction and modification systems that allow bacteria to distinguish between foreign DNA and the bacterial genome.
Here we show that 2′-O-methylation of viral mRNA is biologically important in the context of innate immune responses in the host cell. We demonstrate that human and mouse coronavirus mutants lacking 2′-O-methyltransferase activity induced higher expression of type I interferon and were extremely sensitive to type I interferon treatment. Furthermore, a mouse coronavirus mutant with an inactivated 2′-O-methyltransferase was attenuated in wild-type macrophages but replicated efficiently in the absence of IFNAR or Mda5. The coronavirus 2′-O-methyltransferase mutants were apathogenic in wild-type mice; however, viral replication and spreading was restored in mice lacking IFNAR and in mice lacking TLR7 and Mda5, the two main sensors of coronavirus RNA. Collectively our results show a link between Mda5-mediated sensing of viral RNA and 2′-O-methylation of mRNA and suggest that RNA modifications such as 2′-O-methylation provide a molecular signature for the distinction of self and non-self mRNA.
Results
Deficiency in 2′-O-methyltransferase in human coronavirus infection
Coronaviruses are single-stranded (+) RNA viruses that replicate in the cytoplasm and have evolved N7-methyltransferases and 2′-O-methyltransferases to methylate their viral mRNA 5′ cap structures16,17,18,19. The coronavirus 2′-O-methyltransferase activity is associated with the viral nonstructural protein nsp16, which is highly conserved among coronaviruses (Fig. 1a,b), and an integral subunit of the viral replicase-transcriptase complexes located at virus-induced double-membraned vesicles in the host cell cytoplasm. Coronavirus 2′-O-methyltransferase belongs to the human fibrillarin and Escherichia coli RrmJ-like methyltransferase family20 that includes many viral and cellular homologs (Fig. 1c and Supplementary Table 1).
Figure 1. Conservation of viral 2′-O-methyltransferases.
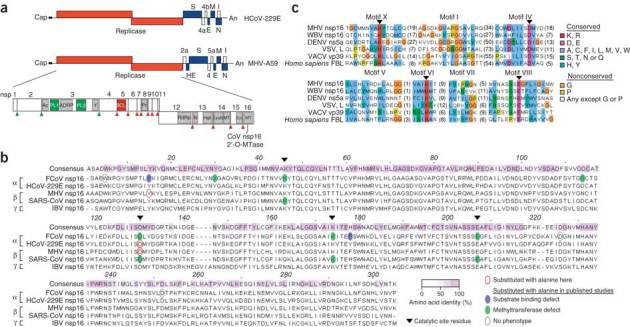
(a) Human and mouse coronavirus genomes, including viral open reading frames (boxes; labels above and below identify gene products). The replicase gene, including conserved domains and viral proteinase cleavage sites (upward arrowheads) that separate nsp1–nsp16, is enlarged. CoV nsp16 MTase, nsp16-associated 2′-O-methyltransferase. (b) ClustalW2 alignment of coronavirus nsp16 amino acid sequences, representative of α-, β- and γ-coronaviruses. Keys (bottom right) identify sequence conservation (amino acid identity), amino acid substitutions and phenotypes of mutant proteins. FCoV, feline coronavirus; MHV, MHV strain A59; SARS-CoV, severe acute respiratory syndrome coronavirus; IBV, infectious bronchitis virus. (c) Comparison of the sequences of viral and cellular methyltransferase motifs. WBV, white bream virus (order Nidovirales); DENV, dengue virus (family Flaviviridae); VSV, vesicular stomatitis virus (order Mononegavirales); VACV, vaccinia virus (family Poxviridae); FBL, fibrillarin (Homo sapiens). Right, key for amino acid similarity (single-letter codes) and conservation according to default ClustalX.
To address the biological importance of 2′-O-methylation of mRNA in the context of host cell innate immune responses, we first used a human model of coronavirus infection. We generated a mutant of the recombinant human coronavirus strain 229E (HCoV-229E) with an inactivated 2′-O-methyltransferase. We produced this mutant by substituting alanine for the aspartic acid at position 129 of the highly conserved catalytic KDKE tetrad of nsp16 (HCoV-D129A; Fig. 1b). Notably, this substitution completely abrogates the 2′-O-methyltransferase activity of recombinant, bacteria-expressed nsp16 proteins of feline coronavirus and severe acute respiratory syndrome coronavirus16,18. The mutant virus had a small plaque phenotype and less replication in the human fibroblast MRC-5 cell line (Fig. 2a,b). We were able to readily methylate poly(A)-containing RNA from HCoV-D129A-infected cells using the vaccinia virus 2′-O-methyltransferase VP39 in vitro21 (Fig. 2c), which confirmed the loss of 2′-O-methyltransferase activity by HCoV-D129A. In contrast, in vitro 2′-O-methylation of poly(A)-containing RNA derived from HCoV-229E-infected cells was indistinguishable from the 2′-O-methylation of poly(A)-containing RNA obtained from mock-infected cells. We observed significantly higher IFN-β expression in blood-derived human macrophages after infection with HCoV-D129A than after infection with HCoV-229E, (Fig. 2d) and complete restriction of HCoV-D129A replication in human macrophages pretreated with IFN-α (Fig. 2e). These results suggest a biological role for 2′-O-methylation of mRNA in the context of the induction of type I interferon and antiviral effector mechanisms stimulated by type I interferon.
