Abstract
Technical advances in mosquito biology are enabling the development of new approaches to vector control. Absent are powerful forward-genetics technologies, such as enhancer and gene traps, that permit determination of gene functions from the phenotypes arising from transposon insertion mutations. We show that the piggyBac transposon is highly active in the germline of the human malaria vector Anopheles stephensi. Up to 6% of the progeny from transgenic A. stephensi containing a single 6-kb piggyBac element with a marker gene expressing EGFP had the vector in new genomic locations when piggyBac transposase was provided in trans from a second integrated transgene. The active transposition of piggyBac resulted in the efficient detection of enhancers, with ∼10% of the progeny with piggyBac in new locations with novel patterns of EGFP expression in third and fourth instar larvae and in adults. The availability of advanced transgenic capabilities such as efficient transposon-based forward-genetics technologies for Anopheles mosquitoes not only will accelerate our understanding of mosquito functional genomics and the development of novel vector and disease transmission control strategies, but also will enable studies by evolutionary developmental biologists, virologists, and parasitologists.
Keywords: transposable element, Plasmodium, Drosophila
Mosquitoes are some of the world's most effective vectors of human pathogens and parasites, including the human malaria parasite Plasmodium falciparum. Controlling vector populations is a key component in ongoing efforts to reduce the burden of malaria, which currently directly results in hundreds of millions of clinical cases and approximately 1 million deaths annually and indirectly produces significant economic losses (1). For example, insecticide-treated bed nets not only reduce human contact with Anopheles mosquitoes, but also reduce vector population sizes, resulting in reduced malaria transmission; however, insecticide-based controls have been difficult to sustain (2–5). The ability to develop new vector-control approaches depends in part on continued growth in the understanding of vector biology. The significant expansion of knowledge of Anopheles mosquitoes has been stimulated by the availability of new resources (e.g., whole genome sequences) and technologies (e.g., transgenics) (6, 7). “Reverse-genetics” approaches are being used in contemporary functional genomics studies, whereby “mutant” phenotypes are created to infer gene function. Although powerful, these approaches when applied to mosquitoes are somewhat crude and limited (8, 9). Almost completely absent from the collection of genetics tools for mosquito studies are “forward-genetics” technologies, whereby mutations are created and used to identify genes of interest based on the resulting phenotypes.
There are a number of transposon-based forward-genetics technologies that enable the identification of genes based on the phenotypes of mutants arising from transposable element insertions (10, 11). These technologies can be quite powerful, because transposons are potent mutagens by virtue of their ability to disrupt gene expression on insertion into genes. When carrying appropriately configured transgenes, transposons can be used not only to disrupt genes, but also to report on the mutated gene's temporal and spatial patterns of expression (12). For example, fluorescent protein reporter genes regulated by weak promoters and contained within transposons are sensitive to the presence of local enhancers, which will result in increased reported gene transcription and permit ready identification of cells in which the enhancer is active. These and other powerful transposon-based forward-genetics technologies have been successfully developed for the insects Drosophila melanogaster, Tribolium castaneum, and Bombyx mori and the crustacean Parhyale hawaiensis (10, 13, 14). Transposon-based enhancer and gene detection technologies in arthropods depend on high-frequency transposition of class II transposons with patterns of transposition that are conducive to the detection of genes and gene regulatory sequences as a result of their insertion site preferences.
Currently there are four broad host-range class II transposons used as primary germline transformation vectors in mosquitoes: Hermes, Mos1, Minos, and piggyBac (15). Hermes is a hobo, Ac, Tam (hAT)-type element originally isolated from Musca domestica (16), and Mos1 and Minos are IS630/Tc1/mariner-type elements originally isolated from Drosophila mauritiana and Drosophila hydei, respectively (17, 18). piggyBac was originally isolated from baculoviruses passaged through Trichoplusia ni cells (19, 20). Transformation rates using these transposons are low when transposons are introduced into germ cells by microinjecting transposon-containing plasmids into preblastoderm embryos (15). Usually only a few percent of the embryos surviving microinjection develop into adults and produce transgenic progeny. Once integrated, none of the transposons tested to date has remobilized in mosquito germlines, despite much effort to detect such movement. Hermes, Mos1, and piggyBac were found to either move rarely or not at all in both the soma and germline of Aedes aegypti in the presence of element-specific transposases (21–23). Minos elements integrated into the genome of Anopheles stephensi were reported to be incapable of remobilizing in the presence of Minos transposase (24). The mechanisms underlying this transposon-silencing phenomenon in mosquitoes are unknown; however, they severely restrict the development and application of many powerful transposon-based functional genomics technologies. Here we report that integrated piggyBac transposons behave quite differently and do not suffer silencing after integration into the genome of A. stephensi, but in fact are extremely active in the germline of this species. For example, ∼6% of the progeny from transgenic A. stephensi with a single 6-kb piggyBac element with a marker gene expressing EGFP had the vector in a new genomic location when piggyBac transposase was provided in trans from a second integrated transgene. The active transposition of piggyBac resulted in the efficient detection of enhancers, with ∼10% of the progeny with piggyBac in new locations with novel patterns of EGFP expression in third and fourth instar larvae that were uncharacteristic of the promoter (3xP3) regulating EGFP expression (25). The efficient movement of piggyBac in the germline of A. stephensi and our ability to use it to detect enhancers demonstrates that this transposon can provide an excellent platform on which to base much needed forward-genetics technologies for Anopheles research.
