Abstract
Although small molecules that modulate amyloid formation in vitro have been identified, significant challenges remain in determining precisely how these species act. Here we describe the identification of rifamycin SV as a potent inhibitor of β2m fibrillogenesis when added during the lag time of assembly or early during fibril elongation. Biochemical experiments demonstrate that the small molecule does not act by a colloidal mechanism. Exploiting the ability of electrospray ionization-ion mobility spectrometry-mass spectrometry (ESI-IMS-MS) to resolve intermediates of amyloid assembly, we show instead that rifamycin SV inhibits β2m fibrillation by binding distinct monomeric conformers, disfavoring oligomer formation, and diverting the course of assembly to the formation of spherical aggregates. The results reveal the power of ESI-IMS-MS to identify specific protein conformers as targets for intervention in fibrillogenesis using small molecules and reveal a mechanism of action in which ligand binding diverts unfolded protein monomers towards alternative assembly pathways.
Keywords: β2-microglobulin, amyloid, rifamycin SV, oligomers, NMR, ESI-MS
Aberrant aggregation of proteins into amyloid fibrils is a characteristic of over twenty-five human disorders. 1 In each case, the precursor protein is dissimilar in terms of its native fold and primary sequence, yet amyloid fibrils share common structural and tinctorial features, such as a cross-β organization of the polypeptide chain and the ability to bind dyes such as thioflavin-T (ThT) and Congo red. 2 The observation of a common structural architecture for amyloid fibrils has motivated efforts to elucidate the molecular mechanisms of fibril formation in vitro with the aim of revealing possible targets for therapeutic intervention. Such investigations have led to the observation of different species populated en route to amyloid fibrils, including oligomers of different size and morphology, as well as protofibrils, annular aggregates and worm-like assemblies. 3-6 The heterogeneity, dynamic properties and transient nature of intermediates in amyloid assembly, however, have hindered the structural analysis of assembly mechanisms and have led to much debate regarding the culprit species of toxicity. 7-9 There is, therefore, an urgent need to develop methods able to separate and characterize individual intermediates present during amyloid assembly, so that small molecules able to inhibit or modulate the aggregation pathway can be identified and their mechanism of action discerned.
The ability of small molecules to impede or modulate fibril formation has been assessed using an array of proteins and peptides in vitro. 5, 10, 11 In studies of transthyretin, stabilization of the native tetramer by ligand binding reduces the concentration of aggregation-competent monomer and has provided an effective therapeutic route. 12 Catecholamines have been shown to inhibit fibril formation of α-synuclein by preventing the conversion of protofibrils to fibrils, 13 while molecules such as Congo red, clioquinol and lacmoid have been shown to act as inhibitors of fibrillation by a variety of specific and non-specific binding mechanisms. 5, 10, 11, 14-16 A recent study which investigated the ability of small molecules to modulate Aβ aggregation indicated several possible mechanisms of intervention, including inhibition of oligomer formation, inhibition of fibril formation, or both.5, 17 Despite this classification and the wealth of studies performed, the molecular mechanisms by which different ligands inhibit or divert the course of amyloid assembly remain unclear. 18
In this study we analyzed the effect of 44 small molecules on the assembly of β2-microglobulin (β2m) into amyloid fibrils at low pH in vitro. Under the conditions used (pH 2.5) β2m has been shown to assemble into long-straight (LS), twisted fibrils that bear all the hallmarks of amyloid. 19, 20 Since β2m has no known natural small molecule ligands and no obvious ligand binding site, discovery of modulators of its aggregation requires screening rather than design. Inspired by the recent discovery that an aromatic-rich region of the sequence of β2m involving residues ~60-70 is important for the nucleation and elongation of LS fibrils 19, 20 the majority of small molecules chosen for screening contained at least one aromatic group. Of all the compounds tested, only one, rifamycin SV, was found to ablate fibrillation of β2m when added in the lag time of assembly or early during fibril elongation. Rather than acting by a colloidal mechanism as has been proposed for several small molecule inhibitors of amyloid formation, 14-16 inhibition was shown to occur by a specific binding mechanism. Using electrospray ionization-ion mobility spectrometry-mass spectrometry (ESI-IMS-MS), combined with NMR and other biophysical methods, we identify different conformers of β2m within the monomeric ensemble able to bind rifamycin SV and determine the effect of ligand binding on the progress of amyloid assembly. The results reveal a powerful inhibitor of β2m fibrillogenesis that exerts its action by binding to specific monomeric conformers within the unfolded ensemble of precursor molecules. As a consequence of ligand binding, new assembly route(s) are opened that result in the formation of off-pathway spherical aggregates that do not contain the cross-β structure of amyloid.
Results
Screening for inhibitors of β2m fibrillogenesis
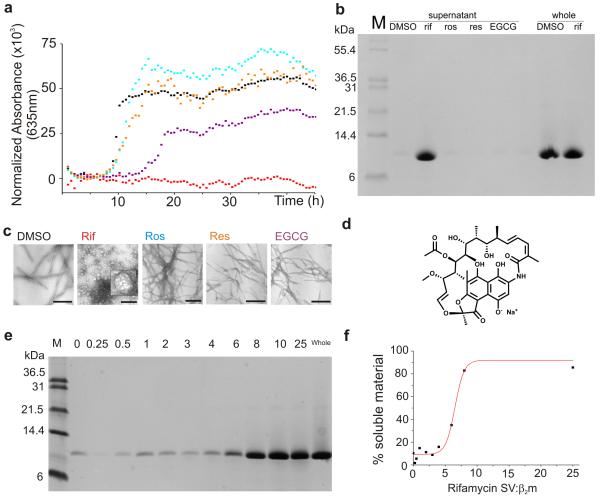
Of the 44 small molecules selected for analysis, ~30 compounds have been shown previously to affect fibrillogenesis of other proteins, or are analogues of such compounds (Supplementary Tables 1 and 2). Molecules chosen for screening included polyphenols, anisoles, stilbenes, naphthoquinones, tetracyclines and anthraquinones. In the screen, 1 mM of each small molecule in the presence of 10% (v/v) DMSO was tested for its ability to inhibit the fibrillation of 45 μM β2m at pH 2.5 (see Methods). Assembly reactions were monitored where possible using thioflavin-T (ThT) fluorescence. However, as has been reported previously 21, several of the small molecules assayed interfere with the fluorescence of ThT, compromising the validity of this assay (Supplementary Table 2). Alternative assays were performed, therefore, for each compound in the test set, including analysis of fibril growth kinetics using turbidity measurements at 635 nm (Figure 1a); assessment of the amount of soluble material remaining after assembly has reached completion (taken as 48 h) using centrifugation (16,300x g) followed by SDS-PAGE (Figure 1b); and negative stain transmission electron microscopy (TEM) of the products of assembly after this time (Figure 1c). Together, these experiments revealed that of the 44 molecules tested, only one, rifamycin SV (Figure 1d), is able to abolish the ability of β2m to assemble into LS fibrils under the conditions employed (Supplementary Table 2). Rather than forming LS fibrils, incubation of β2m in the presence of rifamycin SV results in the formation of spherical aggregates ≈ 36.0 ± 7.0 nm (n = 200) in diameter, with rare examples of toroidal structures (Figure 1c). Quantitative analysis of soluble protein remaining after incubation of the protein with rifamycin SV for 48 h indicated that >90 % of β2m remains soluble in the presence of rifamycin SV, compared with <5 % in the absence of the ligand (Figure 1b), indicating efficient inhibition of fibrillogenesis by this molecule.
Figure 1.