Figure 2. The HCoV 2′-O-methyltransferase mutant has altered replication kinetics and induction of and sensitivity to type I interferon.
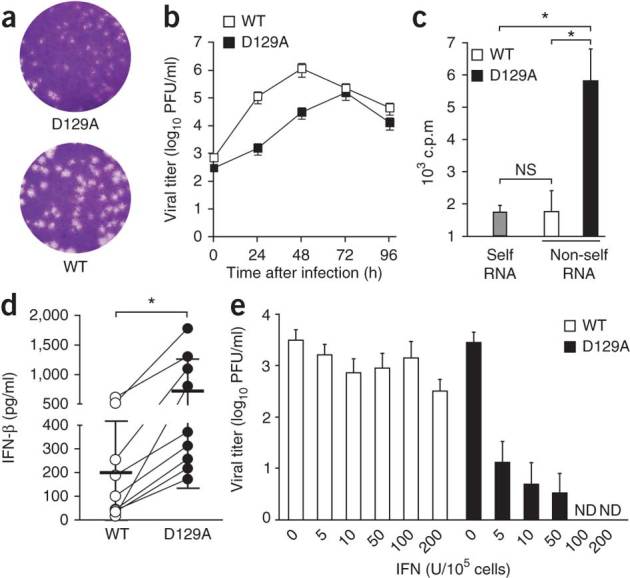
(a) Plaque assay of HCoV-229E and HCoV-D129A. (b) Replication kinetics of wild-type HCoV-229E (WT) and the mutant HCoV-D129A (D129A) in MRC-5 cells infected at an MOI of 0.1, presented as viral titer in plaque-forming units (PFU). (c) Incorporation of 3H into poly(A)-containing RNA derived from mock-infected cells (Self RNA) or cells infected with HCoV-229E or HCoV-D129A (Non-self RNA) after in vitro 2′-O-methylation with VP39. NS, not significant (P > 0.05); *P < 0.01 (unpaired Student's t-test). (d) Enzyme-linked immunosorbent assay of IFN-β in supernatants of human blood–derived macrophages 24 h after infection with HCoV-229E or HCoV-D129A (MOI = 1). Each symbol represents an individual donor (n = 9); thick horizontal lines indicate the mean (thin horizontal lines, s.d.). *P < 0.005 (Wilcoxon matched-pairs test). (e) Plaque assay of viral titers in human blood–derived macrophages pretreated with increasing doses of IFN-α (horizontal axis) 4 h before infection with HCoV-229E or HCoV-D129A (MOI = 1), assessed 24 h after infection. ND, not detected. Data are representative of three experiments (a) or represent two (b,e) or three (c,d) independent experiments (average and s.e.m. of triplicates in b; mean and s.d. in c; error bars, s.d. of six samples in e).
Mda5-dependent induction of type I interferon
To extend our studies on the effect of 2′-O-methylation on coronavirus-induced innate immune responses, we used an animal model of coronavirus infection with mouse hepatitis virus strain A59 (MHV-A59) as a natural mouse pathogen. Plasmacytoid dendritic cells have a unique and crucial role in sensing coronavirus RNA via TLR7 that ensures the swift production of type I interferon after viral encounter22,23. Other target cells such as primary fibroblasts, neurons, astrocytes, hepatocytes and conventional dendritic cells do not have detectable production of type I interferon after infection with MHV23,24. The exceptions to that are macrophages and microglia, which can respond with expression of type I interferon after MHV infection, although only to moderate amounts25,26. The expression of type I interferon detected in macrophages and microglia is dependent on Mda5 (ref. 25).
We generated a recombinant MHV lacking 2′-O-methyltransferase activity by substituting alanine for the aspartic acid at the 2′-O-methyltransferase active site at position 130 of nsp16 (MHV-D130A; Fig. 1b). In addition, we generated a recombinant MHV mutant with a tyrosine-to-alanine substitution at the putative cap 0–binding site of nsp16 (MHV-Y15A; Fig. 1b). This substitution impairs cap 0 binding for the corresponding feline coronavirus nsp16 mutant Y14A18, and we expected that this substitution would result in less 2′-O-methylation of coronaviral mRNA, rather than complete abrogation of this. Indeed, in vitro methylation of mRNA with the vaccinia virus 2′-O-methyltransferase VP39 (ref. 21) confirmed the differences in 2′-O-methylation of mRNA obtained from MHV-infected cells. The transfer of 3H-labeled methyl groups from the methyl donor S-adenosyl-methionine to mRNA was less efficient for mRNA derived from MHV-Y15A-infected cells than for mRNA from MHV-D130A-infected cells but significantly more efficient than for mRNA from MHV-A59-infected cells (Fig. 3a). These results demonstrate the loss of 2′-O-methyltransferase activity by MHV-D130A and show that a considerable proportion of MHV-Y15A mRNA is not methylated at the 2′-O position.