Results
Transgenic A. stephensi lines were created with single inserts of piggyBac vectors from plasmids pXL-BacII-ECFP (lines UMITF-A7ECFP and UMITF-A10ECFP) and pPB-MGshSRPN6-EGFP (line UMITF-M1EGFP), and piggyBac transposase-expressing lines with single inserts of the Minos vector from plasmid pMi[3xP3-DsRed]-hsp70-piggyBac (lines UMITF-PB-F2DsRed and UMITF-PB-M5DsRed). All lines had patterns of enhanced cyan fluorescent protein (ECFP), EGFP, and Discosoma sp. red fluorescent protein (DsRed) protein expression that did not vary during propagation of the lines, as was expected with stable integrations of piggyBac and Minos gene vectors. Remobilization experiments were performed by crossing piggyBac vector-containing individuals to piggyBac transposase-containing individuals and then backcrossing the resulting heterozygotes to WT A. stephensi. The resulting progeny were screened as third and fourth instar larvae. In all of the crosses described below, individuals with patterns of either ECFP or EGFP expression differing from those of the transgenic parental mosquitoes were observed and selected. These individuals had new insertions of piggyBac.
Remobilization of UMITF-A10ECFP
UMITF-A10ECFP contains, based on splinkerette-PCR, a single piggyBac vector with ECFP under the regulatory control of the 3xP3 promoter (Fig. 1). As is regularly observed with 3xP3-regulated gene expression in mosquitoes, ECFP expression was seen in the brain, salivary glands, and anal papillae. However, this line also had ECFP expression in the posterior half of the larval midgut, a phenotype not usually associated with 3xP3-regulated transgenes, caused by the influence of an unknown enhancer or promoter (Fig. 1). Germline transpositions of the piggyBac element in this genome were expected to almost always result in the loss of the position-dependent ECFP phenotype in the midgut. Of the 482 ECFP-expressing progeny arising from the germline of UMITF-A10ECFP;UMITF-PB-F2DsRed males, 13 larvae (2.7%) had phenotypes distinctly different from those of the transgenic parents in that their phenotypes had “reverted” to 3xP3ECFP phenotypes in which only the brain and ventral ganglia expressed the transgene (Fig. 1 and Table 1). No ECFP expression was seen in the anal papillae, salivary glands, or the midgut of these exceptional progeny. Genotyping these individuals confirmed the loss of the piggyBac element from the original integration site, as well as the presence of piggyBac in unique locations (Fig. 1 and Dataset S1).
Fig. 1.
Phenotype and genotype of UMITF-A10ECFP and progeny with remobilized piggyBac vectors. (A) Fourth instar UMITF-A10ECFP larvae with parental phenotype. ECFP expression is seen in the brain, salivary glands (s.g.), anal papillae (a.p.), and midgut (m.g.). (B) Alimentary canal of a fourth instar UMITF-A10ECFP larvae with ECFP in the midgut. (C) Fourth instar larva arising from piggyBac remobilization in UMITF-A10ECFP. ECFP expression is seen only in the brain and ventral ganglia (not visible in this view), not in the salivary glands, anal papillae, or midgut. (D) Splinkerette-PCR. UMITF-A10ECFP individuals (p) had a single piggyBac element. Multiple elements could yield splinkerette-PCR products of exactly the same size, although this is unlikely. Progeny with nonparental phenotypes (lanes 1–13) in most cases showed evidence of the element in new locations (arrows). The gel image is from one gel, and the lanes were rearranged digitally to improve the organization of the image.