Fibrillogenesis of β2m at pH 2.5 in the presence of different small molecules. a Aggregation reactions monitored using turbidity at 635 nm. In each case 45 μM β2m was incubated in the presence of 10 % (v/v) DMSO alone (black), 1 mM rifamycin SV (Rif, red), 1 mM rosmarinic acid (Ros, cyan), 1 mM resveratrol (Res, orange) or 1 mM epigallocatechin gallate (EGCG, purple) each in 10% (v/v) DMSO. One example of each measurement is shown. b Analysis of material remaining in the supernatant after 48 h incubation of each sample in the presence or absence of different ligands. SDS-soluble protein remaining in each sample was measured after centrifugation using SDS-PAGE (labeled ‘supernatant’). The total protein concentration is also shown (labeled ‘whole’). c TEM images of the species formed after incubation of β2m with different small molecules for 48 h (scale bar = 200 nm). The insert in the second image (labeled rif) shows an expanded image (5-fold) of a spherical aggregate. d Structure of rifamycin SV. e Effect of the concentration of rifamycin SV on β2m fibril formation shown by SDS-PAGE. The amount of SDS-soluble, low molecular weight material remaining after 48 h incubation of 45 μM β2m increases in the presence of increasing concentrations of rifamycin SV (numbers above correspond to the molar ratio of rifamycin SV:β2m in the different samples). f Percent soluble material remaining after 48 h incubation versus the molar concentration ratio of rifamycin SV:β2m determined by quantification of the data shown in e.
Rifamycin SV is an antibiotic that contains a naphthohydroquinone moiety attached to an aliphatic ‘ansa’ chain (Figure 1d). Interestingly, the close analogues of rifamcyin SV, rifampicin, rifaximin and rifamycin S (Supplementary Results; Supplementary Figure 1a-d), the latter formed by incubation of rifamycin SV for 24 h at pH 2.5 which results in the oxidation of the dihydroxynaphthalene group to its quinone form (Supplementary Methods, Supplementary Figure 2a,b), did not abolish the formation of fibrils although they did increase lag time of assembly by 1.7-, 2.3- and 3.5-fold, respectively (Supplementary Figure 1a-d and Supplementary Table 2). 5-Hydroxy-naphthoquinone (juglone) and 5,8-dihydroxy-naphthoquinone, each of which contains the naphthohydroquinone functionality of rifamycin SV proposed to convey its anti-amyloid properties, 22-24 also had no effect of fibril assembly (Supplementary Figure 1e,f and Supplementary Table 2). Similarly, suramin and Congo red, which have been shown to bind to β2m at neutral pH 25 have no significant effect on the yield of fibrillar material under the conditions employed here (Supplementary Table 2).
Rifamycin SV binds early assembly intermediates
Whether rifamycin SV prevents fibrillation by inhibiting fibril assembly, by destabilizing fibrils, or both, was next determined by investigating the effect of the small molecule on fibril growth and fibril stability. To test the latter scenario, LS fibrils were assembled from 45 μM β2m at pH 2.5 in the presence of 10 % (v/v) DMSO. After assembly was complete, the sample was incubated with 1 mM rifamycin SV for different lengths of time (5 min to >300 h (Supplementary Methods)) and the effect of the ligand on fibril stability was determined using TEM and SDS-PAGE subsequent to centrifugation of the sample (Supplementary Figure 3). These experiments showed that depolymerization of LS fibrils does not occur in the presence of rifamycin SV under the conditions employed, even when the ligand is added in excess. The ability of rifamycin SV to inhibit fibrillogenesis must occur, therefore, by the small molecule binding to one or more species formed during fibril assembly.
To determine which stage(s) of the assembly reaction is/are affected by the presence of rifamycin SV, β2m fibrillogenesis (45 μM monomer) was initiated at pH 2.5 and rifamycin SV (1 mM) was added at different times during fibril growth. The time course of assembly was subsequently monitored using TEM and by SDS-PAGE of the supernatant after centrifugation of the samples (Supplementary Figure 4a-c). The experiment revealed that rifamycin SV is able to inhibit fibril formation when added during the lag time of assembly, consistent with the ligand binding to monomers and/or one or more of the oligomers known to be populated during the lag phase of β2m assembly under the conditions employed. 26, 27 Addition of rifamycin SV to seeded growth experiments also inhibit fibril elongation, whereas control ligands have no effect (Supplementary Figure 4d,e).
In the assays presented above, excess rifamycin SV (~20-fold over β2m) was employed to ensure complete inhibition of fibrillogenesis. To determine the minimum concentration of rifamycin SV required to inhibit the nucleation events of β2m fibrillation, the protein (45 μM) was incubated at pH 2.5 in the presence of different concentrations of rifamycin SV (0 – 1125 μM) and the effect of the small molecule on fibril formation was monitored using SDS-PAGE (Figure 1e). The results indicated that >95 % of protein remained soluble when a ~5-fold molar excess of ligand is added to monomeric β2m, yielding an IC50 value of ~300 μM for 45 μM β2m (Figure 1f).
Rifamycin SV does not inhibit by a colloidal mechanism
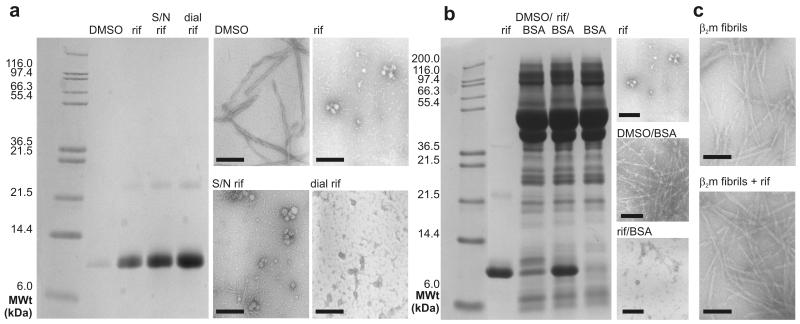
Previous studies of inhibitors of amyloid formation, including Congo red, lacmoid and clioquinol, have suggested that these molecules act by a colloidal mechanism, rather than by specific binding of the small molecule to the amyloidogenic precursor(s). 14-16 Such molecules act via the formation of micellar aggregates which inhibit amyloid assembly by chelating precursor molecules. The ability of rifamycin SV to abolish β2m fibrillogenesis, while close homologues (rifampicin, rifaximin and rifamycin S) are ineffective, suggest that a colloidal mechanism of inhibition is unlikely. Consistent with this view, 97% of rifamycin SV dissolved in aqueous solution containing 10% (v/v) DMSO diffuses freely through a dialysis membrane of molecular weight cut off 3000 Da, ruling out assembly of the ligand into large particles (Supplementary Methods). Material able to pass though the dialysis membrane, along with a sample of rifamycin SV that had been ultracentrifuged for 30 min at 418,000x g prior to addition to the assembly reaction, also inhibit fibril formation (Figure 2a). Inhibition of β2m fibrillogenesis by rifamycin SV is also unaffected by the presence of 5 mg ml−1 of BSA 14 (Figure 2b) and no particulates are seen in samples of rifamycin SV incubated either alone (Supplementary Figure 1g) or after addition to preformed β2m fibrils using TEM (Figure 2c). Finally, rifamycin SV (1 mM) is also unable to inhibit fibrillation of 69 μM α-synuclein at pH 2.5 (Supplementary Methods and Supplementary Figure 5). Together these data rule out a colloidal mechanism of inhibition under the conditions used. Consistent with this conclusion, small-angle neutron scattering experiments have shown that rifamycin does not form colloidal assemblies in the presence of 10% (v/v) organic solvent 28, the solution conditions employed in all experiments presented here.
Figure 2.