Figure 3. MHV 2′-O-methyltransferase mutants induce IFN-β in an Mda5-dependent manner.

(a) Ethidium bromide staining (left) of poly(A)-containing RNA (300 ng) from cells infected with wild-type MHV-A59 (WT), MHV-Y15A (Y15A) or MHV-D130A (D130A), separated by electrophoresis through a 1% agarose gel; right margin (1–7), genomic and subgenomic mRNA; left margin, size in kilobases (kb). Right, incorporation of 3H into poly(A)-containing RNA from mock-infected cells or cells infected with MHV-A59, MHV-Y15A or MHV-D130A after in vitro 2′-O-methylation with VP39. (b) Replication kinetics of MHV-A59, MHV-Y15A and MHV-D130A in 17Cl1 cells after infection at an MOI of 1 or 0.0001. (c–e) Enzyme-linked immunosorbent assay of IFN-β in supernatants of wild-type (WT; c), IFNAR-deficient (Ifnar−/−; d) or Mda5-deficient (Mda5−/−; e) macrophages (MΦ; 1 × 106) infected with wild-type or mutant MHV or Sendai virus (SeV) at an MOI of 1 and assessed 15 h after infection. (f,g) Quantitative RT-PCR analysis of the kinetics of IFN-β mRNA expression in wild-type (f) or IFNAR-deficient (g) macrophages (1 × 106) infected with MHV-A59, MHV-Y15A or MHV-D130A (MOI = 1), presented relative to its expression in uninfected cells. *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired Student's t-test). Data represent seven (a), two (b,f,g) or three (c–e) independent experiments (mean and s.d. of seven (a) or six (c–g) samples, or mean and s.e.m. of six samples (b)).
In cell culture, the replication kinetics of the mutant viruses MHV-D130A and MHV-Y15A differed only slightly from those of MHV-A59 after infection of the mouse fibroblast cell line 17Cl1 at a high multiplicity of infection (MOI) or low MOI (1 or 0.0001, respectively; Fig. 3b). Also, there was no notable difference among cells infected with MHV-A59, MHV-D130A or MHV-Y15A in the formation and morphology of double-membraned vesicles in the cytoplasm, as observed by electron microscopy (Supplementary Fig. 1). That observation is relevant to the cytoplasmic sensing of viral RNA, as coronavirus double-membraned vesicles are known to contain dsRNA. Infection with either of the 2′-O-methyltransferase mutants (MHV-Y15A or MHV-D130A) resulted in more production of type I interferon in wild-type macrophages at 15 h after infection (Fig. 3c and Supplementary Fig. 2a). Likewise, type I interferon was efficiently produced in IFNAR-deficient macrophages infected with MHV-D130A or MHV-Y15A (Fig. 3d and Supplementary Fig. 2b), which indicated that the MHV 2′-O-methyltransferase mutants triggered the production of type I interferon in the absence of IFNAR signaling. Neither MHV-A59 nor the two 2′-O-methyltransferase mutant viruses induced any detectable expression of type I interferon in Mda5-deficient macrophages (Fig. 3e), whereas the production of type I interferon was readily detectable in Mda5-deficient cells after infection with Sendai virus. Detailed analysis of the kinetics of IFN-β mRNA expression showed that infection of wild-type macrophages (Fig. 3f) and IFNAR-deficient macrophages (Fig. 3g) with MHV-D130A or MHV-Y15A resulted in higher expression of the gene encoding IFN-β (with a peak at 12 h after infection) than did MHV-A59 infection. Notably, the induction of IFN-β was greatest after infection with MHV-D130A, which has a mutant 2′-O-methyltransferase active site. These results indicate that deficiency in 2′-O-methylation in viral RNA induces higher IFN-β expression and that IFN-β induction after infection with viruses with mutant 2′-O-methyltransferases is Mda5 dependent.
Interferon-regulatory factor 3 (IRF3) is activated by RIG-I-like receptor signaling and translocates to the nucleus to mediate the transcription of genes encoding type I interferons. We observed more nuclear localization of IRF3 in IFNAR-deficient macrophages infected with MHV-D130A or MHV-Y15A but not in Mda5-deficient macrophages (Fig. 4). Collectively these results demonstrate that 2′-O-methylation of mRNA is linked to the induction of IFN-β expression in an Mda5-dependent manner.
Figure 4. MHV 2′-O-methyltransferase mutants induce the nuclear localization of IRF3 in an Mda5-dependent manner.

