Abstract
The first steps in vertebrate vision take place when light stimulates the rod and cone photoreceptors of the retina 1. This information is then segregated into what are known as the ON and OFF pathways. The photoreceptors signal light information to the bipolar cells (BCs), which depolarize in response to increases (On BCs) or decreases (Off BCs) in light intensity. This segregation of light information is maintained at the level of the retinal ganglion cells (RGCs), which have dendrites stratifying in either the Off sublamina of the inner plexiform layer (IPL), where they receive direct excitatory input from Off BCs, or stratifying in the On sublamina of the IPL, where they receive direct excitatory input from On BCs. This segregation of information regarding increases or decreases in illumination (the On and Off pathways) is conserved and signaled to the brain in parallel.
The RGCs are the output cells of the retina, and are thus an important cell to study in order to understand how light information is signaled to visual nuclei in the brain. Advances in mouse genetics over recent decades have resulted in a variety of fluorescent reporter mouse lines where specific RGC populations are labeled with a fluorescent protein to allow for identification of RGC subtypes 2 3 4 and specific targeting for electrophysiological recording. Here, we present a method for recording light responses from fluorescently labeled ganglion cells in an intact, isolated retinal preparation. This isolated retinal preparation allows for recordings from RGCs where the dendritic arbor is intact and the inputs across the entire RGC dendritic arbor are preserved. This method is applicable across a variety of ganglion cell subtypes and is amenable to a wide variety of single-cell physiological techniques.
Protocol
Animal Use Statement: Animals were cared for in accordance with guidelines described in Guide for the Care and Use of Laboratory Animals, using protocols approved by the University of Minnesota Institutional Animal Care and Use Committee.
1) Preparation of solutions
Before performing electrophysiological recording, intracellular, extracellular, and enzyme solutions need to be prepared.
Extracellular solution: mix one bottle (8.8g) of powdered Ames' medium (Sigma) with 1 l of H2O and 1.9 g of sodium bicarbonate (23 mM). At all points the extracellular solution is saturated with 95% O2/5% CO2 gas mixture. Transfer 200 mL to a beaker for retina storage. Place the remaining solution in the perfusion system container for gravity perfusion once retina is mounted.
Intracellular solution: Intracellular solution should have been made previously and then aliquoted into 500-1000 μL aliquots and stored at -20 °C. Intracellular solution varies depending on the experiment needed, for current clamp recordings shown here we use (in mM): 125 K-gluconate, 2 CaCl2, 2 MgCl2, 10 EGTA, 10 HEPES, 2 Na2-ATP, 0.5 mM NaGTP pH to 7.2 with KOH 5. A fluorescent tracer such as 10 μM Alexafluor-594 hydrazide (Invitrogen) can be added to visualize dendrites during the experiment, and neurobiotin (0.3%) can also be included for analysis of cell anatomy following recording. When ready to record, thaw intracellular solution.
Enzyme solution: Combine 10000 Units Collagenase (Worthington Chemicals) with 83 mg Hyaluronidase (Worthington Chemicals) into 4.150 mL of extracellular solution. Store in 50 μL aliquots at -20°C. Aliquots can be used for several weeks. When ready to treat the retina, dilute aliquot into 500 μL of bubbled extracellular solution.
Phosphate Buffered Solution (PBS): In 800 mL of H2O add 8 g of NaCl, 0.2 g of KCl, 1.4 g of Na2HPO4 and 0.24 g of KH2PO4. Bring the pH to 7.4 with NaOH. Adjust the volume to 1 L.
0.2 M Phosphate Buffer: In 800 ml of H2O add 21.8 g of Na2HPO4 and 6.4 g of NaH2PO4. Bring volume to 1 L.
4 % Paraformaldehyde Solution: Heat 50ml H2O to 60°C. Add 4g paraformaldehyde, stir for several minutes. Add few drops of 1M NaOH (about 5 drops), keep stirring until solution is clear. Add 50 ml 0.2 M phosphate buffer into beaker. Check pH using pH strips (~7.4). Filter and cool.
