Abstract
Biofilms are communities of microbes attached to surfaces, which can be found in medical, industrial and natural settings. In fact, life in a biofilm probably represents the predominate mode of growth for microbes in most environments. Mature biofilms have a few distinct characteristics. Biofilm microbes are typically surrounded by an extracellular matrix that provides structure and protection to the community. Microbes growing in a biofilm also have a characteristic architecture generally comprised of macrocolonies (containing thousands of cells) surrounded by fluid-filled channels. Biofilm-grown microbes are also notorious for their resistance to a range of antimicrobial agents including clinically relevant antibiotics.
The microtiter dish assay is an important tool for the study of the early stages in biofilm formation, and has been applied primarily for the study of bacterial biofilms, although this assay has also been used to study fungal biofilm formation. Because this assay uses static, batch-growth conditions, it does not allow for the formation of the mature biofilms typically associated with flow cell systems. However, the assay has been effective at identifying many factors required for initiation of biofilm formation (i.e, flagella, pili, adhesins, enzymes involved in cyclic-di-GMP binding and metabolism) and well as genes involved in extracellular polysaccharide production. Furthermore, published work indicates that biofilms grown in microtiter dishes do develop some properties of mature biofilms, such a antibiotic tolerance and resistance to immune system effectors.
This simple microtiter dish assay allows for the formation of a biofilm on the wall and/or bottom of a microtiter dish. The high throughput nature of the assay makes it useful for genetic screens, as well as testing biofilm formation by multiple strains under various growth conditions. Variants of this assay have been used to assess early biofilm formation for a wide variety of microbes, including but not limited to, pseudomonads, Vibrio cholerae, Escherichia coli, staphylocci, enterococci, mycobacteria and fungi.
In the protocol described here, we will focus on the use of this assay to study biofilm formation by the model organism Pseudomonas aeruginosa. In this assay, the extent of biofilm formation is measured using the dye crystal violet (CV). However, a number of other colorimetric and metabolic stains have been reported for the quantification of biofilm formation using the microtiter plate assay. The ease, low cost and flexibility of the microtiter plate assay has made it a critical tool for the study of biofilms.
Protocol
1. Growing a Biofilm
Grow a culture of the wild-type Pseudomonas aeruginosa or mutant strain over night in a rich medium (i.e. LB)
Dilute the over night culture 1:100 into fresh medium for biofilm assays. A standard biofilm assay medium for P. aeruginosa is M63 minimal medium supplemented with magnesium sulfate, glucose and casamino acids (see Table). As an alternative biofilm-promoting medium that stimulates less planktonic growth and a more robust biofilm, the glucose and casamino acids can be replaced with arginine as the sole carbon and energy source.
Add 100 μL of the dilution per well in a 96 well dish. For quantitative assays, we typically use 4-8 replicate wells for each treatment.
Incubate the microtiter plate for 4-24 hrs at 37°C.
2. Staining the Biofilm
After incubation, dump out cells by turning the plate over and shaking out the liquid.
Gently submerge the plate in a small tub of water (i.e., use the bottoms of pipette tip boxes for P1000 pipetmen as the tub). Shake out water. Repeat this process a second time. This step helps remove unattached cells and media components that can be stained in the next step, and significantly lowers background staining.
Add 125 μL of a 0.1% solution of crystal violet in water to each well of the microtiter plate. Wear gloves and a lab coat while making the solution. Use caution when weighing out the CV as the powder is hydroscopic and readily stains clothing, skin, etc.
Incubate the microtiter plate at room temperature for 10-15 min.
Rinse the plate 3-4 times with water by submerging in a tub of water as outlined above, shake out and blot vigorously on a stack of paper towels to rid the plate of all excess cells and dye.
Turn the microtiter plate upside down and dry for a few hours or overnight.
For qualitative assays, the wells can be photographed when dry.
3. Quantifying the Biofilm
Add 125 μL of 30% acetic acid in water to each well of the microtiter plate to solubilize the CV.
Incubate the microtiter plate at room temperature for 10-15 min.
Transfer 125 μL of the solubilized CV to a new flat bottomed microtiter dish.
Quantify absorbance in a plate reader at 550 nm using 30% acetic acid in water as the blank.
4. Representative Results:
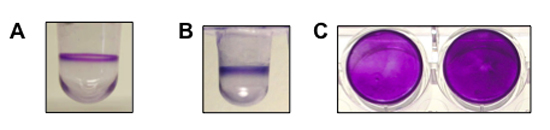 Figure 1. shows a representative result for biofilm formation assays performed for Pseudomonas aeruginosa, Pseudomonas fluorescens and Staphylococcus aureus. (A) A side view of the well with a biofilm of P. aeruginosa (8 hrs, 37°C). (B) A side view of the well with a biofilm of P. fluorescens (6 hrs, 30°C). (C) A top-down view of the biofilm formed by S. aureus in a flat-bottom microtiter plate (two wells, 24 hrs, 37°C). P. aeruginosa and P. fluorescens are both motile organisms and form a biofilm at the air-liquid interface. S. aureus is non-motile and forms a biofilm on the bottom of the well.
Figure 1. shows a representative result for biofilm formation assays performed for Pseudomonas aeruginosa, Pseudomonas fluorescens and Staphylococcus aureus. (A) A side view of the well with a biofilm of P. aeruginosa (8 hrs, 37°C). (B) A side view of the well with a biofilm of P. fluorescens (6 hrs, 30°C). (C) A top-down view of the biofilm formed by S. aureus in a flat-bottom microtiter plate (two wells, 24 hrs, 37°C). P. aeruginosa and P. fluorescens are both motile organisms and form a biofilm at the air-liquid interface. S. aureus is non-motile and forms a biofilm on the bottom of the well.
Discussion
This method can be modified for use with a wide variety of microbial species. Motile microbes typically adhere to the walls and/or bottoms of the wells, while non-motile microbes typically adhere to the bottom of the wells. The optimal conditions for biofilm formation (i.e., growth medium, temperature, time of incubation) must be determined empirically for each microbe. I recommend performing multiple replicates for each strain or condition (4-8), and including a positive control, and if possible, a negative control on each plate.
Disclosures
No conflicts of interest declared.
Acknowledgments
My thanks to Sherry Kuchma, Pete Newell and Robert Shanks for providing the images in Figure 1. This work was supported by NIH grant R01AI083256 to G.A.O.
References
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- O'Toole GA. In: Methods in Enzymology. Doyle RJ, editor. San Diego, CA: Academic Press; 1999. pp. 91–109. [Google Scholar]
- Mah TF. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Kuchma SL, Connolly JP, O'Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, O'Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Sargent JL, Martinez RM, Graber ML, O'Toole GA. Catheter lock solutions influence staphylococcal biofilm formation on abiotic surfaces. Nephrol Dial Transplant. 2006;21:2247–2255. doi: 10.1093/ndt/gfl170. [DOI] [PubMed] [Google Scholar]
- Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Mack D. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intracellular adhesin. Infect. Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal O. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker LM, Clardy J. High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrob Agents Chemother. 2007;51:3582–3590. doi: 10.1128/AAC.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