Inhibition of fibril formation of β2m by rifamycin SV does not occur by a colloidal mechanism. The data presented show the ability of rifamycin SV treated in different ways to inhibit β2m fibril formation monitored by TEM and SDS-PAGE analysis of the supernatant following centrifugation of each sample. a Incubation of 45 μM β2m in the presence of 10 % (v/v) DMSO (DMSO), β2m with 1 mM rifamycin SV (rif), β2m with ultracentrifuged rifamycin SV (S/N rif), β2m with rifamycin SV dialysate (dial rif). b Incubation of β2m with rifamycin SV (rif), β2m with BSA (DMSO/BSA), β2m with BSA and rifamycin SV (rif/BSA) and BSA alone (BSA). c TEM images of preformed β2m fibrils prior to and after addition of rifamycin SV. All samples were dissolved in 25 mM sodium phosphate/sodium acetate, pH 2.5 with a final concentration of 10% (v/v) DMSO and incubated at 37°C, 200 rpm for 48 h before analysis. Where BSA was present, a final BSA concentration of 5 mgml−1 was used (Supplementary Methods). TEM images of the representative lanes are shown alongside (scale bar = 200 nm).
Spherical aggregates lack ordered β-sheet structure
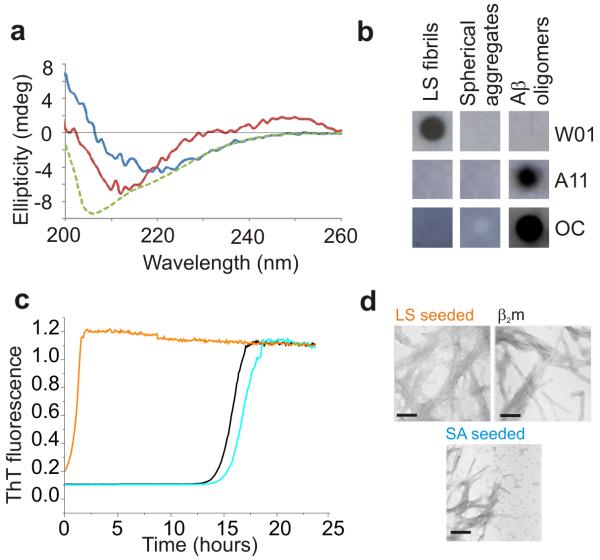
Whether the spherical aggregates formed from β2m in the presence of rifamycin SV contain ordered β-sheet structure characteristic of amyloid was determined using far UV CD, combined with analysis of the ability of these species to bind antibodies able to recognize oligomeric or fibrillar species 29, 30 and to seed fibril growth (Figure 3). The far UV CD spectra of the spherical aggregates suggest that these species contain secondary structure that is distinct from the ordered cross-β structure of amyloid (Figure 3a). Consistent with these observations, the spherical aggregates are not recognized by the antibody WO1, which has previously been shown to bind to amyloid fibrils formed in vitro from a wide variety of proteins, including the LS fibrils of β2m 30 (Figure 3b), nor are they recognized by the antibodies A11 and OC, which identify generic epitopes in pre-fibrillar and fibrillar oligomers, respectively. 29 Akin to short β2m amyloid fibrils (but not their longer counterparts) 31, the spherical aggregates (2.4 μM monomer equivalent) were found to be toxic to RAW 264.7 and SH-SY5Y cell lines (Supplementary Methods, Supplementary Figure 6), although the process by which toxicity is mediated currently remains unknown. Finally, the ability of the spherical aggregates to seed fibril assembly was assessed. Accordingly, spherical aggregates formed by incubation of 45 μM β2m in the presence of 1 mM rifamycin SV were added (5 % (v/v)) to acid unfolded monomeric β2m at pH 2.5 and their ability to seed fibril growth was monitored using ThT fluorescence (Figure 3c). As a control, 5 % (v/v) fibril seeds formed at an identical protein concentration were added to a separate sample of monomeric protein. As expected, addition of fibrillar seeds removes the lag phase required for nucleation of LS fibrils of β2m 19 and results in the rapid assembly of LS fibrils (Figure 3c,d, orange and black lines). By contrast, the addition of the spherical aggregates has no effect on the lag time of assembly and results in the formation of LS fibrils mixed with the initially added spherical aggregates (blue line, Figure 3c,d). Combined with the antibody binding and CD experiments described above, the results suggest that the spherical aggregates formed from β2m in the presence of rifamycin SV lack the ordered cross-β structure of amyloid.
Figure 3.
Structural properties of spherical aggregates of β2m formed in the presence of rifamycin SV. a Far UV CD spectra of monomeric β2m (green), LS fibrils (blue) and spherical aggregates of β2m (red) obtained by incubating 45 μM β2m in the absence or presence of 1 mM rifamycin SV. b Dot blots of LS fibrils (mean length 1440 nm) and rifamycin SV-induced spherical aggregates of β2m with the antibodies WO1, A11 and OC. For positive controls for all antibodies, oligomers of Aβ1-40 were used according to 49. Fibrillar oligomers of biotinylated Aβ1-42 formed according to 50 were used as controls for OC. Control experiments (see Methods) confirmed that rifamycin SV does not prevent binding to each epitope. c Fibril growth of monomeric β2m (45 μM at pH 2.5, 25 °C, 600 rpm) in the presence of 5 % (v/v) fibril seeds (orange), 5 % (v/v) spherical aggregates (SA) formed in the presence of rifamycin SV (blue), or no seeds and no rifamycin SV (black). Seeds (fibrils or spherical aggregates) were taken from the end point (48 h) of incubation of 45 μM monomeric β2m in the absence or presence of 1 mM rifamycin SV. d TEM images (scale bar = 200 nm) of the samples shown in c taken after 48 h incubation using the same color coding as in c.
Specific β2m conformers bind rifamycin SV and rifaximin
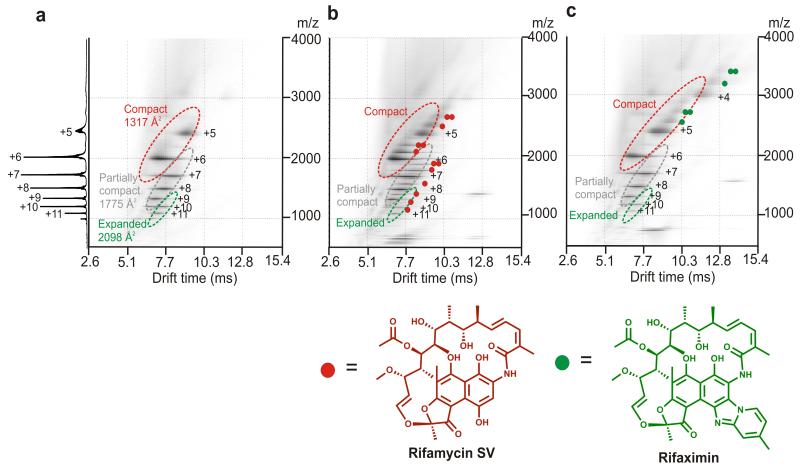
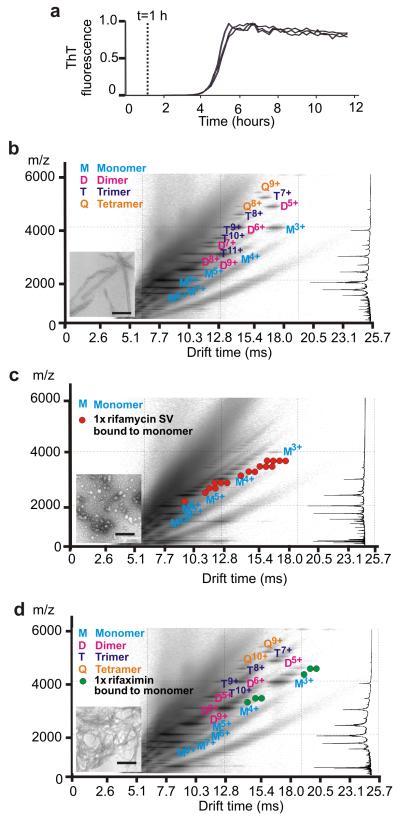
With the aim of deciphering the mechanism(s) by which rifamycin SV inhibits β2m fibril formation and diverts the course of assembly to spherical aggregates, ESI-IMS-MS was used to characterize the ligand binding capability of different monomeric conformers, as well as the oligomers of β2m that form during the lag phase of fibril formation. 26, 32 The power of this technique was exploited to determine the mass, collision cross-section (CCS) and ligand binding capability of each of the transient species within this heterogeneous mixture of assembling species. 32, 33 At the acidic pH employed, monomeric β2m has been shown by CD (Figure 3a) and NMR 20, 34 to be unfolded, lacking ordered secondary structure, but containing non-random interactions involving hydrophobic residues. Analysis of monomeric β2m at pH 2.5 using ESI-IMS-MS reveals that three distinct conformers are co-populated at this pH: a highly expanded monomer (~8% populated, CCS 2098 Å2); a partially compact state (~57% populated, CCS 1775 Å2) and a compact species (~35% populated, CCS 1317 Å2) (see Methods and Figure 4a). 32 To determine which, if any, of these species binds rifamycin SV, β2m (38 μM) was incubated at pH 2.5 with equimolar concentrations of rifamycin SV or, as a control, the non-inhibitor rifaximin, and samples were analyzed immediately (within 2 min of ligand addition) by ESI-IMS-MS. These experiments revealed that either one or two rifamycin SV molecules bind to each of the monomeric β2m conformers resolved by ESI-IMS-MS at this pH, the mass of the bound product indicating that the reduced form of the ligand without its Na+ counter ion binds to the protein (Figure 4b and Supplementary Figure 2a). Strikingly, and in marked contrast with these results, incubation of β2m with equimolar rifaximin, a ligand which is structurally closely related to rifamycin SV yet does not inhibit fibril formation (Figure 4c and Supplementary Figure 1c,h), does not bind to the expanded and partially compact conformers of monomeric β2m. Instead, only the most compact conformer shows binding to this ligand (Figure 4c). Quantitative analysis of the mass spectra showed that rifaximin increases the population of the most compact monomeric species from ~35% without ligand, to ~80% in the presence of rifaximin (Supplementary Figure 2c, Supplementary Methods). By contrast, binding of rifamycin SV has no significant effect on the equilibrium population of different monomeric forms (Figure 4a,b).