Blocking solution (10% goat serum, 0.5% TritonX100): In 1.8 ml of PBS add 0.2 ml of goat serum and 10 μL Triton X-100. TritonX-100 serves to permeabilise the tissue.
Streptavidin solution (5% goat serum, 0.5% TritonX100, 1:500 Streptavidin 594): In 1.9 ml of PBS add 0.1 ml of goat serum, 10 μL Triton X-100 and 4 μL of Streptavidin 594.
2) Isolation of retina from mouse
Dissection tools needed: 1 #2 Forceps, 2 #5 Forceps, ophthalmologic scissors
Euthanize mouse according to approved protocols, in this case CO2 asphyxiation and enucleate eyeball by gently inserting an angled #2 forceps behind the eye and lifting out the eyeball.
Place the eyeball into a 35 mm Petri dish with extracellular solution. Cut the optic nerve and any attached extraocular muscles or connective tissue using ophthalmologic scissors. Cut away the cornea with an ophthalmologic scissor and pull out the lens using #5 forceps. Tear the sclera using a pair of #5 forceps and use the forceps to sever the optic nerve where the retina and sclera meet at the optic disk. Gently peel the retina away from the eyecup. Tip: Do each step on both retinas before moving to next step to minimize time.
Use forceps to remove vitreous attached to the retina. This can be performed by "grabbing" the transparent vitreous above the retina with forceps. The vitreous, if successfully removed, should be visible as a clear, gelatinous substance on the forceps. Tip: Slicing retinas in half following removal of vitreous will maximize the amount of usable tissue and make mounting in the chamber easier.
Keep the retinas in oxygenated extracellular solution at room temperature in a dark environment with minimal ambient light until they are ready to be digested. Retinas can be stored in this way for ~6 hours.
3) Treatment of retina with enzyme mixture and mounting in recording chamber
Dilute an aliquot of collagenase/hyaluronidase in 500 μL extracellular solution, place solution in a 35 mm Petri dish and transfer the retina.
Keep the retina-containing Petri dish, covered and in darkness, in a 95% O2/5% CO2 environment for 15 min with gentle agitation. This step aids in digesting any remaining vitreous and allows for easier access to the ganglion cell layer. Tip: for retinas from younger animals with less vitreous or if adverse effects are observed, time can be shortened to ~5 minutes.
Remove retina from enzyme-containing medium and wash extensively in extracellular solution
Transfer the retina into recording chamber 6 with the photoreceptor layer facing down. Wick away remaining fluid with either a pipette or tissue and roll out sides of retina to flatten tissue being careful to touch only the edges of the retina, particularly avoiding contact with the ganglion cell layer. Place a platinum ring with nylon mesh over retina to anchor the tissue and immediately fill the chamber with extracellular solution.
Mount the recording chamber onto the microscope stage, attach tube for gravity perfusion and vacuum suction and attach ground wire.
4) Localization of EGFP expressing ganglion cells and light stimulation
Perfuse the retina by gravity with extracellular solution at 1 mL/min or faster rates. Tip: If perfusion rate is too slow, the tissue will not be sufficiently oxygenated and synaptic signaling may be disrupted.
Fill a recording pipette with intracellular solution and insert it into the pipette holder. Tip: Pipettes with resistances in the range of 3 to 5MΩ give the best results.
Visualize the retina under infrared differential interference contrast (DIC) optics at low magnification (10X objective, NA 0.3) to localize the recording pipette. Infrared light is used to minimize retina exposure to visible light and minimize light adaptation. Move to higher magnification (40X objective, NA 0.8) and bring electrode into view just above tissue.
At high magnification (40X objective), illuminate the retina with epifluorescence (480 nm for EGFP) and select a fluorescent ganglion cell and mark on computer monitor using tape (Figure 1A).
