Abstract
Cyanophages infecting the marine cyanobacteria Prochlorococcus and Synechococcus encode and express genes for the photosynthetic light reactions. Sequenced cyanophage genomes lack Calvin cycle genes, however, suggesting that photosynthetic energy harvested via phage proteins is not used for carbon fixation. We report here that cyanophages carry and express a Calvin cycle inhibitor, CP12, whose host homologue directs carbon flux from the Calvin cycle to the pentose phosphate pathway (PPP). Phage CP12 was coexpressed with phage genes involved in the light reactions, deoxynucleotide biosynthesis, and the PPP, including a transaldolase gene that is the most prevalent PPP gene in cyanophages. Phage transaldolase was purified to homogeneity from several strains and shown to be functional in vitro, suggesting that it might facilitate increased flux through this key reaction in the host PPP, augmenting production of NADPH and ribose 5-phosphate. Kinetic measurements of phage and host transaldolases revealed that the phage enzymes have kcat/Km values only approximately one third of the corresponding host enzymes. The lower efficiency of phage transaldolase may be a tradeoff for other selective advantages such as reduced gene size: we show that more than half of host-like cyanophage genes are significantly shorter than their host homologues. Consistent with decreased Calvin cycle activity and increased PPP and light reaction activity under infection, the host NADPH/NADP ratio increased two-fold in infected cells. We propose that phage-augmented NADPH production fuels deoxynucleotide biosynthesis for phage replication, and that the selection pressures molding phage genomes involve fitness advantages conferred through mobilization of host energy stores.
Keywords: coevolution, photosynthesis, virus
The use of host-like metabolic genes by viruses is a central theme in the coevolution of viruses and their hosts, and the particular metabolic genes carried by viruses provide clues to the mechanisms of viral replication. Bacteriophages typically carry multiple genes for nucleic acid biosynthesis and replication (1, 2), critical processes for replication of the phage genome. Cyanophages (cyanobacterial dsDNA bacteriophages) infecting the marine cyanobacteria Prochlorococcus and Synechococcus carry a much richer cache of metabolic genes than most sequenced phages, encoding elements of photosynthesis (3), the pentose phosphate pathway (PPP) (4), and phosphate acquisition (5). Such genes have been termed “auxiliary metabolic genes” (6) because they are thought to provide supplemental support to key steps in host metabolism of significance to phage, thereby fostering a more successful infection.
Photosynthetic light reaction genes in phage genomes have attracted particular attention because photosystems are sophisticated macromolecular complexes with functions in core cyanobacterial metabolism, and they are not traditionally associated with the phage infection process. Nevertheless, physiological studies of cyanophage infection have shown that a phage copy of psbA, which encodes the D1 protein essential for photosystem II (PSII), is transcribed and translated during infection (7–9). This result suggested that phage gene products may integrate into host photosystems and contribute to host photosynthesis during infection (7), possibly fueling phage dNTP biosynthesis (10, 11). It has long been recognized that both light and the photosynthetic light reactions are necessary for optimal cyanophage production (7, 12–14). However, in the host, the energy (i.e., ATP) and reducing equivalents (i.e., NADPH) generated by the light reactions are typically used for carbon fixation by the Calvin cycle. The presence of genes encoding Calvin cycle enzymes has not been reported for any sequenced cyanophages (5, 15–21), suggesting that phage use the ATP and NADPH generated by the light reactions of photosynthesis for other functions.
Clues to the role of photosynthesis during infection have been provided by our observation that some cyanophages carry a gene for CP12 (21), an inhibitor of the Calvin cycle. CP12 is an intrinsically unstructured protein that in plants (22) and cyanobacteria (23) binds and inhibits two key enzymes in the Calvin cycle (phosphoribulokinase, PRK, prkB gene; and glyceraldehyde-3-phosphate dehydrogenase, GAPDH, gap2 gene). CP12 binding has the effect of directing carbon flux away from glucose synthesis and toward the PPP, where glucose 6-P is oxidized by NADP to ribose 5-P and NADPH (Fig. 1A). We were prompted to look for CP12 in cyanophages after our studies of the light–dark cycle of Prochlorococcus (24) indicated its probable role in regulating the Calvin cycle and PPP in these cyanobacteria. We showed that host cp12 is maximally expressed at night in Prochlorococcus (24), consistent with its functioning to direct carbon flux away from the Calvin cycle and toward the PPP in the dark.
Fig. 1.
(A) The PPP and Calvin cycle in cyanobacteria showing genes carried by cyanophages. Genes found in phages are boxed and denoted with the number of genomes among 24 in which they are found. CP12 (cp12) is grouped with the PPP, because it shuts off the competing Calvin cycle, inhibiting PRK (prkB) and GAPDH (gap2). Both pathways run clockwise, with the shared reactions (blue) running downward (Calvin cycle) or upward (PPP), and therefore the two pathways are not expected to run concurrently. Transaldolase (talC in phage, talB in host) is the only reaction in the intersecting part of the pathways that is needed only in one direction. Arrows show proposed directionality of the reactions, and one-way arrows do not necessarily indicate irreversibility. P, phosphate; (genes) cbbA, fructose-1,6-bis-P/sedoheptulose-1,7-bis-P aldolase; glpX, fructose-1,6/sedoheptulose-1,7-bisphosphatase; pgi, P-glucose isomerase; pgk, P-glycerate kinase; pgl, 6-P-gluconolactonase; prkB, P-ribulokinase; rbcLS, ribulose-1,5-bis-P carboxylase/oxygenase; rpe, ribulose-5-P epimerase; rpiA, ribulose-5-P isomerase; tktA, transketolase; tpi, triose-P isomerase; (substrates) BPG, 2,3-bis-P-glycerate; DHAP, DHA P; E4P, erythrose 4-P; FBP, fructose 1,6-bis-P; F6P, fructose 6-P; GAP, glyceraldehyde 3-P; G6P, glucose 6-P; PGA, 3-P-glyceric acid; R5P, ribose 5-P; RuBP, ribulose 1,5-bis-P; Ru5P, ribulose 5-P; SBP, sedoheptulose 1,7-bis-P; 6PG, 6-P-gluconate; 6PGL, 6-P-gluconolactone; S7P, sedoheptulose 7-P; and X5P, xylulose 5-P. (B) Relative abundance of T4-like cyanophage genes in the GOS database. For each gene found in a T4-like cyanophage (red circles represent genes found in all sequenced genomes, i.e., core genes, and gray circles represent noncore genes), the number of times it was observed as a sequence read is plotted as a function of gene size. Core genes show a linear increase with gene size in the number of observed reads, as expected, as the likelihood of cloning and sequencing fragments of larger genes is greater than for smaller genes. The linear regression (with 95% confidence intervals) of this pattern is shown for core genes.