Table 1.
Crosses to remobilize piggyBac in the germline of A. stephensi
| Cross | P♂ | P♀ | F1♂ | F1♀ | F2 EC(G)FP+ | F2 EC(G)FP+ nonparental | F2 EC(G)FP+ enhancer |
| I | A10* | F2† | A10/F2 | WT | 482 | 13 | NA‡ |
| II | A7§ | F2 | A7/F2 | WT | 492 | 16 | 3 |
| WT | A7/F2 | 142 | 15 | 6 | |||
| III | M5¶ | M1** | M1/M5 | WT | 454 | 15 | 1 |
| WT | M1/M5 | 541 | 42 | 2 | |||
| IV | M1 | M5 | M1/M5 | WT | 204 | 6 | 1 |
| WT | M1/M5 | 484 | 29 | 4 | |||
| V | F2 | M1 | M1/F2 | WT | 404 | 10 | 0 |
| WT | M1/F2 | 337 | 37 | 2 | |||
| VI | M1 | F2 | M1/F2 | WT | 253 | 8 | 2 |
| WT | M1/F2 | 240 | 24 | 4 |
*piggyBac-containing line UMITF-A10ECFP.
†piggyBac transposase-expressing line UMITF-PB-F2DsRed.
‡Not applicable because this was a “loss of function” screen.
§piggyBac-containing line UMITF-A7ECFP.
¶piggyBac transposase-expressing line UMITF-PB-M5DsRed.
**piggyBac-containing line UMITF-M1EGFP.
Remobilization of UMITF-A7ECFP
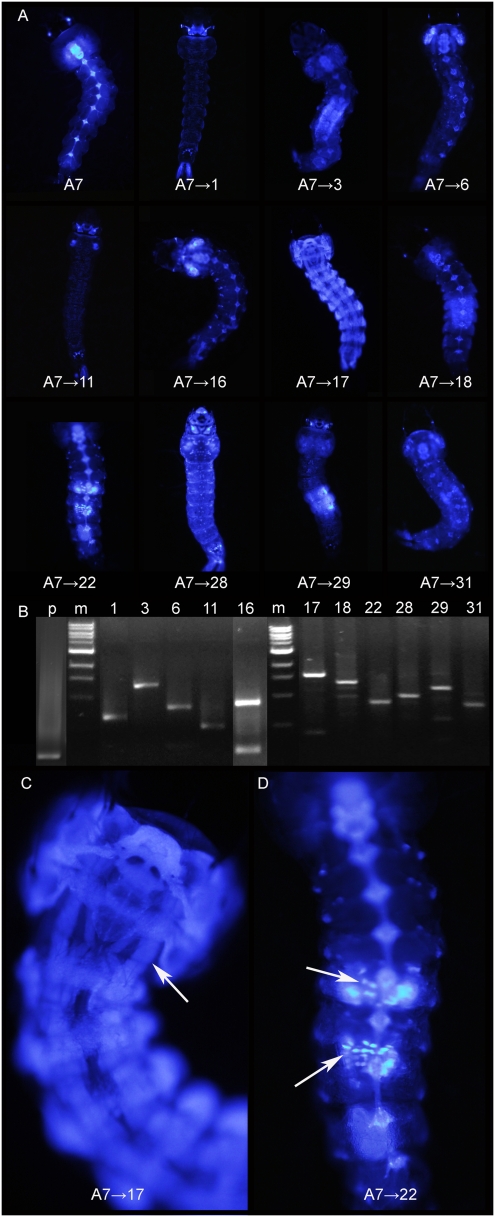
UMITF-A7ECFP is a line arising, based on splinkerette-PCR, from the integration of a single piggyBac vector with a 3xP3ECFP transgene. It exhibited a 3xP3 promoter-mediated pattern of ECFP expression in the brain and ventral ganglia of larvae (Fig. 2). In this line, ECFP expression was not seen in either the anal papillae or the salivary glands (Fig. 2). Crosses to detect element remobilization were designed to allow independent measurement of element activity in male and female germlines (Table 1). Of the 492 ECFP-expressing progeny arising from the germline of UMITF-A7ECFP;UMITF-PB-F2DsRed males, 16 larvae (3.3%) had phenotypes distinctly different from those of the transgenic parents. Notably, due to the influence of local enhancers, these larvae now had ECFP expression in the anal papillae, salivary glands, and, in three individuals, the midgut and other tissues (Fig. 2). Larvae that had ECFP expression phenotypes different from those of the transgenic parents had piggyBac in new locations within the genome (Fig. 2 and Table 1). The reciprocal crosses yielded 142 ECFP-expressing progeny from the germline of UMITF-A7ECFP;UMITF-PB-F2DsRed females, 15 of which (10.6%) had ECFP expression phenotypes uniquely different from the transgenic parents and 6 of which had tissue-specific enhancers (Fig. 2 and Table 1). A variety of tissue-specific enhancers were detected in larvae, resulting in midgut-, muscle-, and epidermal-specific expression of ECFP (Fig. 2). As with UMITF-A10ECFP remobilization, changes in the patterns of ECFP expression were correlated with the presence of piggyBac in new genomic locations, indicating that changes in the ECFP expression phenotype were good indicators of element transposition (Fig. 2 and Dataset S1). Although the detection of tissue-specific enhancers was easiest in larvae, these enhancers also could be detected in adults (Fig. 3). For example, UMITF-H16ECFP and UMITF-A3ECFP were recovered after piggyBac remobilization and displayed adult female salivary gland-specific expression that was readily detected through the cuticle of the proepisternum, and UMITF-A1ECFP had constitutive ECFP expression in the female midgut that was readily detected through the abdominal cuticle (Fig. 3).