Under DIC and infrared illumination, position the recording electrode near the targeted ganglion cell (Figure 1A). Clean area immediately covering cell by applying gentle positive pressure to recording electrode via a 12cc plastic syringe connected to the pipette holder with polyethylene tubing. On the amplifier, zero any voltage offsets. Place electrode onto selected ganglion cell until a dimple appears on cell surface.
With the amplifier in voltage clamp mode, apply a test pulse and monitor the current passing through the pipette. Apply negative pressure to create seal between electrode and cell. Release negative pressure when cell begins to seal and wait until holding current is indicative of a GΩ electrical seal between the cell's membrane and pipette. Apply gentle pulses of negative pressure until capacitive transients appear and then release any application of pressure. Tip: if access (series) resistance is high (>30MΩ), this can often be ameliorated by additional, very gentle, pulses of negative pressure.
Once a stable seal has been reached apply a pulse of negative pressure. After whole cell mode is obtained, allow cell to dark adapt for 5 minutes. Record, in current clamp mode, response to full-field, white light stimulation (Figure 1B).
Following recording, Alexafluor-594 will have diffused to the dendrites. The tracer can be visualized under epifluorescence at 594 nm and used to determine whether a ganglion cell is On-, Off-, or Bi-stratified by switching between epifluorescence and DIC infrared optics 5.
If neurobiotin is included in pipette and retina is going to be processed for immunohistochemistry, then pipette must be gently retracted to provide minimal disruption of the membrane.
5) Analysis of anatomy of recorded ganglion cells
Filling with neurobiotin allows for more detailed morphological analysis of ganglion cells following recording. Tip: If specific correlations between a recorded ganglion cell and detailed morphology is desired, it may be best to record and fill only one RGC per preparation.
If neurobiotin was included in the intracellular solution then place the retina in paraformaldehyde solution and fix overnight at 4°C.
Rinse in PBS, at least 2 hours, at least 6 changes. Transfer the retina to a plate containing blocking solution. Leave in blocking solution for 2 hours, room temperature on shaker.
Replace the blocking solution with the Streptavidin solution. Incubate 2 days at 4°C on shaker. Rinse the retina in PBS, 6 rinses, 20 min each.
Mount the retina with Vectashield in a glass slide. The retina must be placed with the ganglion cells up. Perform confocal microscopy (Figure 1C).
Representative Results
If the retina is healthy and the dissection was successful, individual ganglion cells should be visible under DIC at high magnification (Figure 1A). Unhealthy retinas will have large, granulated nuclei and should be discarded. If the synaptic response of a given ganglion cell is intact, then the cell should respond at light onset or offset with action potentials and, in the case of an intrinsically-photosensitive ganglion cell (Figure 1B) , a sustained depolarization.
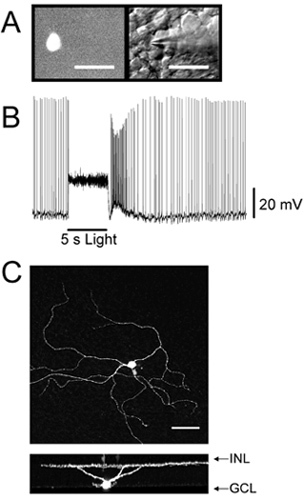 Figure 1. Localization, recording, and staining of a fluorescently labeled retinal ganglion cell. (A) GFP positive RGC visualized under epifluorescent illumination at 40X magnification (left panel) and under DIC illumination (right panel). (B) Synaptic light response of an M2 intrinsically photosensitive retinal ganglion cell recorded in whole-cell current clamp mode from fluorescently labeled RGC. (C) M1 intrinsically photosensitive retinal ganglion cell that was identified under epifluorescence, targeted for whole-cell recording, filled with neurobiotin, and then processed for immunohistochemistry.