Figure 4.
ESI-IMS-MS 3D Driftscope plots (IMS drift time versus m/z versus intensity (z = square root scale)) of β2m monomeric conformers and their ligand binding capabilities at pH 2.5. a β2m alone; b β2m and equimolar rifamycin SV and c β2m and equimolar rifaximin. In b and c the small molecule was initially dissolved in acetonitrile. The ligand was then diluted 10-fold into the protein sample and mass spectra obtained within 2 min of mixing. All spectra were obtained using 38 μM β2m at 25 °C, pH 2.5 with protein dissolved in 10 mM ammonium formate and a sampling cone voltage of 70 V (i.e., conditions optimised to monitor protein-ligand binding). The molecular structures of rifamycin SV and rifaximin are shown below each spectrum. The colored circles represent the number of ligand molecules bound to each charge state of β2m in the different spectra. Note that the mass of the bound product indicated that rifamycin SV binds to β2m in the reduced, Na+-free form and no chemical modification of the protein occurs in the presence of either ligand. Charge states belonging to different conformational states are circled and individually labeled. The summed m/z spectrum for β2m alone is shown on the left-hand side.
The very different effects of rifamycin SV and rifaximin on β2m fibrillation, despite their close structural similarity, suggests that the inhibition of fibrillation results from the unique property of rifamycin SV to bind to the expanded and partially compact conformers within the dynamic ensemble of monomers, while binding to the most compact species is ineffective in preventing fibril formation. The specificity of the interaction between β2m and rifamycin SV was confirmed by analyzing the binding of equimolar rifamycin SV to β2m in the presence of an 10-fold molar excess of rifaximin (Supplementary Figure 2c), or by incubating equimolar β2m, myoglobin (an alternative protein) and rifamycin SV (Supplementary Figure 2d). In each case preferential binding of rifamycin SV to β2m occurred, confirming the specificity of the interaction between this small molecule and β2m.
The highly aromatic-rich region of the human β2m sequence encompassing residues ~60-70 has been proposed as an important region for nucleation of fibril formation at pH 2.5. 20 To determine whether this sequence is involved in ligand binding, rifamycin SV was added to a variant of β2m lacking three aromatic residues in this region (F62A,Y63A,Y67A). 19, 20 Previous experiments have shown that the triple mutant F62A,Y63A,Y67A is only able to form amyloid fibrils at pH 2.5 when incubated for highly extended time periods. 19 Strikingly, addition of an 8-fold molar excess of rifamycin SV to 38 μM F62A,Y63A,Y67A results in only limited binding between the ligand and the variant compared with the extent of ligand binding to wild-type β2m observed by ESI-IMS-MS (Supplementary Figure 7). The data suggest that aromatic residues in the region 60-70 of the sequence of β2m play a role in ligand binding, either directly by being involved in the binding site, or indirectly by affecting the conformational properties of the unfolded ensemble. 20
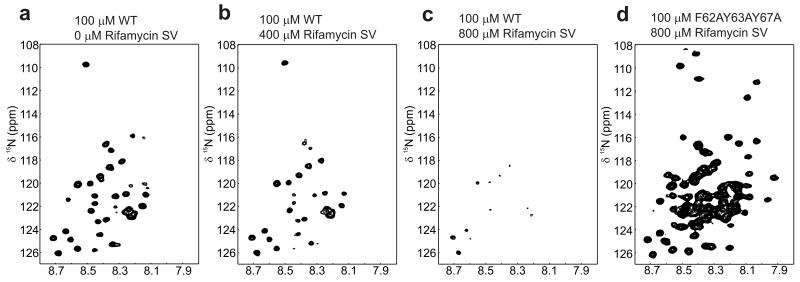
To investigate further how rifamycin SV affects the conformational properties of wild-type β2m at pH 2.5, rifamycin SV (0 – 1500 μM) was added to 15N-labeled protein and the effect of ligand binding on the chemical shift and intensity of resonances arising from the protein was measured by NMR (see Methods). The 1H-15N HSQC spectrum of monomeric β2m obtained at pH 2.5 in the absence of ligand shows limited chemical shift dispersion (Figure 5a), consistent with previous observations that this species is primarily a dynamic acid-unfolded polypeptide chain that is monomeric under the conditions employed. 27, 34 Nonetheless, several resonances display significant line broadening such that these peaks are not observed or are have low intensity in the 1H-15N HSQC spectrum, specifically those arising from resonances in regions enriched in hydrophobic and aromatic resonances (residues 29-51 and 58-79), suggestive of substantial non-random conformations within the acid-unfolded protein. 34
Figure 5.
1H-15N HSQC NMR spectra of β2m and its variant F62A,Y63A,Y67A in the presence of rifamycin SV. a 100 μM β2m in the absence of ligand or in the presence of b 400 μM or c 800 μM rifamycin SV. In b and c the spectra were acquired immediately upon addition of the small molecule to the protein. d 1H-15N HSQC NMR spectra of 100 μM F62A,Y63A,Y67A in the presence of 800 μM rifamycin SV. Note that increased conformational dynamics in this variant gives rise to sharper lines for several resonances compared with the wild-type protein, which are seen as the additional peaks in the spectrum of this variant. 34 Direct overlays of 1H-15N HSQC NMR spectra of wild-type b2m in the presence of rifamycin SV and rifaximin and F62A,Y63A,Y67A bound to rifamycin SV are shown in Supplementary Figure 8a-c.