Cyanophages that encode CP12 also encode proteins for the PPP, the light reactions, and dNTP biosynthesis (21). Of particular interest among these phage-encoded functions is a putative transaldolase, widely represented in cyanophage genomes (5), which could boost the host PPP during infection. As a whole, the set of carbon metabolism genes carried by cyanophages is consistent with the hypothesis that, under phage infection, the ATP and NADPH produced by photosynthesis are not used by the Calvin cycle but rather are used to fuel phage dNTP biosynthesis. We now report our genomics and metagenomics studies, our enzymological studies of transaldolase, and our analyses of gene expression and cellular redox balance in uninfected and infected cells, which support this hypothesis.
Results and Discussion
CP12 and PPP Genes in Cultured and Wild Cyanophages.
We first examined the genetic potential of all sequenced cyanophages for photosynthesis and carbon metabolism and then measured the frequency of these genes in wild cyanophage populations. Twenty-four cultured cyanophage genomes representing the three major morphotypes (T4-like myoviruses, T7-like podoviruses, and siphoviruses) were analyzed for the presence and frequency of all host photosynthesis and carbon metabolism genes (Table S1). Notably, a focused search (Methods) for all Calvin cycle genes confirmed their absence in all 24 cyanophage genomes, supporting the suspicion that cyanophage do not exploit this part of host metabolism. The sequence analysis did reveal, however, the presence of the Calvin cycle inhibitor gene, cp12, in 18 of 24 cyanophages (Fig. 1A and Table S1), including all three major morphotypes. All cyanophage CP12 sequences (Fig. S1) contain a conserved C-terminal CxxxPxxxxC motif found in all cyanobacterial and nearly all plant CP12 sequences (25) and a pattern of hydrophilic residues, characteristic of intrinsically unstructured proteins (26). Unequivocal determination of the function of CP12 in marine cyanobacteria and cyanophage, however, awaits in vitro characterization of these proteins.
In addition to CP12, whose presence in phage has implications for promoting PPP flux in the host, three genes for PPP enzymes reported previously (4, 18) were found: transaldolase (talC), glucose-6-phosphate dehydrogenase (zwf), and 6-phosphogluconate dehydrogenase (gnd; Fig. 1A). We reveal here that these genes and cp12 display a hierarchical pattern of gene abundance (Table S1), which, as we discuss later, provides hints to the selection pressures on phage. Most abundant are talC and cp12, found in 20 and 18 of the 24 genomes, respectively, followed by gnd and zwf, found in eight and six, respectively (Fig. 1A and Table S1). cp12 is found in most T4-like phages and only once in available T7-like phages and siphoviruses; talC is widespread in both T4-like and T7-like phages; and gnd and zwf are found only in T4-like phages isolated on Synechococcus (Table S1).
The abundance of phage DNA fragments in environmental metagenomic databases, such as the Global Ocean Sampling (GOS) database (27), allowed us to analyze the prevalence of CP12 and PPP genes in wild phage populations, focusing on T4-like cyanophages, for which there are the most reference genomes (17) (Table S1). Interestingly, the prevalence pattern of cp12 and the PPP genes among cultured phages was largely recapitulated in the GOS metagenome (Fig. 1B). cp12 and talC (found in 16/17 T4-like cyanophage genomes) had abundances similar to “core genes,” i.e., genes found in every T4-like cyanophage genome, whereas gnd and zwf (found in 8/17 and 6/17) were less abundant than most core genes (Fig. 1B). This trend was not dependent on the number of different orthologues used to recruit sequences from the GOS database (SI Methods). As a side note, our metagenomic analysis was designed to probe only for PPP/Calvin cycle-related genes identified in cultured cyanophage genomes, and therefore we did not find evidence of fructose-1,6-bisphosphate aldolase, shared by the two pathways, which was recently reported in GOS scaffolds of suspected cyanophage origin (28) and represents an additional phage enzyme for the host PPP.
The emergent prevalence pattern suggests not only that the host PPP is important to infecting phage, but that some steps in the PPP may be bottlenecks more often than others, and therefore phage genes for those steps are under stronger selection. Transaldolase especially may be a key metabolic bottleneck under infection. Transaldolase, although reversible, is the only reaction in the nonoxidative PPP (blue plus transaldolase, Fig. 1A) that is not also shared by the Calvin cycle, required only in the direction that regenerates glucose 6-P for NADPH production (blue and red, clockwise, Fig. 1A). In our previous studies over the light–dark cycle of host Prochlorococcus MED4, all of the shared Calvin cycle/PPP genes had maximal mRNA levels in the morning, whereas the host transaldolase gene had maximal mRNA in the evening, with the other PPP genes (24). By 10:00 AM, the level of host transaldolase protein had decreased to half its maximum level, whereas all other proteins in the PPP and Calvin cycle remained relatively constant over this period (29). Thus, if phage exploit host photosynthetic light reactions by infecting during the day, expression of their own transaldolase may be critical to overcoming low levels of the host enzyme at this time.
Coordinated Transcription of Phage CP12 and Metabolic Enzymes.