Figure 1. Localization, recording, and staining of a fluorescently labeled retinal ganglion cell. (A) GFP positive RGC visualized under epifluorescent illumination at 40X magnification (left panel) and under DIC illumination (right panel). (B) Synaptic light response of an M2 intrinsically photosensitive retinal ganglion cell recorded in whole-cell current clamp mode from fluorescently labeled RGC. (C) M1 intrinsically photosensitive retinal ganglion cell that was identified under epifluorescence, targeted for whole-cell recording, filled with neurobiotin, and then processed for immunohistochemistry.
Discussion
This technique is applicable to any retinal ganglion cell recordings from an isolated retina. Though in the mouse line used above intrinsically photosensitive, melanopsin-expressing RGCs are labeled with EGFP2, 5, this protocol is readily transferable to other fluorescently labeled RGC lines or can be generalized to record from randomly selected RGCs as well. This technique is particularly valuable because the dendritic arbor of each RGC and its respective input neurons are left entirely intact. Recordings can readily be performed in current or voltage clamp modes, cell-attached or loose-patch configuration, or can even be used for nucleated patch, inside- or outside-out patch configurations from retinal ganglion cell membranes. This technique could be readily adapted to record from fluorescently labeled displaced amacrine cells 7 or even non-displaced amacrine cells 8 One limitation of the utilization of fluorescence to localize ganglion cells is that the retina must always be exposed to light for RGC identification prior to recording. Thus, this preparation may not be suitable for recording rod-mediated responses unless exogenous chromophore is included in the bath solution and/or the eyecup and retinal pigmented epithelium are left attached to the retina 9. If the goal is to record rod-mediated light responses, then special precautions must be taken to avoid bleaching of the photopigment rhodopsin. Dissection must be performed under infrared light conditions (i.e. 10).
To perform a successful dissection, it is important that the dissection be performed as rapidly as possible. It is especially important that the vitreous be successfully removed with the forceps. If the vitreous is remaining, then following enzyme digestion the retina will be covered with a gooey film making successful patching difficult. To obtain good recordings, it is particularly critical that the retina is well-oxygenated, meaning that the extracellular solution is being vigorously bubbled and the flow rate is sufficiently high through the chamber. It is also important to minimize light exposure of the tissue once the retina is isolated from the eyecup.
Acknowledgments
This work was supported in part by grants from the NIH R01EY012949, R21EY018885, T32 EY0707133. We thank Darwin Hang for his technical assistance.
References
- Wassle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5:747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Taniguchi K, Kofuji P. Intrinsic and extrinsic light responses in melanopsin-expressing ganglion cells during mouse development. J Neurophysiol. 2008;100:371–384. doi: 10.1152/jn.00062.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Kofuji P. Functional and morphological differences among intrinsically photosensitive retinal ganglion cells. J Neurosci. 2009;29:476–482. doi: 10.1523/JNEUROSCI.4117-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EA, Bartosch R. An eyecup preparation for the rat and mouse. J Neurosci Methods. 1999;93:169–175. doi: 10.1016/s0165-0270(99)00138-7. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Inta D, Monyer H, Wassle H. Expression analysis of green fluorescent protein in retinal neurons of four transgenic mouse lines. Neuroscience. 2009;160:126–139. doi: 10.1016/j.neuroscience.2009.01.081. [DOI] [PubMed] [Google Scholar]
- Zhang DQ, Zhou TR, McMahon DG. Functional heterogeneity of retinal dopaminergic neurons underlying their multiple roles in vision. J Neurosci. 2007;27:692–699. doi: 10.1523/JNEUROSCI.4478-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu EH, Dacheux RF, Bloomfield SA. A flattened retina-eyecup preparation suitable for electrophysiological studies of neurons visualized with trans-scleral infrared illumination. J Neurosci Methods. 2000;103:209–216. doi: 10.1016/s0165-0270(00)00319-8. [DOI] [PubMed] [Google Scholar]
- Wu SM, Gao F, Pang JJ. Synaptic circuitry mediating light-evoked signals in dark-adapted mouse retina. Vision Res. 2004;44:3277–3288. doi: 10.1016/j.visres.2004.07.045. [DOI] [PubMed] [Google Scholar]