(a) IRF3 in IFNAR-deficient or Mda5-deficient mouse macrophages infected with MHV-A59, MHV-Y15A or MHV-D130A at an MOI of 1 and stained at 3 h after infection for IRF3 (red) and with the DNA-intercalating dye DAPI (blue). Original magnification, ×20. Data are representative of three experiments. (b) Frequency of cells (infected as in a) with nuclear IRF3. *P < 0.01 and **P < 0.001 (unpaired Student's t-test). Data are representative of three experiments (mean and s.d. of five random fields with approximately 50–250 cells each).
Two distinct antiviral mechanisms affected by 2′-O-methylation
As HCoV-D129A, the human coronavirus with a mutant 2′-O-methyltransferase active site, showed greater sensitivity to treatment with type I interferon, we assessed whether type I interferon–induced restriction of viral replication was also effective against the MHV 2′-O-methyltransferase mutants. We investigated the viral replication kinetics of MHV-D130A and MHV-Y15A in primary macrophages, which represent the most important target cells for MHV22,27. MHV-D130A replication was considerably impaired in wild-type macrophages (even after infection at a high MOI of 1), whereas the replication of MHV-Y15A was similar to that of MHV-A59 (Fig. 5a). MHV-D130A replication was fully restored in Mda5-deficient macrophages, even after infection at a low MOI of 0.0001 (Fig. 5b). This demonstrates that Mda5-dependent expression of type I interferon is a prerequisite for the induction of effective restriction of MHV-D130A replication. In agreement with the idea that MHV-D130A replication was impaired in wild-type macrophages, but MHV-Y15A replication was not, we observed much less replication of MHV-D130A in macrophages pretreated with IFN-α. Thus, in contrast to MHV-A59, MHV-D130A was not detectable at 24 h after infection after 4 h of pretreatment of wild-type macrophages with 50–200 units of IFN-α, whereas MHV-Y15A replication was not restricted much (Fig. 5c). In Mda5-deficient macrophages, pretreatment with at least 200 units of IFN-α was needed to restrict MHV-D130A replication to undetectable amounts (Fig. 5d), which suggested that endogenous expression of type I interferon mediated by Mda5 affected the MHV-D130A restriction in wild-type macrophages. These data indicate that the D130A and Y15A substitutions of MHV nsp16 had different effects on the induction of type I interferon and viral replication. The D130A substitution led to more production of type I interferon as well as to considerable sensitivity to pretreatment with type I interferon. In contrast, the Y15A substitution induced more production of type I interferon but not more sensitivity to type I interferon. Thus, we conclude that in addition to triggering Mda5-dependent induction of type I interferon, loss of 2′-O-methylation triggers a second antiviral mechanism that is induced by type I interferon and accounts for the restriction of viral replication during the host cell antiviral state.
Figure 5. Differences in the effect of type I interferon on the replication of MHV 2′-O-methyltransferase mutants.

(a,b) Kinetics of the replication of MHV-A59, MHV-Y15A or MHV-D130A in wild-type and Mda5-deficient mouse macrophages (1 × 106) after infection at an MOI of 1 (a) or 0.0001 (b). Data represent two independent experiments (mean ± s.e.m. of five samples). (c,d) Titer of MHV-A59, MHV-Y15A or MHV-D130A at 24 h after infection (MOI = 1) of wild-type (c) or Mda5-deficient (d) macrophages (1 × 105) pretreated with IFN-α (dose, horizontal axis) 4 h before infection. Data represent two independent experiments (mean and s.d. of four samples).
The replication of a mutant of West Nile virus (family Flaviviridae; genus Flavivirus) lacking 2′-O-methylation is considerably inhibited by members of the IFIT family28, which are encoded by interferon-stimulated genes linked to translational regulation. To assess whether this molecular mechanism is also relevant to coronavirus infection, we analyzed the replication kinetics of MHV-A59, MHV-D130A and MHV-Y15A in primary macrophages derived from wild-type or IFIT1-deficient mice. MHV-D130A replication was almost completely restored in IFIT1-deficient macrophages (Fig. 6), analogous to the restoration of MHV-D130A replication in Mda5-deficient macrophages (Fig. 5a,b). These results suggest that the Mda5-dependent induction of type I interferon and the IFIT-1-mediated restriction of viral replication are two distinct antiviral mechanisms based on the distinction between 2′-O-methylated mRNA and nonmethylated mRNA.
Figure 6. Restoration of MHV-D130A replication in IFIT1-deficient macrophages.

Kinetics of the replication of MHV-A59, MHV-Y15A or MHV-D130A after infection (MOI = 0.01) of wild-type mice (a) or IFIT1-deficient (b) macrophages (5 × 105), assessed by plaque assay of viral titers in supernatants. Data represent two independent experiments (mean ± s.e.m. of four samples).