In support of our hypothesis, the expression of talC has already been demonstrated for the T7-like cyanophage P-SSP7 (9) during infection. We now report the expression of phage cp12, zwf, and gnd during infection, together with psbA, encoding PSII protein D1, and nrdA/nrdB, encoding ribonucleotide reductase (RNR), the enzyme responsible for synthesis of deoxynucleotides from nucleotides. Our hypothesis about the coordinated activity of auxiliary metabolic genes for dNTP biosynthesis suggests that all these genes should have similar expression profiles and be expressed early in the infection cycle. To explore this, we examined gene expression patterns in two yet-unstudied T4-like myoviruses: Syn9, infecting Synechococcus WH8109 (Fig. 2); and P-HM2, infecting Prochlorococcus MED4 (Fig. S2 A and C). Both phages carry cp12, and in both phages it was coexpressed with talC, psbA, and nrdA/nrdB. In Syn9, these genes were also coexpressed with zwf and gnd (Fig. 2). As expected, each of these metabolic genes was expressed with the T4-like “early genes,” which tend to encode enzymes for phage replication, rather than with T4-like “late genes,” which usually encode structural proteins (2), thus positioning the gene products to participate in the takeover of host metabolism.
Fig. 2.
Cyanophage Syn9 infection of Synechococcus WH8109, showing expression of a phage gene for the Calvin cycle inhibitor CP12 alongside other phage metabolic genes. Error bars represent SDs of three technical and three biological replicates. (A) Degradation of host gDNA and replication of phage gDNA (free and intracellular) as determined by qPCR of chromosomal genes in host (rnpB) and phage (g20). (B) mRNA of T4-like early genes g61 (DNA primase) and g43 (DNA polymerase) and late genes g20 (portal protein) and g23 (major coat protein). (C) mRNA of PSII gene psbA (D1 subunit) and RNR gene nrdA (α subunit). (D) mRNA of PPP genes zwf (glucose-6-P dehydrogenase), gnd (6-P-gluconate dehydrogenase), talC (transaldolase), and cp12 (Calvin cycle inhibitor CP12).
The coordinated early expression of genes involving host photosynthesis, central carbon metabolism, and dNTP biosynthesis in these T4-like phages is reminiscent of the pattern observed in the T7-like phage P-SSP7 (9). However, in contrast to T4-like phages, in T7-like phages, gene expression is dictated by gene order rather than by early and late promoters (30). In P-SSP7, talC is the last gene in the genome but was expressed much earlier than expected based on its terminal position, so that it was coexpressed with psbA and nrdJ (i.e., RNR) (9). Thus, although the mechanisms that dictate timing of gene expression in these two phage types are different, the resultant coordination of expression is similar.
What differs among cyanophages, then, is not the timing of metabolic gene expression—evidence suggests they are generally expressed early—but which metabolic genes they carry. For example, P-SSP7 and, in total, five of six T7-like cyanophages do not carry cp12, and no T7-like phages have zwf or gnd (Table S1). The PPP gene that emerges as most important for phage is transaldolase (talC). Although still not universal, talC is found in more cyanophages (20 of 24) than any other PPP gene. We therefore conducted a detailed enzymological study of phage and host transaldolases to confirm their functions and measure their kinetic parameters. We asked whether talC encodes a functional transaldolase, and if so, whether it is kinetically more efficient than the host transaldolase.
Comparative Kinetics of Phage and Host Transaldolases.
The cyanophage transaldolase (TalC) differs significantly in sequence from the host transaldolase (TalB), which led us to question not only why phages encode transaldolase, but why they encode one so different from the host enzyme. Multiple sequence alignments reveal that TalC has a number of deletions relative to TalB (Fig. S3A) but no change in the active site residues essential for catalysis (Fig. S3A) (31, 32). TalC more closely resembles the Escherichia coli fructose-6-phosphate (F6P) aldolase, FsaA (33), than host TalB (Fig. S3 A and B). Therefore, we sought to confirm that phage TalCs have transaldolase activity, not F6P aldolase activity. We then compared their kinetic properties to those of their corresponding host TalBs, as their structural differences might be accompanied by kinetic differences. We anticipated that phage transaldolase might possess kinetic advantages relative to host transaldolase and thus augment the host's PPP during infection.
Purification and assay.
As a first step in TalC and TalB characterization, the genes from three phages and their corresponding hosts were cloned and the proteins expressed and purified to approximately 90% homogeneity (Fig. S4A). Many different vectors and expression systems were investigated before settling on N- or C-terminal 6×His-tagged constructs (Tables S3 and S4) to facilitate purification and a cold-adapted expression strain wherein small amounts of soluble protein could be produced (SI Methods). The first task was to establish whether TalC is a transaldolase or a F6P aldolase. As shown in Fig. 1A and 1, transaldolase is central to the PPP and catalyzes the reversible, three-carbon transfer of dihydroxyacetone (DHA) from F6P (C1-3 of F6P) to erythrose 4-phosphate (E4P) to generate sedoheptulose 7-phosphate (S7P) and glyceraldehyde 3-phosphate (GAP). In contrast, F6P aldolase catalyzes the conversion of F6P to GAP and DHA. Thus, for transaldolase activity, both F6P and E4P are required, whereas for F6P aldolase, only the former is required (33).
The assay for transaldolase activity (34) involves two coupling enzymes: triosephosphate isomerase (TPI) and glycerol-3-phosphate dehydrogenase (G3PDH). TPI converts GAP (as shown in 1) to DHA phosphate, which is then reduced to glycerol 3-phosphate by G3PDH using an NADH cofactor, whose oxidation is monitored spectrophotometrically by a decrease in A340. The results of these experiments (Fig. S4B) reveal that all three TalCs and all three TalBs required E4P for turnover and are thus transaldolases. This result is consistent with a role for TalC in the host PPP (Fig. 1A), with 1 proceeding from right to left.
Structures of TalC and TalB.