Fig. 2.
piggyBac remobilization and enhancer detection in line UMITF-A7ECFP. (A) Fourth instar larva of UMITF-A7ECFP (A7). ECFP expression is seen in the brain and ventral ganglia only. There are 11 exceptional progeny with nonparental phenotypes (A7→X). (B) Splinkerette-PCR; parental (p) and progeny from A. The parental line contains one element, although multiple elements yielding exactly the same splinkerette-PCR products are possible. The gel images are from three gels (p, 1–16, and 17–31), with the lanes rearranged digitally to improve organization. (C) A7→17 with muscle-specific expression of ECFP (arrow). (D) A7→22 with ECFP expression in midgut cells (arrows).
Fig. 3.
Female adult phenotypes associated with piggyBac remobilization. (A) Ventral prothroax and head of UMITF-H16ECFP. (Inset) ECFP in all cells of the salivary gland. Glands are visible through the proepisternum cuticle (arrow). (B) Ventral prothroax and head of UMITF-A3ECFP. (Inset) ECFP in ommatidia and proximal lateral lobes of the salivary glands. Salivary glands can be seen through the proepisternum cuticle (arrow). (C) UMITF-A1ECFP alimentary canal and ovaries; ECFP in the midgut.
Remobilization of UMITF-M1EGFP.
The piggyBac vectors in UMITF-A7ECFP and UMITF-A10ECFP were small (∼2 kb), consisting of a few hundred base pairs of the 3′ and 5′ terminal sequences of piggyBac and the 3xP3ECFP marker gene. We tested the ability of a larger piggyBac vector to remobilize. UMITF-M1EGFP contains, based on splinkerette-PCR, a single ∼6-kb piggyBac vector composed of not only the terminal sequences of piggyBac and the 3xP3EGFP marker gene, but also a ∼3-kb transgene (Fig. 4). In UMITF-M1EGFP individuals, EGFP was expressed only in the brain and ventral ganglia and was not detected in the anal papillae or salivary glands. Although the pattern of EGFP expression in UMITF-M1EGFP was within the spectrum of phenotypes expected from 3xP3EGFP gene expression in this species, EGFP fluorescence was not very intense, likely due to the reduced gene expression related to the location of the insertion site. Thus, we expected to readily detect movements of this element within UMITF-M1EGFP, given that most changes in position can be expected to result in the acquisition of a 3xP3EGFP expression pattern more readily detected in the brain and ventral ganglia, as well as expression in the anal papillae and/or salivary glands. Genetic crosses were performed so that remobilization could be measured in male and female germlines using two piggyBac transposase-expressing lines, UMITF-PB-F2DsRed and UMITF-PB-M5DsRed. Of the 2,917 EGFP-positive larval progeny screened in these experiments, 171 (5.9%) had a phenotype distinctly different from that of the transgenic parents (Table 1 and Fig. 4). Genotyping confirmed the presence of at least one piggyBac vector in a new location in many of these insects (Fig. 4 and Datasets S1 and S2). There were no significant differences in remobilization frequencies between measurements using UMITF-PB-F2DsRed and those using or UMITF-PB-M5DsRed as a transposase source; however, progeny with nonparental phenotypes were detected at a higher frequency after piggyBac remobilization in the germline of females compared with males (Table 2). Cell- and tissue-specific enhancers also were detected frequently in this experiment. Of the 171 progeny with nonparental patterns of EGFP expression, 16 (9.4%) had cell- or tissue-specific expression, reflecting the influence of local enhancers or promoters (Fig. 4).