Effect of 2′-O-methylation on innate immune recognition in vivo
Next we examined the effect of 2′-O-methylation of viral mRNA on innate immune recognition and virulence in vivo. We compared the phenotype of MHV-A59 with that of each MHV 2′-O-methyltransferase mutant in C57BL/6 mice after intraperitoneal infection with 500 plaque-forming units of virus (Fig. 7). In contrast to MHV-A59, neither of the MHV 2′-O-methyltransferase mutants was detectable in spleen or liver of C57BL/6 mice at 48 h after infection, which demonstrated the importance of 2′-O-methylation of viral mRNA for efficient replication and spread in the host. However, the two MHV 2′-O-methyltransferase mutants replicated and spread in IFNAR-deficient mice. MHV-D130A was not detectable in spleens or livers of mice lacking either Mda5 or TLR7, which suggested that induction of the expression of type I interferon via either RNA sensor suffices to completely restrict the replication and spread of mutants with altered 2′-O-methyltransferase active sites. However, we were able to detect small amounts of the cap 0–binding mutant MHV-Y15A in the spleens of TLR7-deficient or Mda5-deficient mice, which suggested that lower induction of type I interferon in these mutant mice did not fully restrict replication of the virus with partially impaired 2′-O-methyltransferase. The replication and spread of the two MHV 2′-O-methyltransferase mutants in mice deficient in both TLR7 and Mda5 was indistinguishable from that in IFNAR-deficient mice. These observations confirm that TLR7 and Mda5 represent the main sensors for recognition of coronavirus RNA and demonstrate that 2′-O-methylation of viral mRNA serves as a mechanism by which viruses evade the recognition of non-self RNA by the host innate immune system in vivo.
Figure 7. Deficiency in 2′-O-methyltransferase affects the recognition of virus by the innate immune system in vivo.
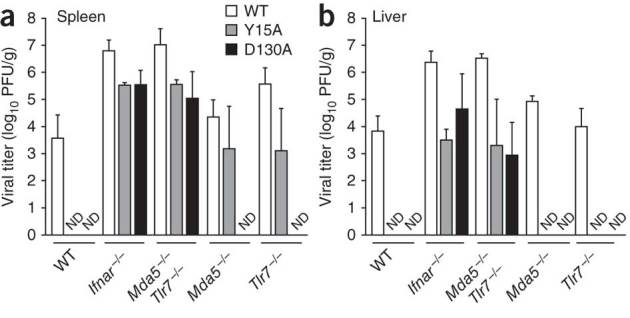
Viral titers in the spleen (a) and liver (b) of wild-type mice and mice deficient in IFNAR, Mda5 or TLR7 or both Mda5 and TLR7, assessed 48 h after infection with MHV-A59, MHV-Y15A or MHV-D130A (500 plaque-forming units injected intraperitoneally). Data represent two independent experiments (mean and s.d. of six samples).
Discussion
The correct functioning of host innate immune responses is based on reliable pathogen detection and is essential in limiting pathogen replication and spread. Using human and mouse coronavirus models of infection, we have shown here that 2′-O-methylation of mRNA provides a molecular signature with a dual role in the interaction with host innate immune responses. First, 2′-O-methylation of mRNA protects viral RNA from recognition by Mda5 and thus prevents Mda5-dependent production of type I interferon in virus-infected cells. Second, 2′-O-methylation of viral mRNA contributes to evasion of the interferon-mediated restriction of viral replication. These distinct effects can be uncoupled either in the absence of type I interferon signaling (such as in IFNAR-deficient cells or mice) or through a genetic approach that targets the cap 0–binding residue Tyr15 of MHV nsp16. The lack of 2′-O-methylation on a proportion of MHV-Y15A mRNA was sufficient to trigger the Mda5 pathway of the production of type I interferon while allowing the virus to evade the IFIT1-mediated restriction of viral replication. In contrast, the absence of 2′-O-methylation on the MHV-D130A viral RNA activated the Mda5 pathway and resulted in restriction of viral propagation.
Our study has described the effect of 2′-O-methylation of mRNA on Mda5-dependent induction of type I interferon. The use of coronavirus greatly facilitated this analysis because coronaviruses encode their own 5′ mRNA cap-methylation machinery, which allowed us to study the phenotype of recombinant viruses with mutant 2′-O-methyltransferase proteins, and because expression of type I interferon is almost undetectable in infected cells other than plasmacytoid dendritic cells, with the notable exception of macrophages, which produce small amounts of Mda5-mediated type I interferon after infection23,24,25,29. In contrast, most other RNA viruses that replicate in the cytoplasm induce considerable amounts of type I interferon, which may mask the specific effect of 2′-O-methylation of mRNA on Mda5 activation2,30. It will be important to clarify whether viral mRNA lacking 2′-O-methylation is directly recognized by Mda5, resulting in its activation, or whether 2′-O-methylation represents an activation signal for Mda5, possibly in combination with dsRNA regions. The generation of recombinant viruses with defined substitutions in RNA-processing enzymes, combined with biochemical approaches, will thus be useful in the identification of naturally occurring Mda5 ligands.