Further comparison of host and phage enzymes has been possible by structural determination of Prochlorococcus MIT9312 TalB (1.90-Å resolution by X-ray crystallography; Table S5) and homology modeling of cyanophage P-SSP7 TalC using sequence alignments and the Thermotoga maritima TalC structure [Protein Data Bank (PDB) ID no. 1VPX; 41% identity]. An overlay of the two structures (SI Methods) reveals a superimposable α8/β8-barrel including the conserved active site residues (Fig. S3C) previously demonstrated to be important in catalysis (31, 32). The major distinguishing feature between these transaldolases is the number and orientation of exterior α-helices, with TalB containing more external α-helices relative to TalC's more compact structure. To reveal the quaternary structures of TalB and TalC, the same two representatives (MIT9312 TalB and P-SSP7 TalC) were examined by size-exclusion chromatography (Fig. S3D). Comparison of the elution profiles with a standard curve generated from globular proteins of known molecular weight revealed that TalB appears to form a monomer and TalC a pentamer. Most other TalBs are reported to be dimers in solution (35), but it is interesting to note that a monomeric state is also observed in our crystal structure. Most other TalCs form decamers in solution (35), consisting of a dimer of doughnut-shaped pentamers (31). Thus, both host and phage transaldolases have unusual quaternary structures.
Kinetic properties of TalC and TalB.
Finally, to assess the potential of phage transaldolase to substitute for or even outperform the host transaldolase, we compared the kinetic parameters of the three phage TalCs and three host TalBs by using the coupled assay. The results are summarized in Table 1. In general, the kcat and kcat / Km values for host transaldolases are approximately threefold higher than for phage transaldolases. Our hypothesis was that the TalCs might have higher efficiencies than the TalBs. However, in all three phage–host pairs of TalC and TalB, this was shown not to be the case. We consider three explanations for why a phage would carry a kinetically less efficient enzyme than its host. First, for phage, the acquisition and use of host-like metabolic genes must always involve fitness tradeoffs: the gene may contribute to a more successful infection, but it also increases the size of the genome that must be replicated (11). Larger gene size could reduce phage fitness, as phage genome size is partially limited by capsid size (36). The decreased length of the phage talC gene (average, 659 bp) relative to the host talB gene (average, 1,098 bp) may be a result of this tradeoff. Supporting this argument, many of the phage metabolic genes with host orthologues are shorter than their host orthologues (Fig. 3 and Table S6). More specifically, comparing phage and host gene sizes separately for each of 24 shared orthologues (SI Methods), 14 orthologues were shorter in phages than in hosts, one orthologue was longer in phages than in hosts, and nine orthologues were not statistically different in size (Fig. 3 and Table S6). This is consistent with the general trend that phage genes are, on average, shorter than host genes (37, 38), but this pairwise comparison between phage and host genes with the same functions is more directly relevant to the evolutionary tradeoffs in encoding a particular gene function. Second, phage transaldolase gene dosage may exceed that of the host and compensate for a less active enzyme. Many copies of the phage genome are synthesized during the latent period of infection, each encoding and potentially expressing transaldolase. Third, as we have already mentioned, changes in host TalB levels over the light–dark cycle could render the host PPP less active during the day, and a separate phage TalC could potentially circumvent this regulation, regardless of its activity. Unfortunately, the genetic tools required to address the effects of differing enzyme properties in vivo are not yet available for these phages and hosts. Whatever the underlying pressures, phage transaldolase is functional and has been conserved across a wide range of cyanophages, consistent with a key role in host metabolism under infection.
Table 1.
Comparison of kinetic parameters of three TalBs and three TalCs
|
Km, mM |
kcat/Km, s−1 mM−1 |
||||
| TalB/TalC | kcat, s−1 | Fructose 6-P | Erythrose 4-P | Fructose 6-P | Erythrose 4-P |
| Prochlorococcus NATL2A TalB | 15.2 ± 0.7 | 1.1 ± 0.2 | 0.11 ± 0.02 | 13.6 ± 2.5 | 134 ± 29 |
| Prochlorococcus MED4 TalB | 14.9 ± 0.4 | 1.5 ± 0.2 | 0.15 ± 0.02 | 9.8 ± 1.2 | 103 ± 14 |
| Prochlorococcus MIT9312 TalB | 20.8 ± 1.3 | 1.0 ± 0.1 | 0.10 ± 0.04 | 20.8 ± 2.3 | 206 ± 75 |
| Cyanophage P-SSM2 TalC | 3.6 ± 0.3 | 0.7 ± 0.1 | 0.08 ± 0.01 | 5.4 ± 0.7 | 48 ± 9 |
| Cyanophage P-SSM4 TalC | 5.6 ± 0.4 | 1.3 ± 0.2 | 0.20 ± 0.05 | 4.5 ± 0.9 | 29 ± 7 |
| Cyanophage P-SSP7 TalC | 5.9 ± 0.2 | 1.6 ± 0.2 | 0.07 ± 0.01 | 3.6 ± 0.5 | 86 ± 18 |
Kinetic parameters (kcat, Km, and kcat / Km) were measured at 25 °C in 50 mM Gly-Gly (pH 8.0), 15 mM MgCl2, 200 μM NADH, 0.6 U TPI, 0.06 U G3PDH, 0.1–10.0 mM F6P, and 0.01–1.00 mM E4P. Prochlorococcus TalB assays also contained 10 mM DTT.
Fig. 3.
Comparison of phage and host gene sizes for shared orthologues. For each gene orthologue found in at least three cyanophage genomes and at least three Prochlorococcus or Synechococcus genomes, the median gene size across all phages and across all hosts is plotted. Bars represent the range of sizes observed. The line shows a 1:1 ratio, whereby phage and host gene sizes are identical. Median gene sizes for each host–phage pair were compared by using the Mann–Whitney–Wilcoxon test (P < 0.05) with correction for false discovery rate (SI Methods). Blue circles indicate genes that were shorter in phages than in hosts, red circles those that were shorter in hosts than in phages, and unfilled circles those that were statistically the same size in phages and hosts.
Proposed Model of Cyanobacterial Metabolism Under Phage Infection.
Expression of phage genes for the PPP, the light reactions, and inhibition of the Calvin cycle early in the infection process—combined with evidence that at least one of the gene products (transaldolase) is functional in vitro—implies that these metabolic pathways are augmented in infected cells (Fig. 1A). We hypothesize that, when a cyanophage infects a host cell, the Calvin cycle is down-regulated by phage-encoded CP12, preventing energy and reducing power produced by the phage-augmented light reactions from being used to fix carbon dioxide, such that phage infection decouples the light reactions from the Calvin cycle. Thus, two pathways that under the light–dark cycle are normally offset by 12 h—the light reactions and the PPP—potentially occur simultaneously in the host cell. We propose that the combined ATP and NADPH from these processes (along with ribose 5-P from the PPP) fuels phage dNTP biosynthesis, particularly via the phage-encoded RNR, which requires NADPH as the terminal electron donor to reduce NDPs to dNDPs.