While the chemical shifts of resonances observed in the 1H-15N HSQC spectrum of acid-unfolded β2m remain unchanged upon the addition of rifamycin SV (Supplementary Figure 8a) the intensity of all resonances decreases substantially when the ligand is added (Figure 5a-c, Supplementary Figure 8a). At 800 μM rifamycin SV (an 8-fold molar excess over protein), few resonances remain detectable in the spectrum, consistent with rapid and quantitative formation of large oligomeric species that have a long correlation time and hence give rise to resonances too broad to detect using 1H-15N HSQC NMR. Control experiments in which rifaximin was added to 15N-labeled β2m in similar concentrations to those of rifamycin SV (Supplementary Figure 8b), or in which rifamycin SV was added to the triple mutant F62A,Y63A,Y67A (Figure 5d and Supplementary Figure 8c)) showed little effect on the chemical shift and no significant change in intensity of resonances arising from β2m, reinforcing the view that the interaction of β2m with rifamycin SV is highly specific and that aromatic residues in the region 60-70 are required for binding. In a separate experiment resonances arising from rifamycin SV were also monitored as β2m was titrated into the sample. These experiments also showed a loss of signal for rifamycin SV in the presence of β2m, consistent with the ligand binding to species of β2m with correlation times larger than would be expected for a ligand:monomer complex or intermediate exchange from weak binding of the ligand to the monomer (Supplementary Figure 8d). Consistent with this observation, analysis of the time-course of assembly of the spherical aggregates of β2m in the presence of rifamycin SV using ESI-MS showed depletion of the monomer without the observation of higher order oligomeric species, presumably because the spherical oligomers either do not ionize sufficiently well, or are too heterogeneous or too large in mass to detect using ESI-MS (Supplementary Figure 8d). Overall, therefore, the combination of ESI-IMS-MS and NMR show that binding of rifamycin SV to specific expanded conformers of β2m prevents assembly of the monomer into fibrils by diverting the course of assembly into large spherical aggregates.
Rifamycin SV disassembles early assembly intermediates
The mechanism by which rifamycin SV diverts β2m assembly towards spherical aggregates was explored further using ESI-IMS-MS. In these experiments, β2m (38 μM) was incubated under fibril-forming conditions in the presence or absence of rifamycin SV or rifaximin (each in a 10-fold molar excess). The presence of oligomeric species after incubation for 1 h was subsequently determined using ESI-IMS-MS under conditions optimized for the detection of non-covalently bound protein oligomers (see Methods). ESI-IMS-MS spectra of samples after incubation for 1 h at 37 °C (corresponding to the mid-lag time in the absence of ligand (Figure 6a)) revealed the presence of dimers, trimers and tetramers, but no higher order oligomers (Figure 6b), consistent with previous results using ESI-MS and analytical ultracentrifugation that these species develop rapidly (within 2 min of dissolution of β2m at pH 2.5) and remain populated throughout the lag phase. 26, 27 By contrast with these results, analysis of samples in which rifamycin SV was added to the assembly reaction at its onset showed the absence of oligomeric species after incubation for 1 h, while monomers that bind up to four rifamycin SV molecules remain after this time (Figure 6c). Addition of rifaximin to the fibril growth assays, by contrast, has little effect on the oligomers formed: dimers, trimers and tetramers all remain visible in the ESI-IMS-mass spectra after 1 h, with the ligand remaining bound to the most compact monomeric conformers (Figure 6d).
Figure 6.
β2m assembly at pH 2.5, 37 °C monitored using ThT fluorescence and ESI-IMS-MS. a Progress of assembly of β2m (with no ligand) monitored by ThT fluorescence with the 1 h time-point marked with a dotted line. The trace is included to show the progress of assembly in the absence of ligand. b ESI-IMS-MS Driftscope plot of β2m 1 h after incubation without ligand. c ESI-IMS-MS Driftscope plot of β2m in the presence of a 10-fold molar excess of rifamycin SV 1 h after ligand addition. d ESI-IMS-MS Driftscope plot of β2m in the presence of a 10-fold molar excess of rifaximin 1 h after ligand addition. All ESI-IMS-MS Driftscope plots show IMS drift time versus m/z versus intensity (z = log scale) and the corresponding mass spectrum for each experiment is shown on the right hand side. Data were acquired at a sampling cone voltage of 170 V, conditions optimized for the detection of protein oligomers. The oligomers in all plots are colored monomer (cyan), dimer (pink), trimer (dark blue) and tetramer (orange). The red circles in c indicate one or more molecules of rifamycin SV bound to different protein species. The green circles in d indicate one or more molecules of rifaximin bound to different protein species. TEM images (scale bar = 200 nm) were taken for each sample 48 h after incubation was commenced.
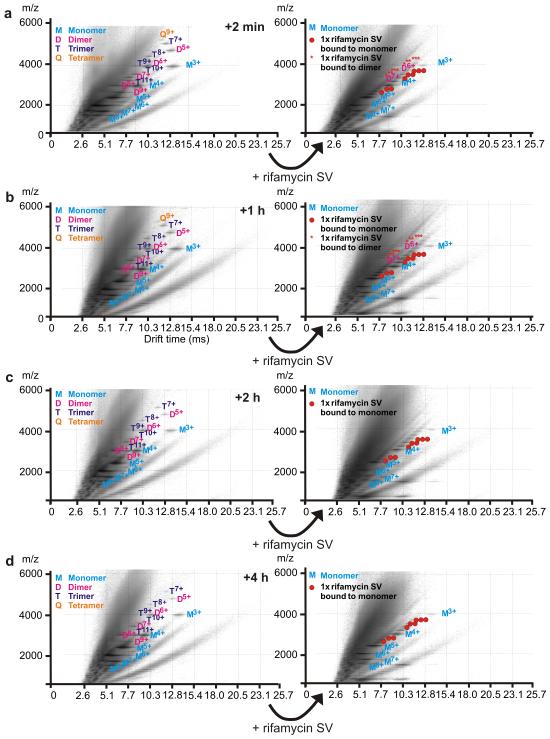
To determine whether rifamycin SV is able to disassemble oligomers formed at different times during the lag phase, a second assembly reaction was monitored in which β2m fibrillation was initiated in the absence of rifamycin SV. The ligand (10-fold molar excess) was added to the reaction at different times during the lag phase and the population of oligomeric species was monitored immediately prior to ligand addition, and 2 min thereafter, using ESI-IMS-MS (Figure 7a-d). The experiment revealed that dimeric, trimeric and tetrameric species form within 2 min of dissolution of β2m and are populated to ~ 8%, ~0.5% and <0.5% of the total protein, respectively after this time. Irrespective of the time during the lag phase at which rifamycin SV is added, rapid disassembly of all oligomeric species is observed, resulting in ligand-bound monomers (Figure 7b-d). At early time-points, where the population of oligomers is maximal, rifamycin SV-bound dimers are observed after addition of the inhibitor, their population changing from ~ 8% immediately before ligand addition to < 0.5% immediately after rifamycin SV is added (Figure 7a,b). At later time-points, ligand-bound dimers are not observed after rifamycin SV is added, presumably because their population is too low to detect using ESI-IMS-MS. These experiments confirm the ability of rifamycin SV to disassemble oligomers, presumably by thermodynamically favoring the monomer bound state. Such an activity is consistent with the rapid equilibration of oligomeric species with monomers determined previously using subunit exchange experiments monitored using ESI-IMS-MS. 32 Taken together, therefore, the ESI-IMS-MS and NMR data suggest a mechanism of action of rifamycin SV in which ligand binding prevents fibril formation by disfavoring oligomer formation and by binding to expanded and partially compact monomers that are crucially involved in the nucleation step of fibrillation. As a consequence of this interaction the course of monomer assembly is diverted to alternative route(s) that result in the formation of spherical aggregates.
Figure 7.