The evasion of Mda5-dependent RNA recognition and IFIT-dependent restriction of viral replication provide a reasonable explanation for the conservation of 2′-O-methyltransferases in many viruses that replicate in the cytoplasm of higher eukaryotes. Other viruses, such as bunyaviruses and arenaviruses, that replicate in the cytoplasm but have not acquired the ability to autonomously generate and modify their 5′ cap structures have evolved means for snatching the cap structure from cellular mRNA31,32. Moreover, structural and functional analyses have shown that the cap-binding nucleoprotein of Lassa virus (family Arenaviridae) can antagonize type I interferon through its associated 3′-to-5′ exoribonuclease activity, probably by cleaving RNAs that function as pathogen-associated molecular patterns33. This protein has an unusually deep cap-binding pocket proposed to accommodate an entire cap 1 structure33 and could potentially discriminate between 2′-O-methylated and nonmethylated capped RNAs. Finally, members of the Picornavirales order and related viruses, which replicate in the cytoplasm but do not encode methyltransferases, have evolved alternative 5′ ends of their RNA. These viruses covalently attach a small viral protein (VPg) to the genomic 5′ terminus and have an internal ribosomal entry site at the 5′ untranslated region34. The replication of encephalomyocarditis virus (family Picornaviridae) seems to be not restricted by IFIT proteins28; however, infection with this virus is sensed through the Mda5 pathway35. Thus, we speculate that the use of internal ribosomal entry allows encephalomyocarditis virus to evade host restriction by IFIT family members, whereas the covalent attachment of VPg to the 5′ end of picornavirus RNA does not prevent Mda5-dependent recognition and induction of type I interferon.
The idea that RNA modification influence host cell innate immune responses is supported by the observation that activation of RIG-I and PKR is diminished when 5′-triphosphate RNA contains modified nucleotides9,10,36. Similarly, nucleoside modifications diminish the potential of RNA to trigger TLRs37. Although most of those observations were made in vitro (for example, by transfection of short synthetic RNA), it seems that RNA modifications may affect sensing by the innate immune system on a wider scale38. Therefore, it will be important to extend the knowledge of naturally occurring RNA modifications and their effect on innate immune responses. Other molecular RNA signatures that function as pathogen-associated molecular patterns will probably be discovered.
In summary, our study has identified 2′-O-methylation of eukaryotic mRNA cap structures as a molecular pattern of self mRNA and has demonstrated that there are at least two cellular mechanisms that allow the distinction of 2′-O-methylated mRNA versus nonmethylated mRNA. Consequently, many viruses that replicate in the cytoplasm, without access to the nuclear host cell machinery for mRNA capping and modification, have evolved their own RNA-modifying enzymes as means of mimicking cellular mRNA. Our data should encourage further studies to evaluate the full spectrum and functional importance of mRNA modifications as an additional layer of information 'imprinted' on eukaryotic mRNA.
Methods
Mice, viruses, cells and viral infection.
C57BL/6 mice were from Charles River Laboratories. Immunodeficient mice deficient in IFNAR, Mda5 or TLR7 or both Mda5 and TLR7 on the C57BL/6 background were bred in the animal facilities of the Kantonal Hospital St. Gallen. IFIT1-deficient mice were bred in the animal facilities of the Washington University School of Medicine. All mice were maintained in individually ventilated cages and were used between 6 and 9 weeks of age. All animal experiments were done in accordance with the Swiss Federal legislation and with approval from the animal studies committees of the Cantonal Veterinary Office (St. Gallen, Switzerland) and St. Louis University.
The HCoV-229E, HCoV-D129A, MHV strain A59, MHV-D130A and MHV-Y15A recombinant viruses were generated with a vaccinia virus–based reverse-genetic system as described39 and were propagated on Huh-7 hepatocarcinoma cells (HCoV) or 17Cl1 mouse fibroblasts (MHV). BHK-21 baby hamster kidney cells, L929 mouse fibroblasts, NIH3T3 mouse fibroblasts, MRC-5 human fetal lung cells and CV-1 monkey kidney fibroblasts were from the European Collection of Cell Cultures. Huh-7 cells were a gift from V. Lohmann; D980R human epithelial cells were a gift from G.L. Smith; and 17Cl1 cells were a gift from S.G. Sawicki. All cells were maintained in minimal essential medium supplemented with 5–10% (vol/vol) FBS and antibiotics. Thioglycollate-elicited mouse macrophages were generated as described40. Human macrophages were isolated from peripheral blood of normal donors as described41. Experiments using human cells were in compliance with the Swiss federal legislation and institutional guidelines of the Kantonsspital St.Gallen (including informed consent) and were appoved by the ethical committee of Canton St. Gallen, Switzerland.
Mice were injected intraperitoneally with 500 plaque-forming units of MHV. Organs were stored at −70 °C until further analysis. Human blood-derived macrophages and thioglycolate-elicited mouse macrophages (0.5 × 106 to 1 × 106) were infected at the appropriate MOI in a 24-well format. MHV titers were determined by standard plaque assay with L929 cells. HCoV titers were determined by plaque assay with Huh-7 cells overlaid at 1 h after infection with 1.2% (wt/vol) Avicel microcrystalline cellulose in 10% (vol/vol) DMEM and stained with crystal violet 3 d after infection.