The hypothesis that cyanophage need to boost dNTP synthesis because dNTPs are limiting during infection is supported by differences in phage and host molecular composition and by host physiology. As described in Table S7, the protein/DNA ratio of a cyanobacterial host cell is 20 times the protein/DNA ratio of a T4 phage; therefore, the metabolic fluxes required for phage reproduction relative to host reproduction are biased toward DNA synthesis. However, the host chromosome (2 Mbp), even if completely digested, would be insufficient to supply all of the dNTPs needed by progeny phage: assuming an average burst size of 100 phage cell−1 [range of 40–150 phage cell−1 (39, 40)] and a phage genome size of 200 kbp phage−1 (Table S1), the host chromosome could supply only 10% of the dNTPs in the progeny phage. Phytoplankton living in low-nutrient environments are thought to have limited dNTP pools (10, 41). Indeed, for many marine viruses, the majority of phage dNTPs are derived from the host chromosome (41) and host genome size limits burst size in dNTP equivalents (10). Modeling of the phage infection process (11) suggests that phage infecting hosts with small genomes like Prochlorococcus are more likely to encode auxiliary metabolic genes for dNTP synthesis. Cyanophages are able to yield significantly higher burst sizes than predicted by their host genomes (10), possibly resulting from an ability to help the host produce large quantities of dNTPs de novo.
Dynamics of NADPH and Its Derivatives.
In our model of phage-augmented nucleotide biosynthesis, the shared Calvin cycle/PPP reactions are proposed to run in the direction of the PPP (clockwise in Fig. 1A) such that ribose 5-P provides the sugars for new dNTPs. However, much of the pentose phosphate produced is recycled back to glucose 6-P for additional NADPH production. Meanwhile, the light reactions of photosynthesis produce ATP and additional NADPH for making dNTPs. Our model therefore makes the following predictions: first, phage-infected cells produce more NADPH than uninfected cells; second, light is required for optimal NADPH production under infection; and third, phage-encoded proteins are partially responsible for this increase in NADPH production. To test these predictions, we grew Prochlorococcus MED4 cultures in constant light, inoculated with cyanophage P-HM2 or spent medium, and returned to constant light or shifted to darkness. At regular intervals following inoculation, we measured ratios of NADPH and its derivatives (NADP, NADH, NAD) by using an enzymatic assay (23), and we measured gene expression and phage replication dynamics by quantitative PCR (qPCR). Additionally, to better understand what phage in the wild might encounter when infecting cells during the day or night, we synchronized Prochlorococcus MED4 on a 24-h light–dark cycle, took samples at regular intervals over a period of 48 h, and measured ratios of NADPH and its derivatives by using the enzymatic assay. We now describe the results of these experiments and compare them with those predicted by our model.
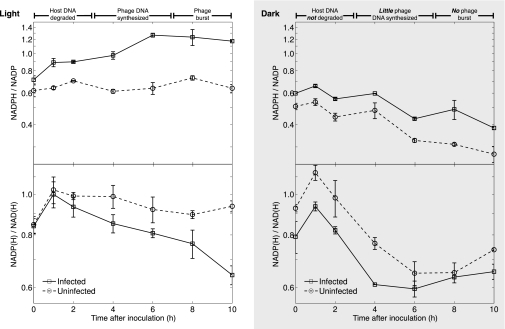
First, our model predicts that, in order for infected cells to increase de novo production of dNTPs, they require increased NADPH, which provides reducing equivalents for RNR (42). The NADPH/NADP ratio should therefore increase in infected cells, as phage shift metabolism to provide more NADPH for nucleotide reduction. In accordance with our prediction, the NADP(H) pool became more reduced during infection in the light relative to uninfected cells, with the NADPH/NADP ratio increasing from approximately 0.7 to approximately 1.3 over the first 6 h and then remaining unchanged (Fig. 4). The elevated NADPH/NADP ratio indicates an even greater increase in NADPH production than apparent from the ratio, as the added demand for dNTPs under infection requires increased NADPH consumption.
Fig. 4.
Dynamics of the intracellular NADPH/NADP ratio (Top) and NADP(H)/NAD(H) ratio (Bottom) during infection of Prochlorococcus MED4 by cyanophage P-HM2 in the light or in the dark. Error bars represent SDs of two technical and two biological replicates.
Second, as a major source of NADPH in cyanobacteria is the light reactions of photosynthesis, which are required for maximal phage production (7, 12), we predicted that the increase in NADPH/NADP ratio would be dampened or nullified under infection in the dark, accompanied by decreased phage production and gene expression. Indeed, the NADPH/NADP ratio decreased in infected and uninfected cultures in the dark (Fig. 4). The decrease in NADPH/NADP ratio in infected cultures in the dark coincided with stunted phage genomic DNA (gDNA) replication and host gDNA degradation in the dark (Fig. S2B) relative to infection in the light (Fig. S2A). We also observed more than 100-fold lower phage gene expression (on average for cp12, talC, psbA, nrdA/nrdB) in the dark (Fig. S2D) than in the light (Fig. S2C). Our results show that phage infecting in the dark are unable to degrade the host chromosome, transcribe their genes fully, or make progeny phage. The oxidizing effect of darkness on the NADPH/NADP ratio may help explain why photosynthesis is necessary for optimal phage production: without the photosynthetic light reactions to generate NADPH, glucose stores could quickly become exhausted, leaving no source of NADPH to fuel phage production.
To assess the applicability of these results to wild cyanophage populations, we measured ratios of NADPH and its derivatives in uninfected cultures synchronized on a 24-h light–dark cycle, which simulates the light conditions of the surface ocean. In our measurements over two 24-h light–dark cycles, uninfected cells were more reduced in the light and more oxidized in the dark (Fig. S5). Interestingly, the change was observed in the NADH/NAD ratio rather than in the NADPH/NADP ratio, and thus further work is needed to fully explain these redox dynamics. Nevertheless, it is clear that darkness causes Prochlorococcus to become more oxidized, and this may be an important factor limiting phage replication.