Rifamycin SV dissociates β2m oligomers when added at different times throughout the lag phase. ESI-IMS-MS Driftscope plots of wild-type β2m (38 μM) acquired after a 2 min, b 1 h, c 2 h and d 4 h incubation at pH 2.5 in 100mM ammonium formate at 37 °C, 200 rpm in the absence of ligand (left hand plots). Rifamycin SV (400 μM) was added to an aliquot taken from the same aggregating β2m stock solution at different times and Driftscope plots were acquired both immediately before and after (within 2 min) rifamycin SV addition. The red circles indicate one or more molecules of rifamycin SV bound to monomeric β2m and red asterisks indicate one or more molecules of rifamycin SV bound to dimeric β2m. All ESI-IMS-MS Driftscope plots show IMS drift time versus m/z versus intensity (z = log scale) and data were acquired with a sampling cone voltage of 170 V (i.e., conditions optimized for the detection of protein oligomers).
Discussion
The results presented above describe a screen for inhibitors of β2m fibrillogenesis that builds on previous findings that aromatic residues play a key role in fibril nucleation and elongation of this protein. 19, 20 Using different methods of analysis we identify the antibiotic rifamycin SV as a potent inhibitor able to block fibril formation of β2m in a specific and quantitative manner when added early during assembly. Rifamycins (including rifamycin SV, rifamycin S, rifamycin B and rifampicin) have been shown to inhibit amyloid formation of a variety of proteins by diverse mechanisms involving stabilization of the monomer and/or destabilization of oligomers or fibrils, although the precise mechanisms involved remain unclear. 18, 22, 23, 35 Here, by combining a screen for inhibition of β2m fibrillation with analyses using NMR and ESI-IMS-MS and comparing the behavior of structurally closely related ligands that have profoundly different effects on the course of assembly, we identify specific conformers within the dynamic ensemble of unfolded monomers that are able to bind rifamycin SV and, as a consequence, reveal the role of specific conformeric states in the fibril assembly mechanism.
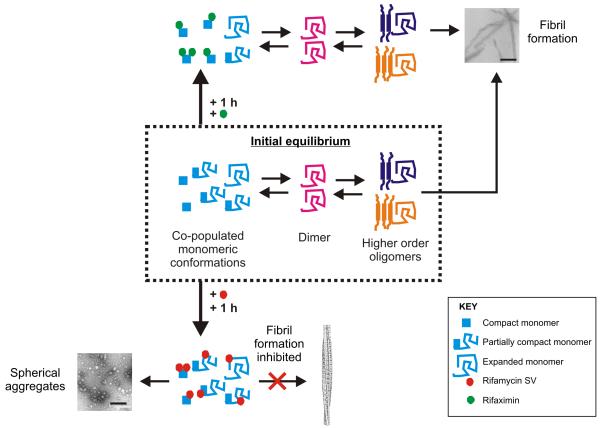
The results of the ESI-IMS-MS and NMR experiments support a model for the inhibition of β2m fibrillation in which rifamycin SV binding to the expanded and/or partially compact forms of β2m monomer prevents fibril nucleation (Figure 8) and demonstrate the ability of the approach developed to identify one or more specific conformers as the crucial agent(s) in fibril nucleation even within a rapidly interconverting ensemble of unfolded species. These conclusions are consistent with our previous findings that the rate of fibril formation increases at low pH and correlates with an increased population of the most expanded species within the monomeric ensemble. 36, 37 In addition, we demonstrate that rifamycin SV disassembles dimers, trimers and tetramers formed in the lag phase of assembly; species shown previously to be on-pathway to fibril nucleation, 38 further precluding fibril formation. By contrast, rifaximin binds only to the most compact conformers and, as a consequence, remodels the equilibrium between monomeric conformers. The result is a decreased population of the more expanded conformers which increases the lag time but allows fibril formation to proceed (Figure 8).
Figure 8.
Model of the inhibition of β2m aggregation by rifamycin SV at pH 2.5. Acid-unfolded β2m forms an interconverting ensemble of expanded, partially compact and compact monomers. These species assemble into amyloid fibrils (TEM image, scale bar = 200 nm) via dimers, trimers and tetramers that accumulate prior to nucleation of assembly. 27, 38 Previous analysis of the on-pathway oligomers using ESI-IMS-MS has enabled the collisional cross section and stability of these species to be discerned. 27 Studies here using ESI-IMS-MS reveal that binding of rifamycin SV (shown as a red circle) to the expanded and partially compact monomeric conformers disfavors the formation of the dimers, trimers and tetramers known to be on-pathway intermediates of amyloid formation. 38 As a consequence, protein self-assembly is re-routed towards the formation of large spherical aggregates that lack the cross-β structure of amyloid and are incapable of further assembly under the conditions employed (TEM image, scale bar = 200 nm). The closely related structural analogue, rifaximin (shown as a green circle), by contrast, binds only to compact monomers within the unfolded ensemble, such that the population of these conformers is increased. As a consequence, the formation of dimers, trimers and tetramers that are crucial intermediates in fibril nucleation can still occur, but the lag time of assembly is increased compared with assembly in the absence of the ligand.
At low pH, β2m aggregates on a complex energy landscape that involves different monomeric conformations (expanded, partially compact and compact species) as judged by ESI-MS 36 and ESI-IMS-MS. 33 These species are in dynamic equilibrium with each other as well as with the dimers, trimers and tetramers that are on-pathway to fibril assembly (Figure 8). 26, 27, 34 Previous studies have shown that altering the experimental conditions perturbs the equilibration between different monomeric conformers 6, 37 and switches the assembly pathway from the formation of LS fibrils that are produced via a nucleated mechanism,38 to the formation of worm-like fibrils that develop without a lag phase and are less well-ordered assemblies. 39, 40 These pathways are in direct kinetic competition, such that careful balancing of the rates of fibrillation along each route can result in pure products (LS or WL fibrils) or a mixture of both. 6, 37 Preliminary experiments suggest that rifamycin SV is also able to block the assembly of WL fibrils of β2m formed at pH 3.6, suggestive of common precursors in each pathway, although the mechanism of inhibition requires further investigation. Despite exploring a wide range of conditions, the formation of alternative assemblies of β2m have not been observed in the absence of small molecule additives, suggesting a bifurcation in the assembly landscape that leads to two well-defined energy minima occupied by LS or WL fibrils. Addition of rifamycin SV during β2m assembly thus allows access to a new assembly pathway that results in a thermodynamic minimum occupied by spherical aggregates with structural properties distinct from those of both WL and LS fibrils. This assembly route is presumably kinetically or thermodynamically disfavored in the absence of ligand and is made possible by ligand binding to unfolded monomers of β2m that are known to be crucial for fibril assembly. 19, 20 The result is the formation of spherical aggregates that do not possess the cross-β structure of amyloid.
Many proteins and peptides have been shown to utilize aromatic stacking interactions to promote fibril nucleation and elongation and to stabilize amyloid fibrils. 41 Accordingly, small molecules enriched in aromatic moieties have been shown to inhibit fibrillation in other systems including α-synuclein, Aβ, Tau, amylin, insulin, calcitonin and human prions. 5, 10, 14, 42-45 While in some cases inhibition has been shown to occur via a non-specific colloidal mechanism, 5, 10, 11, 14-16 in many cases the mechanism of inhibition, the target(s) of ligand binding and the manner by which ligand binding modulates assembly, remain unclear. 18 Here, using β2m assembly at pH 2.5 as a model system, we show that ESI-IMS-MS is able to resolve rapidly interconverting conformers of protein molecules with the same mass and to determine their ligand binding capabilities. Furthermore, this technique is able to separate and analyze protein assemblies of different mass without prior isolation, so that the ligand binding capability of individual conformers, the consequences of ligand binding on their structural properties, and the effect of ligand binding on the progress of assembly can be discerned in real-time within a single experiment. As well as becoming an increasingly utilized structural tool for the determination of assembly mechanisms 3, 46-48, we show here that ESI-IMS-MS offers a new capability to identify specific, even rarely populated conformers, able to bind ligands and to determine the effect of binding on an assembly mechanism. The approach can be applied to the plethora of macromolecular assembly reactions involving proteins, nucleic acids and their co-factors that are known to play central roles in cellular homeostasis as well as in disease. ESI-IMS-MS, therefore, is not only of generic utility for developing new fundamental understanding of the mechanisms of protein self-assembly into the cross-β structure of amyloid, but as a screening method in the development of therapeutics, it also will enable the exploration and refinement of inhibitors of amyloid formation to a level of precision that would not be possible without the unique separating and resolving powers of this approach.