Fig. 4.
Remobilization and enhancer detection in UMITF-M1EGFP. (A) Third and fourth instar larvae; parental (M1) and nonparental phenotypes. (B) Splinkerette-PCR. Parents (p) contain one element, although multiple elements yielding exactly the same splinkerette-PCR products are possible. Evidence for piggyBac in locations different from the parents is seen in lanes 3, 16, 32, 51, and 116. A faint nonspecific band is seen in some of the lanes (denoted by *). The gel image is from one gel, and the lanes have been rearranged digitally to improve organization.
Table 2.
Analysis of UMITF-M1EGFP genetic data
| Comparison | f* | z† | P‡ |
| F2♂♀§ vs. M5♂♀¶ | 0.0640 | 1.0626 | 0.2879 |
| 0.0547 | |||
| All♂♂ vs. All♀♀ | 0.0297 | 6.0332 | <0.0001 |
| 0.0824 | |||
| F2♂♂ vs. F2♀♀ | 0.0274 | 5.6080 | <0.0001 |
| 0.1057 | |||
| M5♂♂ vs. M5♀♀ | 0.0319 | 3.2894 | 0.0010 |
| 0.0693 |
*Proportions of EGFP-positive progeny with nonparental phenotypes. The upper value is associated with the first class being compared; the lower value is associated with the second class.
†Test statistic.
‡Probability that a value of z or greater would be observed under the null hypothesis of equal proportions.
§piggyBac transposase-expressing line UMITF-PB-F2DsRed.
¶piggyBac transposase-expressing line UMITF-PB-M5DsRed.
Discussion
We have reported data showing that the class II transposon piggyBac is highly active in the germline of the mosquito A. stephensi in the presence of piggyBac transposase. This transposition activity resulted in ∼6% of the progeny arising from germlines in which piggyBac was active, with the element in a new genomic location. Although this is likely a slight overestimation of the transposition rate within the germline, because it does not correct for clusters of progeny arising from the same transposition event, these rates of recovery of exceptional progeny compare favorably with similar data reported for piggyBac movement in D. melanogaster, T. castaneum, and B. mori (13, 26–28). In most instances where piggyBac underwent transposition in these experiments, elements integrated into TTAA sites, the preferred target site of piggyBac (Datasets S1 and S2); however, in two cases piggyBac was found in noncanonical target sites (CTAC and CTAA). Overall, these observations are consistent with previous reports on piggyBac's preferred target site in various permissive host genomes (29, 30). The absence of a complete genome sequence for A. stephensi prevents analysis of the spatial patterns of transposition within the genome at this time; thus, questions regarding regional insertion preferences, intragenic insertion site preferences, and preferences for integrating into linked chromosomal sites must await the availability of more extensive genomic resources, which are expected to be forthcoming.
The absence of specific data on the insertion preferences of piggyBac in A. stephensi notwithstanding, this element was highly active in this species and proved to be a sensitive detector of local cis-regulatory elements responsible for controlling transcription. The screens conducted here focused on third and fourth instar larvae, because these larvae are particularly convenient for screening large numbers of individuals and have a transparent cuticle; however, unique enhancer-influenced marker gene expression patterns also could be detected in adults. Enhancer detection was efficient, and we were able to detect the presence of a unique enhancer or regulatory sequence in 1 of every 200 progeny arising from germlines containing piggyBac vectors and a source of piggyBac transposase. Given the high reproductive potential of A. stephensi in the laboratory, detecting genes, enhancers, and regulatory sequences will be quite practical. The biology of adult mosquitoes is of particular relevance to those interested in their roles, given that vectors of pathogens and genetic screens specifically designed to detect novel patterns of reporter gene expression in adult A. stephensi will benefit from the use of fluorescent proteins that are brighter than ECFP and EGFP (with brightness the product of quantum yield and molar extinction coefficient/1,000; ECFP = 13; EGFP = 34) and with spectral properties better suited for deep tissue detection and localization (31). Because light absorption and scattering decrease with increasing wavelengths, fluorescent proteins with spectral emissions in the red and far-red regions of the spectrum are desirable for whole adult mosquito imaging. The fluorescent protein tdTomato has spectral emission and brightness properties that are useful for imaging within whole adult mosquitoes (emission peak = 554 nm; brightness = 95) (31).