Finally, because phage auxiliary metabolic genes are conserved in the phage and have roles in the PPP and its production of NADPH, the encoded proteins are predicted to be directly involved in NADPH production. A corollary of this prediction is that intracellular conditions under infection should be compatible with high activity of the phage-encoded enzymes and regulatory proteins. This is particularly relevant to CP12, which not only affects NADPH levels via its action on the Calvin cycle, but is itself regulated by levels of NADPH and its derivatives. Although the detailed regulatory mechanisms in cyanophage have not yet been determined, in plants and cyanobacteria, the complex of CP12 with PRK and GAPDH (Fig. 1A) is stabilized by NAD or NADH [i.e., NAD(H)] and destabilized by NADP or NADPH [i.e., NADP(H)] (22, 23, 43). [The ratio NADP(H)/NAD(H) therefore refers to the phosphorylation state of the pool comprising NADPH and its derivatives, whereas the ratios NADPH/NADP and NADH/NAD refer to the redox state of this pool.] Thus, we examined not only the ratio of NADPH/NADP but also the ratio of NADP(H)/NAD(H).
Based on our understanding of CP12 regulation in other systems (22, 23, 43), for phage CP12 to be active during infection, NAD(H) is predicted to increase relative to NADP(H) in infected cells, such that the NADP(H)/NAD(H) ratio would decrease. The NADP(H)/NAD(H) ratio is predicted to decrease likewise during nighttime of the day–night cycle of uninfected cells, as host CP12 is putatively active at night. As shown in Fig. 4, infected cells in the light indeed showed a decrease in NADP(H)/NAD(H), which was also seen in both infected and uninfected cells in the dark. Similarly, during the dark portion of the light–dark cycle, uninfected cells displayed a decrease in their NADP(H)/NAD(H) ratio (Fig. S5). Phage infection in the light therefore appears to mimic the effect of darkness on light–dark-synchronized Prochlorococcus, leading to conditions that favor CP12 activity [low NADP(H), high NAD(H)] and therefore dark metabolism, whereby the PPP is activated and the Calvin cycle is deactivated, resulting in NADPH production.
Further Considerations and Conclusions.
The evidence presented here supports our hypothesis that cyanophage gene products redirect host metabolism to increase dNTP biosynthesis. Demonstrating the causal role of phage auxiliary metabolic genes in this process, however, will require genetic tools as yet unavailable for this host–phage system. Even when these tools are in hand, however, multipronged approaches like the one described here will be essential for developing a complete understanding of the forces that shape the coevolution of host and phage in the oceans.
Our analysis has focused on the need for NADPH to fuel dNTP biosynthesis for phage replication, but clearly, additional raw materials for making dNTPs could be limiting, such as ATP, carbon skeletons, nitrogen for purine and pyrimidine bases, and phosphate linkages. There appear to be cyanophage adaptations for many of these other dNTP inputs. ATP is needed at various steps in dNTP biosynthesis and could be produced via the photosynthetic electron transport chain, assisted by well documented phage-encoded gene products for photosystem II along with more recently discovered phage proteins for photosystem I (44) and NAD(P)H dehydrogenase (45). Ribose 5-P, the sugar skeleton for both purine and pyrimidine deoxynucleotides, is also produced by the PPP, and phage augmentation of this pathway as argued here likely produces ribose 5-P in parallel with NADPH. Nitrogen is needed for both purine and pyrimidine bases, and phage-induced nitrogen stress is suggested by the presence of multiple NtcA promoters in T4-like cyanophages, with phage possibly exploiting the host N-stress response system to drive gene expression (21). Finally, phage carry genes involved in host phosphate acquisition, which has clear implications for dNTP production in P-limited oceanic regions (21).
A phage reproductive strategy reflects selective pressures on both phage and host. As viewed by the genes phage carry, this strategy is the result of opposing selective forces: the cost of maintaining a gene in a size-limited genome versus the benefit of encoding a novel metabolic function. However, this strategy also reflects the physiology and environment of the host—in this case a photoautotroph living in low-nutrient waters. The fact that all three cyanophage types carry genes for the PPP despite having evolved from completely separate phage lineages (46) points to the pressures brought on by their common host type and its austere habitat. This remarkable example of convergent evolution in phage demonstrates the unifying pressures of infecting an obligate photoautotrophic host with a very small genome, a host that alone may not be able to supply the DNA building blocks a phage needs to replicate effectively.
Methods
Sequences and Gene Annotation.
Twenty-four genomes from cyanophages infecting Prochlorococcus and marine Synechococcus were included in the analyses (Table S1), three of which are being introduced in the present study. We searched the 24 cyanophage genomes for each Calvin cycle and PPP gene, along with known cyanophage genes in photosynthetic electron transport and nucleotide biosynthesis, using the genome annotations. To check for the possibility of uncalled or miscalled genes in the annotations, we also searched the 24 genomes by using TBLASTN with default parameters, an E-value cutoff of 10-5, and using all Prochlorococcus or Synechococcus Calvin cycle and PPP genes as queries. In cases of positive hits for genes not reported in the annotations, multiple sequence alignments were used to confirm the presence of key conserved residues. Hydrophobicity plots were used to compare hydrophobicity patterns of CP12 sequences. More details are provided in SI Methods.
Metagenomic Analyses.
Sequence reads from the GOS database (27) were recruited to cyanophage gene clusters from 17 T4-like cyanophage genomes defined previously (21). Reads were kept only if the top five BLAST hits were to the same gene cluster and only if the read did not have a better hit when blasted against an exhaustive set of marine genomes and the GenBank database. To test whether the size (i.e., number of orthologues) of a gene cluster adversely affected the number of sequence reads that could be recruited, we randomly excluded different orthologues from talC, cp12, gnd, and zwf gene clusters and repeated the recruitment; we found that as few as one orthologue of each gene was sufficient to recruit most of the reads recruited by the full gene cluster. Additional details are provided in SI Methods.