Methods
Fibril growth experiments
Thioflavin-T (ThT) fluorescence was recorded using either a FLUOstar Optima or FLUOstar Omega reader (BMG) and 96-well blackwall plates (Costar) sealed with clear sealing film (Axygen). Samples had a volume of 100 μl containing 100 μM ThT in 25 mM sodium acetate / 25 mM sodium phosphate (pH 2.5), 0.04 % (w/v) NaN3, 10 % (v/v) DMSO, 1 mM test compound and 45 μM β2m (purified as described in 19). All assays contained 45 μM β2m and 1 mM small molecule and were incubated at 25 °C with agitation (600 rpm). Measurements were acquired simultaneously for three or more replicates for 48 h. Seeded experiments were performed under identical conditions but with the addition of 5 % (v/v) preformed LS β2m fibrils or rifamycin SV-induced β2m oligomers at the start of the reaction. Turbidity measurements were acquired by monitoring the apparent absorbance at 635 nm using a NEPHALOstar Galaxy reader (BMG) and 96-well clear plates (Greiner) sealed with clear sealing film (Axygen). For these experiments the samples were prepared as described above, except that ThT was not included in the reaction. The data were normalized to the signal at ~ 2 h after the initiation of each reaction.
SDS-PAGE
The effect of small molecules on fibril formation and disassembly was monitored by SDS-PAGE. In each experiment 50 μl from each of the replicate samples for each test compound was removed and pooled. The pooled sample was centrifuged at 16,300x g for 30 min and 20 μl of supernatant analysed by SDS-PAGE. A similar sample was analysed without centrifugation to reveal the total protein within each experiment. Gel band intensities were quantified using SnapGene software (Syngene).
Electron microscopy
Samples from the growth or depolymerisation assays were examined undiluted on freshly ionized formvar- and carbon-coated TEM grids (Agar) using negative staining with uranyl acetate as previously described. 19
Circular dichroism (CD)
CD spectra were acquired using a Jasco J-715 spectropolarimeter using 45 μM β2m incubated for 48 h in the presence or absence of 1 mM rifamycin SV (initially dissolved in acetonitrile) in 25 mM sodium acetate / 25 mM sodium phosphate (pH 2.5). Spectra were acquired at 25 °C using a 2 nm band width and 20 nm min−1 scanning speed. A total of five scans were averaged for each dataset. Spectra of the buffer constituents were then subtracted from that of the protein sample.
ESI-IMS-MS
Samples for ESI-IMS-MS analysis were infused into a Synapt HDMS (Waters UK Ltd.) quadrupole-travelling wave IMS-oaTOF mass spectrometer, equipped with a Triversa Nanomate (Advion Biosciences) automated nano-ESI interface. Positive ESI was used with a capillary voltage of 1.7 kV and a nitrogen nebulising gas pressure of 0.7 p.s.i.. The ion accelerating voltages into the trap and transfer T-wave devices were set at 6 and 20 V, respectively. Mass calibration was performed by an infusion of CsI cluster ions and the raw data were processed using MassLynx v.4.1 software (Waters UK Ltd.).
Analysis of ligand binding to monomeric β2m
β2m (38 μM) was dissolved in 10 mM ammonium formate (pH 2.5) containing 40 μM or 400 μM of test compound in 10 % (v/v) acetonitrile. A sampling cone voltage of 70 V was used to preserve protein/ligand interactions and a backing pressure of 3.8 mbar was applied. Data were acquired over the range m/z 500-6000. For IMS-MS experiments, the wave height was ramped from 4-10 V at a speed of 300 ms−1.
Analysis of oligomers of β2m
To initiate fibril assembly lyophilised β2m (38 μM) was dissolved in 100 mM ammonium formate (pH 2.5) to form a stock solution. The sample was then sterile filtered, and the concentration determined using the A280. Fibril growth was allowed to progress by incubation at 37 °C in a FLUOstar Optima (BMG) and aliquots were removed and analysed in real time using ESI-IMS-MS. To determine the effect of test compound (final concentration 400 μM in 10 % (v/v) acetonitrile) an identical stock solution of β2m was used so that detailed quantitative comparisons of oligomer populations could be made (described in the Supplementary Methods.). Ligand was either added at the start of assembly or at different times during the lag phase. A backing pressure of 3.8 mbar was applied and data were acquired over the range m/z 500-8000 with a sampling cone voltage of 170 V applied in order to observe larger oligomeric species. For IMS-MS experiments, the wave height was ramped from 4-20 V with a speed of 200 ms−1.
NMR analyses
NMR experiments were carried out using 100 μM 15N-labeled β2m in 25 mM sodium acetate / 25 mM sodium phosphate (pH 2.5), 10 % (v/v) D2O, 0.04 % (w/v) NaN3 and 10 % (v/v) DMSO containing the required amount of test compound.. Rifamycin SV (1 mM) and other ligand samples were prepared in the same buffer and β2m was added to the desired final concentration.
NMR experiments were performed at 25 °C using a Varian Unity Inova spectrometer operating at a 1H frequency of 500 MHz. Gradient enhanced 1H-15N HSQC spectra were acquired as previously described using 128 complex points and 16 scans per increment with spectral widths of 4508 Hz and 1200 Hz in the 1H and 15N dimensions, respectively. 19
Immunoreactivity assays
Dot blots of the β2m samples were probed with WO1 30, OC 29 or A11 49 antibodies as previously described 6 except that phosphate-buffered saline (with 0.01% (v/v) Tween 20 for A11 and OC antibodies and 0.2 % (v/v) Tween 20 for WO1) was used as buffer. Control experiments in which LS fibrils or monomers were immobilised onto nitrocellulose and incubated with 1 mM rifamycin SV prior to the addition of each antibody confirmed that rifamycin SV does not prevent binding to the A11 epitope. Similarly, incubation of each sample with or without 1 mM rifamycin SV in solution prior to immobilisation gave identical results on immunoblotting with WO1 and OC antibodies.
Supplementary Material
Acknowledgements
We thank Arnout Kalverda for advice on NMR spectroscopy, David Smith for advice about α-synuclein, Keith Ainley for technical assistance, Charlie Glabe for the gift of OC and A11 antibodies and Aβ1-40 oligomers, Jo Humphrey for creating fibrillar oligomers of Aβ1-42, Ronald Wetzel for the gift of WO1 antibodies and the Radford and Ashcroft groups for helpful discussions and advice. We acknowledge the Wellcome Trust (grant numbers 075675 and 062164) and BBSRC (Grant Number BB/526502/1) for funding. The Synapt HDMS mass spectrometer was purchased with funding from the Biotechnology and Biological Sciences Research Council (Research Equipment Initiative grant BB/E012558/1).