Our finding of high piggyBac activity in the A. stephensi germline has some important implications. First, advances in our understanding of mosquito molecular genetics over the last 2 decades have depended heavily on the development of new technologies; for example, whole genome sequencing and parallel-transcription profiling (microarrays) have been important enabling technologies that have led to detailed descriptions of global patterns of gene expression in mosquitoes. More recently, reverse-genetics technologies based on RNAi have provided opportunities to link descriptions of gene expression and structure to gene function and phenotypes. In mosquitoes, this has been done primarily through the massive introduction of long dsRNA molecules into the hemocoel of adults, where the dsRNA disseminates, is taken up by cells, and is processed into small interfering RNAs that suppress gene expression. Although useful, this particular reverse-genetics approach lacks the precision needed for silencing gene expression in space and time in mosquitoes to permit more detailed analysis of phenotypes and gene function. Transgenic approaches to RNAi-based gene silencing in mosquitoes through the in vivo expression of shRNAs have the potential to add more precision, but have been applied only occasionally (32). Current approaches to the creation of transgenic mosquitoes are inefficient and technically demanding, and investigators have very few options for regulating shRNA expression with respect to temporal and spatial patterns within mosquitoes because of the dearth of well-characterized promoters needed for such studies. The absence of a robust collection of well-characterized promoters that can be used to regulate transgene expression in mosquitoes is a widespread problem that greatly limits the utility of transgenic mosquito technology. The discovery of piggyBac's abilities to be efficiently remobilized in A. stephensi and to readily detect enhancers and regulatory sequences will enable the creation of a number of powerful genetic technologies that will greatly enhance our abilities to probe the genetic basis of important mosquito biology. piggyBac's efficient detection of enhancers can now be exploited in conjunction with the use of binary transcriptional regulatory systems (e.g., Gal4/UAS) to create genetic resources (Gal4-expressing lines) that will greatly expand the number of options available to investigators for expressing transgenes as it has in D. melanogaster (33). A particular advantage of transposon-based enhancer/promoter detection methods is that regulatory sequences do not need to be isolated and molecularly characterized before they can be productively used to regulate transgene expression.
The efficient movement of piggyBac in A. stephensi also will enable the creation of a number of powerful forward-genetics technologies for A. stephensi. For example, gene- and protein-trap systems can now be created that will permit genes to be identified based on their patterns of expression, providing immediate opportunities to study these patterns in great detail as well as to analyze mutant phenotypes associated with transposon-induced insertion mutations. Although global transcription-profiling and protein-profiling methods have been put to excellent use in the study of mosquitoes, forward-genetics technologies will provide opportunities to add much-needed temporal and spatial details, as well as the study of mutant phenotypes, to efforts to understand the basis of important mosquito phenotypes.
The significance of this work is not confined to studies of mosquito functional genomics. Evolutionary developmental biology (evo/devo) uses interspecific transgenic approaches, and advanced transgenic technologies in mosquitoes will increase the opportunities to use these insects in evo/devo studies (34). Virology and parasitology studies investigating virus and parasite biology in mosquitoes also will benefit from advanced transgenic technologies in these insects (35, 36). Finally, applied entomology programs focused on the genetic control of mosquito populations or their pathogen transmission characteristics will benefit from the advanced transgenic technologies based on transposon remobilization (37).
Methods
Mosquitoes.
A. stephensi (SDA 500) larvae were grown at 29 °C and provided with TetraMin Tropical Flakes ad libitum, whereas adults were grown at 29 °C and 80% relative humidity and provided with 10% sucrose continuously (38). Adult females were provided with blood from live mice in accordance with the University of Maryland, College Park's Institutional Animal Care and Use Committee operating under the National Institutes of Health's Office of Laboratory Animal Welfare guidelines. Protocols involving mice were not terminal, and animal pain and distress were minimized.
Vectors.
pXL-BacII-ECFP contains a piggyBac vector with functional terminal sequences and a marker gene consisting of the synthetic promoter 3xP3 regulating the expression of ECFP (30, 39). The plasmid pMi[3xP3-DsRed]-hsp70-piggyBac contains a functional Minos vector carrying the 3xP3 promoter regulating the expression of DsRed and the piggyBac transposase ORF regulated by the promoter from the hsp70 gene in D. melanogaster (26). The plasmid pPB-MGshSRPN6-EGFP was constructed by inserting a 3.1-kb transgene into the AscI site on plasmid pBac(3xP3-EGFPaf) (25). The transgene in this plasmid was unrelated to piggyBac remobilization and contained an Anopheles midgut-specific promoter regulating the expression of shRNA containing part of an Anopheles gambiae gene encoding a serine protease inhibitor.