Infection of Synechococcus WH8109 by Cyanophage Syn9.
Choice of strains was based on the fact that cyanophage Syn9 carries all four of the PPP genes of interest (cp12, talC, zwf, gnd; Table S1), and it readily infects Synechococcus WH8109, which has a sequenced genome. Log-phase Synechococcus WH8109 in the light was inoculated with cyanophage Syn9 at a multiplicity of infection (MOI) of 3. Three infected and three uninfected replicates were maintained. Uninfected controls were given spent medium instead of phage lysate. Samples were taken at regular intervals to be analyzed for gDNA and RNA. For phage and host gDNA quantification, samples were filtered with 0.2-μm polycarbonate filters, and the filtrate was used for extracellular phage gDNA quantification, whereas the retentate was used for intracellular phage gDNA quantitation and host gDNA quantification. For RNA, Synechococcus culture was harvested by centrifugation, flash-frozen in liquid nitrogen, and stored at −80 °C. Detailed descriptions of materials and methods are provided in SI Methods.
Infection of Prochlorococcus MED4 by Cyanophage P-HM2.
Choice of strains was based on the fact that the transcriptional (24) and proteomic (29) dynamics of MED4 grown on a light–dark cycle have been studied extensively, and cyanophage P-HM2 was isolated on MED4 and carries the most prevalent auxiliary metabolic genes (cp12, talC, psbA; Table S1). Procedures were identical to those described above except that the MOI was 1, duplicate experiments were done in which bottles were moved to the light or the dark after inoculation (two infected and two uninfected replicates for each treatment), and culture aliquots were centrifuged for pyridine nucleotide extraction. Detailed descriptions of materials and methods are provided in SI Methods.
qPCR and RT-PCR.
qPCR primers (Table S2) were designed from the genomes of cyanophages P-HM2 and Syn9, Prochlorococcus MED4, and Synechococcus WH8109, and were tested by using gDNA and shown to have specific and concentration-dependent amplification of target DNA. For mRNA quantification, RNA was extracted from lysed Prochlorococcus or Synechococcus cell pellets, gDNA degraded with DNase I, and first-strand cDNA synthesized from the RNA. For both gDNA and mRNA quantitation, copies were measured by using the QuantiTect SYBR Green PCR Kit (Qiagen) with a LightCycler 480 Real-Time PCR System (Roche Diagnostics). Relative copy numbers of each cDNA over the time course were determined by the ΔΔCT method, with rnpB (RNA component of ribonuclease P) as the internal calibrator gene. Detailed descriptions of materials and methods are provided in SI Methods.
Cloning, Expression, and Purification of Transaldolases.
Transaldolases from Prochlorococcus and cyanophages were PCR-amplified, cloned into pET vectors (Tables S3 and S4) encoding N- or C-terminal 6×His-tags, and transformed into BL21 (DE3) expression strains. Large-scale cultures of these strains were induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated for 15 to 35 h at 25 °C or 13 °C. The cells were harvested and lysed by French press or sonication. The recombinant proteins were then purified to homogeneity by Ni-NTA affinity chromatography (elution with imidazole) and dialyzed into 50 mM Gly-Gly. Detailed descriptions of materials and methods are provided in SI Methods.
Transaldolase Assay and Determination of Kinetic Parameters.
Transaldolase activity was measured by using a coupled assay as described previously (34) and in SI Methods. Enzymes and reagents were from Sigma-Aldrich. Kinetic data were fitted to the Michaelis–Menten equation to determine kinetic parameters as described in SI Methods.
Crystal Structure Determination.
Prochlorococcus MIT9312 TalB was crystallized and its structure solved by X-ray crystallography with molecular replacement as described in SI Methods. The atomic coordinates and structure factors for Prochlorococcus MIT9312 TalB have been deposited in the PDB (accession no. 3HJZ).
Structure Homology Modeling and Alignment.
A homology model was built for cyanophage P-SSP7 TalC using the structure of T. maritima TalC (PDB accession no. 1VPX) with SWISS-MODEL software, and this structure was overlaid with the structure of Prochlorococcus MIT9312 TalB by using UCSF Chimera software, both as described in SI Methods.
SEC Determination of Transaldolase Quaternary Structure.
SEC was performed with a Superose 12 column (10 × 300 mm; GE Healthcare) attached to a Waters 2487 HPLC detector. Gel filtration molecular weight standards were run at the beginning of each experiment and used to determine the molecular weights of Prochlorococcus MIT9312 TalB and cyanophage P-SSP7 TalC as described in SI Methods.
Measurement of Pyridine Nucleotides in Prochlorococcus.
Pyridine nucleotides NAD, NADH, NADP, and NADPH were extracted fresh from Prochlorococcus cultures and measured enzymatically as described previously (23) and in SI Methods.
Supplementary Material
Acknowledgments
The authors thank Jacob Waldbauer and Erik Zinser for valuable discussions on carbon metabolism and its regulation; Matthew Sullivan for providing phages for genome sequencing; Debbie Lindell for providing Synechococcus WH8109 gene sequences; Brianne Holmbeck and Sara Roggensack for assistance in maintaining cultures; Hong Cui and Xiaohui Xu for help in crystallization and data collection; Allison Ortigosa, Daniela Hristova, Mohammad Seyedsayamdost, Joseph Cotruvo, Kenichi Yokoyama, Sumit Chakraborty, Alexa Price-Whelan, Arne Materna, Katya Frois-Moniz, Sébastien Rodrigue, Kolea Zimmerman, Daniel Sher, and Nadav Kashtan for advice on technical issues; James Calvin for statistical assistance; Marcia Osburne for comments on the manuscript; and Jeffrey Palm for assistance in manuscript preparation. This research was funded in part by the Gordon and Betty Moore Foundation (GBMF) through a grant to the Broad Institute. Samples G1170, G1171, and G1172 were sequenced and assembled at the Broad Institute. This study was supported by GBMF, the Department of Energy (Genomics:GTL Program), the National Science Foundation (Center for Microbial Oceanography: Research and Education), and a National Institutes of Health Training Grant (to L.R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HJZ). The sequences reported in this paper have been deposited in the GenBank database [GU071107 (P-SSP2), GU071104 (P-HP1), and GU071102 (P-RSP5)].