In vitro 2′-O-methylation of poly(A)-containing RNA.
Poly(A)-containing RNA was isolated from 1 × 107 mock- or HCoV-infected Huh-7 cells (MOI = 1; isolated 48 h after infection) or 1 × 107 mock- or MHV-infected NIH-3T3 cells (MOI = 1; isolated 24 h after infection) with the Dynabeads mRNA DIRECT kit according to the manufacturer's recommendations (Invitrogen). RNA was precipitated after the addition of 0.1 volume of 4 M ammonium acetate and 1 volume of isopropanol, then was washed with 70% ethanol and dissolved in 10 mM TRIS-HCl, pH 7.5, to a final concentration of 150 ng/μl. In vitro 2′-O-methylation reactions were incubated for 1 h at 37 °C and included 300 ng poly(A)-containing RNA derived from virus-infected cells or the corresponding amount of poly(A)-containing RNA from uninfected cells (as determined by quantitative RT-PCR with primers specific for mouse GAPDH and human β-actin; data not shown) plus ScriptCap 2′-O-methyltransferase (Epicentre Biotechnologies) in 0.5 μM S-adenosyl-methionine and 1.4 μM 3H-labeled S-adenosyl-methionine (78 Ci/mmol; Perkin Elmer). Reactions were purified with SigmaSpin Post-Reaction Clean-Up columns (Sigma-Aldrich) and eluates were mixed with 2 ml Ultima Gold scintillation fluid for measurement of 3H incorporation with a Packard Tri-Carb liquid scintillation counter (Perkin Elmer).
Immunofluorescence, IFN-β enzyme-linked immunosorbent assay and IFN-α pretreatment.
IRF3 was detected in mouse macrophages (2 × 105 cells per well in 200 μl) 3 h after infection with MHV at an MOI of 1. Cells were stained with antibody to IRF3 (FL-425; Santa Cruz Biotechnology) and with DAPI (4,6-diamidino-2-phenylindole). Images were acquired with a DMRA microscope (Leica). The concentration of mouse and human IFN-β in cell culture supernatants was measured by enzyme-linked immunosorbent assay (PBL Biomedical Laboratories). Universal type I interferon (IFN-α A/D; Sigma) was used for pretreatment of cells with IFN-α before viral infection.
Bioassay for type I interferon.
Total type I interferon in supernatants was measured with LL171 cells (a gift from M. Pelegrin), which are L929 cells stably transfected with a luciferase reporter plasmid under control of the interferon-stimulated response element42. Recombinant IFN-α A/D (Sigma) was used as a cytokine standard. Before measurement, virus was removed by centrifugation of supernatants through AMICON spin columns with a cutoff of 100 kilodaltons (Millipore). LL171 cells grown in 96-well plates were treated for 6 h with column-filtered supernatants, and luciferase activity was detected by a GloMax 96 Microplate Luminometer (Promega) after the addition of Bright-Glo Luciferase substrate (Promega).
Quantitative RT-PCR.
Total cellular RNA was isolated with a NucleoSpin RNA II kit according to the manufacturer's instructions (Macherey-Nagel) and was used as template for cDNA synthesis with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). A LightCycler FastStart DNA Master SYBR Green I kit (Roche) and a LightCycler 1.5 (Roche) were used for measurement of mRNA for IFN-β and TATA box–binding protein with the following primers: IFN-β, 5′-GGTGGAATGAGACTATTGTTG-3′ and 5′-AGGACATCTCCCACGTC-3′; TATA box–binding protein, 5′-CCTTCACCAATGACTCCTATGAC-3′ and 5′-CAAGTTTACAGCCAAGATTCAC-3′. Measurements were made in duplicate and the expression of IFN-β was normalized to that in uninfected cells by the comparative cycling threshold method (ΔΔCT).
Phylogenetic analysis of viral methyltransferase domains.
Regions of methyltransferase homology have been identified as members of the RrmJ-like superfamily (InterPro accession code IPR002877). For viruses with no RrmJ-like domain, secondary structure–assisted alignment of amino acids was done to determine whether a distant homolog might be present. Viruses without identification of a primary or secondary structure match to RrmJ-like proteins are designated 'not detected' (Supplementary Table 1). Amino acid sequences were aligned with ClustalW2 and were manually adjusted on the basis of published structural data and protein secondary structure predictions of the PSIPRED server43. Motif nomenclature was as published44. Amino acids were assigned colors according the conventions of ClustalX for amino acid similarity and conservation as implemented in the JalView alignment editor45.
Electron microscopy.
Ultrathin sections of mouse L929 cells were stained with osmium tetroxide and uranyl acetate 5 h after infection with MHV at an MOI of 1 and images were obtained by transmission electron microscopy as described46.