Mosquito Transformation.
Transgenic A. stephensi were created by injecting preblastoderm embryos with plasmids containing vectors and the vector-specific transposase. The vector-containing plasmids pXL-BacII-ECFP and pMi[3xP3-DsRed]-hsp70-piggyBac were injected at 50 ng/μL, and pPB-MGshSRPN6-EGFP was injected at 75 ng/μL. The plasmid pHSS6hsILMi20 and phsp-pBac were the sources of Minos and piggyBac transposase during mosquito transformation, respectively, and were injected at 200 ng/μL (40, 41). Surviving insects were mated to noninjected insects, and the progeny were screened and selected for the expression of the vector-specific marker gene. Transgenic individuals were either genotyped (splinkerette-PCR) or used to establish lines.
Remobilization Crosses.
Approximately 20 piggyBac-containing individuals (male or female depending on the cross) were mated en masse with ∼20 piggyBac transposase-expressing individuals. Approximately 20 heterozygous individuals containing both piggyBac and piggyBac transposase were mated to ∼20 WT individuals en masse, and the resulting progeny were screened for nonparental patterns of ECFP and EGFP expression. Although piggyBac transposase was under the regulatory control of the promoter from the hsp70 gene from D. melanogaster, heterozygous individuals containing both piggyBac and piggyBac transposase were not heat-shocked, and the effects of heat shock during development on the frequency of remobilization were not tested.
Splinkerette-PCR.
The splinkerette-PCR genotyping method is based on amplification of genomic DNA containing the 5′ end of the piggyBac element and a variable amount of adjoining genomic DNA. This method was used to confirm the integration of piggyBac into the genome, to compare genotypes of transgenic individuals, and to sequence the genomic DNA flanking integrated piggyBac elements. Splinkerette-PCR was performed as described previously using genomic DNA isolated from individual third or fourth instar larvae or adults (42).
Microscopy.
All microscopic observations were made with an Olympus MVX10 microscope equipped with a DP70 digital camera.
Supplementary Material
Acknowledgments
We thank Drs. Steven Hoffman and Peter Billingsley, Sanaria Inc, for providing mosquitoes; Dr. Malcolm Fraser, University of Notre Dame, for providing the pXL-BacII-ECFP plasmid; Dr. Alfred Handler, United States Department of Agriculture, Agriculture Research Service, Center for Medical and Veterinary Entomology for providing the pMi[3xP3-DsRed]-hsp70-piggyBac, pHSS6hslLMi20, and phsp-pBac plasmids; Dr. Ernst Wimmer, Georg-August-Universität Göttingen, for providing the pBac3xP3EGFP plasmid; and Dr. Ada Rafaeli, Dr. Caroline Esnault, and Mr. Kasim George for helpful discussions. Financial support was provided by National Institutes of Health Grant R01AI070812.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110628108/-/DCSupplemental.