See Author Summary on page 16147.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102164108/-/DCSupplemental.
References
- 1.De Waard A, Paul AV, Lehman IR. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci USA. 1965;54:1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller ES, et al. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 4.Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart M, Thompson LR, Suttle CA, Sullivan MB. Exploring the vast diversity of marine viruses. Oceanography (Wash DC) 2007;20:135–139. [Google Scholar]
- 7.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 8.Clokie MRJ, et al. Transcription of a ‘photosynthetic’ T4-type phage during infection of a marine cyanobacterium. Environ Microbiol. 2006;8:827–835. doi: 10.1111/j.1462-2920.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- 9.Lindell D, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 10.Brown C, Lawrence J, Campbell D. Are phytoplankton population density maxima predictable through analysis of host and viral genomic DNA content? J Mar Biol Assoc UK. 2006;86:491–498. [Google Scholar]
- 11.Bragg JG, Chisholm SW. Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS ONE. 2008;3:e3550. doi: 10.1371/journal.pone.0003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackenzie JJ, Haselkorn R. Photosynthesis and the development of blue-green algal virus SM-1. Virology. 1972;49:517–521. doi: 10.1016/0042-6822(72)90503-x. [DOI] [PubMed] [Google Scholar]
- 13.Sherman LA. Infection of Synechococcus cedrorum by the cyanophage AS-1M. III. Cellular metabolism and phage development. Virology. 1976;71:199–206. doi: 10.1016/0042-6822(76)90105-7. [DOI] [PubMed] [Google Scholar]
- 14.Kao CC, Green S, Stein B, Golden SS. Diel infection of a cyanobacterium by a contractile bacteriophage. Appl Environ Microbiol. 2005;71:4276–4279. doi: 10.1128/AEM.71.8.4276-4279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Lu J. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol. 2002;68:2589–2594. doi: 10.1128/AEM.68.5.2589-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann NH, et al. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol. 2005;187:3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pope WH, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J Mol Biol. 2007;368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigele PR, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- 19.Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol. 2009;11:2370–2387. doi: 10.1111/j.1462-2920.2009.01966.x. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan MB, et al. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome’. Environ Microbiol. 2009;11:2935–2951. doi: 10.1111/j.1462-2920.2009.02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan MB, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–3056. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 24.Zinser ER, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE. 2009;4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groben R, et al. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynth Res. 2010;103:183–194. doi: 10.1007/s11120-010-9542-z. [DOI] [PubMed] [Google Scholar]
- 26.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 27.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharon I, et al. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 2011;5:1178–1190. doi: 10.1038/ismej.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldbauer JR. Molecular biogeochemistry of modern and ancient marine microbes. 2009 PhD Thesis (Massachusetts Institute of Technology and Woods Hole Oceanographic Institution, Woods Hole, MA) [Google Scholar]
- 30.Bräutigam AR, Sauerbier W. Transcription unit mapping in bacteriophage T7. I. In vivo transcription by Escherichia coli RNA polymerase. J Virol. 1973;12:882–886. doi: 10.1128/jvi.12.4.882-886.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorell S, Schürmann M, Sprenger GA, Schneider G. Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family. J Mol Biol. 2002;319:161–171. doi: 10.1016/S0022-2836(02)00258-9. [DOI] [PubMed] [Google Scholar]
- 32.Schneider S, Sandalova T, Schneider G, Sprenger GA, Samland AK. Replacement of a phenylalanine by a tyrosine in the active site confers fructose-6-phosphate aldolase activity to the transaldolase of Escherichia coli and human origin. J Biol Chem. 2008;283:30064–30072. doi: 10.1074/jbc.M803184200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schürmann M, Sprenger GA. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J Biol Chem. 2001;276:11055–11061. doi: 10.1074/jbc.M008061200. [DOI] [PubMed] [Google Scholar]
- 34.Bergmeyer HU, Gawehn K, Grassl M. Enzymes as biochemical reagents. In: Bergmeyer HU, editor. Methods of Enzymatic Analysis. Vol. 1. New York: Academic; 1974. pp. 425–522. [Google Scholar]
- 35.Samland AK, Sprenger GA. Transaldolase: From biochemistry to human disease. Int J Biochem Cell Biol. 2009;41:1482–1494. doi: 10.1016/j.biocel.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Russell PW, Müller UR. Construction of bacteriophage φX174 mutants with maximum genome sizes. J Virol. 1984;52:822–827. doi: 10.1128/jvi.52.3.822-827.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath S, van Sinderen D. Bacteriophage: Genetics and Molecular Biology. Norfolk, UK: Caister; 2007. [Google Scholar]
- 38.Hatfull GF, et al. Comparative genomic analysis of 60 Mycobacteriophage genomes: Genome clustering, gene acquisition, and gene size. J Mol Biol. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson W, Carr N, Mann N. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH 7803. J Phycol. 1996;32:506–516. [Google Scholar]
- 40.Thompson LR. Auxiliary metabolic genes in viruses infecting marine cyanobacteria. 2010 PhD Thesis (Massachusetts Institute of Technology, Cambridge, MA) [Google Scholar]
- 41.Wikner J, Vallino J, Steward G, Smith DC, Azam F. Nucleic acids from the host bacterium as a major source of nucleotides for three marine bacteriophages. FEMS Microbiol Ecol. 1993;12:237–248. [Google Scholar]
- 42.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 43.Wedel N, Soll J. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc Natl Acad Sci USA. 1998;95:9699–9704. doi: 10.1073/pnas.95.16.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharon I, et al. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alperovitch-Lavy A, et al. Reconstructing a puzzle: Existence of cyanophages containing both photosystem-I and photosystem-II gene suites inferred from oceanic metagenomic datasets. Environ Microbiol. 2010;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]
- 46.Glazko G, Makarenkov V, Liu J, Mushegian A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol Direct. 2007;2:36. doi: 10.1186/1745-6150-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]