Supplementary information
Supplementary Figures 1–2 and Supplementary Table 1 (PDF 394 kb)
Acknowledgements
We thank V. Lohmann (University of Heidelberg) for Huh-7 cells; G.L. Smith (Imperial College, London) for D980R cells; S.G. Sawicki (Medical University of Ohio) for 17Cl1 cells; M. Pelegrin (Institut de Génétique Moléculaire de Montpellier) for LL171 cells; L. Onder for assistance with fluorescence microscopy; and R. de Giuli, B. Schelle and N. Karl for technical assistance. This study was supported by the Swiss National Science Foundation, the European Commission (TOLERAGE), the Novartis Foundation for Biomedical Research, Switzerland, the German Ministry of Education and Research (V.T.), the Austrian Science Fund (FWF J3044 to M.H.), Deutsche Forschungsgemeinschaft (J.Z.), the National Institutes of Health (AI060915 and AI085089 to S.C.B.; U54 AI081680 (Pacific Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research to M.S.D), the Medical Research Council (B.W.N.) and the Wellcome Trust (S.G.S.).
Author Contributions
R.Z., L.C.-B., M.H., R.M. and K.J.S. did most of the experiments; B.W.N. did phylogenetic analyses; B.W.N. and S.C.B. did electron microscopy; J.Z., S.C.B., W.B., M.S.D., S.G.S. and B.L. contributed reagents and expertise; and S.G.S., B.W.N., B.L. and V.T. conceived of and designed the project and wrote and edited the manuscript.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Competing interests
The authors declare no competing financial interests.
Footnotes
Roland Züst and Luisa Cervantes-Barragan: These authors contributed equally to this work.
References
- 1.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loo YM, Gale M., Jr. Viral regulation and evasion of the host response. Curr. Top. Microbiol. Immunol. 2007;316:295–313. doi: 10.1007/978-3-540-71329-6_14. [DOI] [PubMed] [Google Scholar]
- 4.Haller O, Weber F. Pathogenic viruses: smart manipulators of the interferon system. Curr. Top. Microbiol. Immunol. 2007;316:315–334. doi: 10.1007/978-3-540-71329-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui DJ, Terenzi F, Merrick WC, Sen GC. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 2005;280:3433–3440. doi: 10.1074/jbc.M406700200. [DOI] [PubMed] [Google Scholar]
- 6.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 7.Fensterl, V. & Sen, G.C. The ISG56/IFIT1 gene family. J. Interferon Cytokine Res. published online, doi:10.1089/jir.2010.0101 (25 October 2010).
- 8.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornung V, et al. 5′-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 10.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 11.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh A, Lima CD. Enzymology of RNA cap synthesis. WIREs RNA. 2010;1:152–172. doi: 10.1002/wrna.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 14.Nomoto A, Detjen B, Pozzatti R, Wimmer E. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature. 1977;268:208–213. doi: 10.1038/268208a0. [DOI] [PubMed] [Google Scholar]
- 15.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 16.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decroly E, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′-O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/S0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- 21.Schnierle BS, Gershon PD, Moss B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc. Natl. Acad. Sci. USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cervantes-Barragán L, et al. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 2009;182:1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 23.Cervantes-Barragan L, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose KM, Elliott R, Martinez-Sobrido L, Garcia-Sastre A, Weiss SR. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J. Virol. 2010;84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth-Cross JK, Bender SJ, Weiss SR. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H, Zhao J, Perlman S. Autocrine interferon priming in macrophages but not dendritic cells results in enhanced cytokine and chemokine production after coronavirus infection. MBio. 2010;1:e00219–10. doi: 10.1128/mBio.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bocharov G, et al. A systems immunology approach to plasmacytoid dendritic cell function in cytopathic virus infections. PLoS Pathog. 2010;6:e1001017. doi: 10.1371/journal.ppat.1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiel V, Weber F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 2008;19:121–132. doi: 10.1016/j.cytogfr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin B, et al. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010;6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, X. et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature advance online publication, doi:10.1038/nature09605 (17 November 2010). [DOI] [PMC free article] [PubMed]
- 34.Le Gall O, et al. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008;153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 35.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eriksson KK, Makia D, Thiel V. Generation of recombinant coronaviruses using vaccinia virus as the cloning vector and stable cell lines containing coronaviral replicon RNAs. Methods Mol. Biol. 2008;454:237–254. doi: 10.1007/978-1-59745-181-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Züst R, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wünschmann S, Becker B, Vallbracht A. Hepatitis A virus suppresses monocyte-to-macrophage maturation in vitro. J. Virol. 2002;76:4350–4356. doi: 10.1128/JVI.76.9.4350-4356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uzé G, et al. Domains of interaction between α interferon and its receptor components. J. Mol. Biol. 1994;243:245–257. doi: 10.1006/jmbi.1994.1651. [DOI] [PubMed] [Google Scholar]
- 43.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fauman, E.B., Blumenthal, R.M. & Cheng, X. Structure and Evolution of AdoMet-Dependent Methyltransferases. in S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions (eds. Cheng, X. & Blumenthal, R.M.) 1–38 (World Scientific, Singapore, 1999).
- 45.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–2 and Supplementary Table 1 (PDF 394 kb)