Footnotes
Financial Statement I declare that the authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Sipe JD, et al. Amyloid fibril protein nomenclature: 2010: Recommendations from the nomenclature committee of the international society of amyloidosis. Amyloid. 2010;17:101–104. doi: 10.3109/13506129.2010.526812. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Ann. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein SL, et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glabe CG. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that Aβ oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 6.Gosal WS, et al. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J. Mol. Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 7.Carrell RW. Cell toxicity and conformational disease. Trends Cell Biol. 2005;15:574–580. doi: 10.1016/j.tcb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Martins IC, et al. Lipids revert inert Aβ amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 2008;27:224–233. doi: 10.1038/sj.emboj.7601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HG, et al. Challenging the amyloid cascade hypothesis: Senile plaques and amyloid-β as protective adaptations to Alzheimer disease. Ann. N. Y. Acad. Sci. 2004;1019:1–4. doi: 10.1196/annals.1297.001. [DOI] [PubMed] [Google Scholar]
- 10.Porat Y, Abramowitz A, Gazit E. Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol.Drug Des. 2006;67:27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 11.Regazzoni L, et al. A combined high-resolution mass spectrometric and in silico approach for the characterization of small ligands of β2-microglobulin. Chem. Med. Chem. 5:1015–1025. doi: 10.1002/cmdc.201000082. [DOI] [PubMed] [Google Scholar]
- 12.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 13.Conway KA, Rochet JC, Bieganski RM, Lansbury PT. Kinetic stabilization of the α-synuclein protofibril by a dopamine-α-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 14.Feng BY, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lendel C, et al. On the mechanism of non-specific inhibitors of protein aggregation: Dissecting the interactions of α-synuclein with Congo red and lacmoid. Biochemistry. 2009;48:8322–8334. doi: 10.1021/bi901285x. [DOI] [PubMed] [Google Scholar]
- 16.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 17.Ladiwala AR, Dordick JS, Tessier PM. Aromatic small molecules remodel toxic soluble oligomers of amyloid β through three independent pathways. J. Biol. Chem. 286:3209–3218. doi: 10.1074/jbc.M110.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong AH, Chen J, McKoy AF, Hecht MH. Mutations that replace aromatic side chains promote aggregation of the Alzheimer’s Aβ peptide. Biochemistry. 2011;50:4058–4067. doi: 10.1021/bi200268w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt GW, Routledge KE, Homans SW, Radford SE. Fibril growth kinetics reveal a region of β2-microglobulin important for nucleation and elongation of aggregation. J. Mol. Biol. 2008;378:251–263. doi: 10.1016/j.jmb.2008.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Routledge KE, Tartaglia GG, Platt GW, Vendruscolo M, Radford SE. Competition between intra-molecular and inter-molecular interactions in an amyloid forming protein. J. Mol. Biol. 2009;389:776–786. doi: 10.1016/j.jmb.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng FL, Marek P, Potter KJ, Verchere CB, Raleigh DP. Rifampicin does not prevent amyloid fibril formation by human islet amyloid polypeptide but does inhibit fibril thioflavin-T interactions: Implications for mechanistic studies β-cell death. Biochemistry. 2008;47:6016–6024. doi: 10.1021/bi702518m. [DOI] [PubMed] [Google Scholar]
- 22.Lieu VH, Wu JW, Wang SSS, Wu CH. Inhibition of amyloid fibrillization of hen egg-white lysozymes by rifampicin and p-benzoquinone. Biotech. Prog. 2007;23:698–706. doi: 10.1021/bp060353n. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Zhu M, Rajamani S, Uversky VN, Fink AL. Rifampicin inhibits α-synuclein fibrillation and disaggregates fibrils. Chem. Biol. 2004;11:1513–1521. doi: 10.1016/j.chembiol.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 24.Tomiyama T, Kaneko H, Kataoka K, Asano S, Endo N. Rifampicin inhibits the toxicity of pre-aggregated amyloid peptides by binding to peptide fibrils and preventing amyloid-cell interaction. Biochem. J. 1997;322:859–865. doi: 10.1042/bj3220859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carazzone C, et al. Sulfonated molecules that bind a partially structured species of β2-microglobulin also influence refolding and fibrillogenesis. Electrophoresis. 2008;29:1502–1510. doi: 10.1002/elps.200700677. [DOI] [PubMed] [Google Scholar]
- 26.Smith AM, Jahn TR, Ashcroft AE, Radford SE. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionisation mass spectrometry. J. Mol. Biol. 2006;364:9–19. doi: 10.1016/j.jmb.2006.08.081. [DOI] [PubMed] [Google Scholar]
- 27.Smith DP, Radford SE, Ashcroft AE. Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Nat. Acad. Sci.USA. 2010;107:6794–6798. doi: 10.1073/pnas.0913046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong DW, Schneiderheinze J, Nair U, Magid LJ, Butler PD. Self-association of rifamycin B: Possible effects on molecular recognition. J. Phys. Chem. B. 1999;103:4338–4341. [Google Scholar]
- 29.Kayed R, et al. Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J. Biol. Chem. 2009;284:4230–4237. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Nuallain B, Wetzel R. Conformational abs recognizing a generic amyloid fibril epitope. Proc. Nat. Acad.Sci. USA. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue WF, et al. Fibril fragmentation enhances amyloid cytotoxicity. J. Biol. Chem. 2009;284:34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DP, et al. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur. J. Mass Spectrom. 2009;15:113–130. doi: 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- 33.Smith DP, Giles K, Bateman RH, Radford SE, Ashcroft AE. Monitoring copopulated conformational states during protein folding events using electrospray ionization-ion mobility spectrometry-mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18:2180–2190. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt GW, McParland VJ, Kalverda AP, Homans SW, Radford SE. Dynamics in the unfolded state of β2-microglobulin studied by NMR. J. Mol. Biol. 2005;346:279–294. doi: 10.1016/j.jmb.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 35.Tomiyama T, et al. Rifampicin prevents the aggregation and neurotoxicity of Aβ protein in vitro. Biochem. Biophys. Res. Commun. 1994;204:76–83. doi: 10.1006/bbrc.1994.2428. [DOI] [PubMed] [Google Scholar]
- 36.Borysik AJ, Radford SE, Ashcroft AE. Co-populated conformational ensembles of β2-microglobulin uncovered quantitatively by electrospray ionisation mass spectroscopy. J. Biol. Chem. 2004;279:27069–27077. doi: 10.1074/jbc.M401472200. [DOI] [PubMed] [Google Scholar]
- 37.Smith DP, Jones S, Serpell LC, Sunde M, Radford SE. A systematic investigation into the effect of protein destabilization on β2-microglobulin amyloid formation. J. Mol. Biol. 2003;330:943–954. doi: 10.1016/s0022-2836(03)00687-9. [DOI] [PubMed] [Google Scholar]
- 38.Xue WF, Homans SW, Radford SE. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Nat. Acad. Sci. USA. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladner CL, et al. Stacked sets of parallel, in-register beta-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J. Biol. Chem. 285:17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Magic angle spinning NMR analysis of β2-microglobulin amyloid fibrils in two distinct morphologies. J. Am. Chem. Soc. 132:10414–10423. doi: 10.1021/ja102775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gazit E. A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi S, et al. Inhibition of heparin-induced tau filament formation by phenothiazines, polyphenols, and porphyrins. J. Biol. Chem. 2005;280:7614–7623. doi: 10.1074/jbc.M408714200. [DOI] [PubMed] [Google Scholar]
- 43.Ehrnhoefer DE, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 44.Bieschke J, et al. EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Nat. Acad. Sci. USA. 2010;107:7710–7715. doi: 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ladiwala ARA, et al. Resveratrol selectively remodels soluble oligomers and fibrils of amyloid Aβ off-pathway conformers. J. Biol. Chem. 2010;285:24228–32427. doi: 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabenauer M, Wu C, Soto P, Shea JE, Bowers MT. Oligomers of the prion protein fragment 106-126 are likely assembled from beta-hairpins in solution, and methionine oxidation inhibits assembly without altering the peptide’s monomeric conformation. J. Am. Chem. Soc. 132:532–539. doi: 10.1021/ja905595k. [DOI] [PubMed] [Google Scholar]
- 47.Dupuis NF, Wu C, Shea JE, Bowers MT. Human islet amyloid polypeptide monomers form ordered beta-hairpins: A possible direct amyloidogenic precursor. J. Am. Chem. Soc. 2009;131:18283–18292. doi: 10.1021/ja903814q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashcroft AE. Mass spectrometry and the amyloid problem--how far can we go in the gas phase? J. Am. Soc. Mass Spectrom. 2010;21:1087–1096. doi: 10.1016/j.jasms.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 49.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 50.Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.