References
- 1.Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- 2.Collins FH, Besansky NJ. Vector biology and the control of malaria in Africa. Science. 1994;264:1874–1875. doi: 10.1126/science.8009215. [DOI] [PubMed] [Google Scholar]
- 3.Rieckmann KH. The chequered history of malaria control: Are new and better tools the ultimate answer? Ann Trop Med Parasitol. 2006;100:647–662. doi: 10.1179/136485906X112185. [DOI] [PubMed] [Google Scholar]
- 4.Alonso PL, et al. A research agenda for malaria eradication: Vector control. PLoS Med. 2011;8:e1000401. doi: 10.1371/journal.pmed.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enayati A, Hemingway J. Malaria management: Past, present, and future. Annu Rev Entomol. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- 6.Beaty BJ, et al. From Tucson to genomics and transgenics: The Vector Biology Network and the emergence of modern vector biology. PLoS Negl Trop Dis. 2009;3:e343. doi: 10.1371/journal.pntd.0000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terenius O, Marinotti O, Sieglaff D, James AA. Molecular genetic manipulation of vector mosquitoes. Cell Host Microbe. 2008;4:417–423. doi: 10.1016/j.chom.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellés X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- 9.Blandin S, et al. Reverse genetics in the mosquito Anopheles gambiae: Targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quiñones-Coello AT, et al. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-López M, García-Pérez JL. DNA transposons: Nature and applications in genomics. Curr Genomics. 2010;11:115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzen MD, et al. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol Biol. 2007;16:265–275. doi: 10.1111/j.1365-2583.2007.00727.x. [DOI] [PubMed] [Google Scholar]
- 14.Uchino K, Imamura M, Shimizu K, Kanda T, Tamura T. Germ line transformation of the silkworm, Bombyx mori, using the transposable element Minos. Mol Genet Genomics. 2007;277:213–220. doi: 10.1007/s00438-006-0176-y. [DOI] [PubMed] [Google Scholar]
- 15.Handler AM, O'Brochta DA. Transposable elements for insect transformation. In: Gilbert LI, editor. Insect Biochemistry and Molecular Biology. Oxford, UK: Elsevier; 2012. pp. 90–133. [Google Scholar]
- 16.Atkinson PW, Warren WD, O'Brochta DA. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc Natl Acad Sci USA. 1993;90:9693–9697. doi: 10.1073/pnas.90.20.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz G, Savakis C. Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res. 1991;19:6646. doi: 10.1093/nar/19.23.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medhora MM, Maruyama K, Hartl DL. Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics. 1991;128:311–318. doi: 10.1093/genetics/128.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cary LC, et al. Transposon mutagenesis of baculoviruses: Analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 20.Fraser MJ, Cary L, Boonvisudhi K, Wang H-GH. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995;211:397–407. doi: 10.1006/viro.1995.1422. [DOI] [PubMed] [Google Scholar]
- 21.O'Brochta DA, et al. Gene vector and transposable element behavior in mosquitoes. J Exp Biol. 2003;206:3823–3834. doi: 10.1242/jeb.00638. [DOI] [PubMed] [Google Scholar]
- 22.Sethuraman N, Fraser MJ, Jr, Eggleston P, O'Brochta DA. Post-integration stability of piggyBac in Aedes aegypti. Insect Biochem Mol Biol. 2007;37:941–951. doi: 10.1016/j.ibmb.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson R, et al. Post-integration behavior of a Mos1 mariner gene vector in Aedes aegypti. Insect Biochem Mol Biol. 2003;33:853–863. doi: 10.1016/s0965-1748(03)00044-4. [DOI] [PubMed] [Google Scholar]
- 24.Scali C, et al. Post-integration behavior of a Minos transposon in the malaria mosquito Anopheles stephensi. Mol Genet Genomics. 2007;278:575–584. doi: 10.1007/s00438-007-0274-5. [DOI] [PubMed] [Google Scholar]
- 25.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210:630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 26.Horn C, Offen N, Nystedt S, Häcker U, Wimmer EA. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics. 2003;163:647–661. doi: 10.1093/genetics/163.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trauner J, et al. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 2009;7:73. doi: 10.1186/1741-7007-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchino K, et al. Construction of a piggyBac-based enhancer trap system for the analysis of gene function in silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1165–1173. doi: 10.1016/j.ibmb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- 31.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 32.Franz AWE, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci USA. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy JB. GAL4 system in Drosophila: A fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 34.Sommer RJ. The future of evo-devo: Model systems and evolutionary theory. Nat Rev Genet. 2009;10:416–422. doi: 10.1038/nrg2567. [DOI] [PubMed] [Google Scholar]
- 35.Khoo CCH, et al. The RNA interference pathway affects midgut infection and escape barriers for Sindbis virus in Aedes aegypti. BMC Microbiol. 2010;10:130. doi: 10.1186/1471-2180-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saenz FE, et al. The transmembrane isoform of Plasmodium falciparum MAEBL is essential for the invasion of Anopheles salivary glands. PLoS One. 2008;3:e2287. doi: 10.1371/journal.pone.0002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catteruccia F. Malaria vector control in the third millennium: Progress and perspectives of molecular approaches. Pest Manag Sci. 2007;63:634–640. doi: 10.1002/ps.1324. [DOI] [PubMed] [Google Scholar]
- 38.Feldmann AM, Ponnudurai T. Selection of Anopheles stephensi for refractoriness and susceptibility to Plasmodium falciparum. Med Vet Entomol. 1989;3:41–52. doi: 10.1111/j.1365-2915.1989.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 39.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 40.Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- 41.Klinakis AG, Loukeris TG, Pavlopoulos A, Savakis C. Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
- 42.Potter CJ, Luo LQ. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.