Abstract
Mounting evidence suggests that PERIOD (PER) proteins play a central role in setting the speed (period) and phase of the circadian clock. Pharmacological and genetic studies have shown that changes in PER phosphorylation kinetics are associated with changes in circadian rhythm period and phase, which can lead to sleep disorders such as Familial Advanced Sleep Phase Syndrome in humans. We and others have shown that casein kinase 1δ and ε (CK1δ/ε) are essential PER kinases, but it is clear that additional, unknown mechanisms are also crucial for regulating the kinetics of PER phosphorylation. Here we report that circadian periodicity is determined primarily through PER phosphorylation kinetics set by the balance between CK1δ/ε and protein phosphatase 1 (PP1). In CK1δ/ε-deficient cells, PER phosphorylation is severely compromised and nonrhythmic, and the PER proteins are constitutively cytoplasmic. However, when PP1 is disrupted, PER phosphorylation is dramatically accelerated; the same effect is not seen when PP2A is disrupted. Our work demonstrates that the speed and rhythmicity of PER phosphorylation are controlled by the balance between CK1δ/ε and PP1, which in turn determines the period of the circadian oscillator. Thus, our findings provide clear insights into the molecular basis of how the period and phase of our daily rhythms are determined.
Keywords: dynamic regulation of phosphorylation, stoichiometry, period determination
The circadian clock controls so much of mammalian physiology that dysregulation of the clock contributes to medical conditions such as jet lag, shift work sleep disorder, metabolic diseases, and mood disorders (1, 2). In most of the body's cells, from the neurons of the suprachiasmatic nucleus (SCN) to hepatocytes, lung epithelial cells, and even fibroblasts, the clock is present and its molecular mechanism is essentially the same (3–6). The backbone of the oscillator mechanism is a transcriptional negative feedback loop driven by positive and negative elements (7–10). The positive elements are three bHLH/PAS-containing transcription factors, CLOCK, NPAS2, and BMAL1; the CLOCK (or NPAS2):BMAL1 heterodimer binds to E-box enhancer motifs and activates transcription of the negative element genes, Per1 and 2, and Cryptochrome (Cry)1 and 2. The PER:CRY complexes complete the feedback loop by inhibiting CLOCK:BMAL1.
Although all of these clock proteins are essential, PERIOD (PER) is of special importance for clock regulation. PER is the rate-limiting component for PER:CRY complex formation, and thus the timing and duration of PER nuclear entry and accumulation dictate the phase and period of the molecular clock (11–13). Of all of the core clock proteins, only PER's expression is absolutely required to oscillate for the clock to function; constitutive high expression of PER completely disrupts circadian rhythms in cells and mice (11). Phosphorylation of PER is a vital part of clock regulation, as it affects PER's nuclear translocation, interaction with other clock proteins, and timely degradation (14–18). In vivo, PER phosphorylation—detectable as a mobility shift in SDS/PAGE—occurs progressively over several hours (12, 19), which is critical for stretching the feedback loop to ∼24 h. However, PER2 can be maximally phosphorylated by CK1ε in vitro kinase reactions within 30 min (20, 21), suggesting that PER phosphorylation must be counterbalanced by phosphatases in vivo.
Because the phase and period of the clock are primarily determined by temporal regulation of PER phosphorylation (12, 15, 21–26), the characterization of PER kinases and phosphatases is vital to understanding the circadian clock mechanism. CK1δ and ε have been shown to be PER kinases and are important for clock function, but PER phosphorylation is largely intact in several CK1δ or CK1ε mutants (12, 21, 23, 27). Furthermore, PER can be phosphorylated by several other kinases in vitro, including CK1α and γ, CK2, and GSK3β (28–32). Thus, it remains unclear which kinases are responsible for the majority of mobility-shifting, progressive phosphorylation of PER in vivo. In this study, we show that CK1δ and ε are the major PER kinases and that they are largely redundant with each other, such that PER phosphorylation and the circadian clock's function can endure the loss of just one kinase. Moreover, we identify PP1 as the major PER phosphatase that counterbalances CK1δ/ε and other PER kinases in vivo.
Results
PER Phosporylation Is Severely Compromised and the Molecular Clock Is Not Functional in CK1δ/ε-Deficient Fibroblasts.
We previously provided evidence for the importance of CK1δ/ε for clock function by combining CK1δ gene deficiency with dominant negative CK1ε expression (21). However, the dominant negative CK1ε may have interfered not only with wild-type CK1ε (wtCK1ε) activity, but also with other isoforms of CK1 as suggested by Hirota et al. (29). To definitively test how CK1δ/ε contribute to temporal PER phosphorylation in vivo, we assessed circadian rhythms and the molecular clock in mouse fibroblasts deficient in both CK1δ and ε. Because CK1δ deletion results in embryonic lethality, we derived the fibroblasts from mice with floxed CK1δ/ε genes, so that the genes could be deleted conditionally using the Cre-loxP recombination system (27). Cre recombinase was expressed using an adenoviral vector (11). Neither adenoviral infection nor Cre expression affected circadian rhythms in WT cells (Fig. S1). In the double-floxed mutant cells, CK1δ and ε were both deleted in almost 100% of cells within 4 d after the adenoviral cre infection, as confirmed by genotyping PCR (Fig. S2) and immunoblots (Fig. 1A).
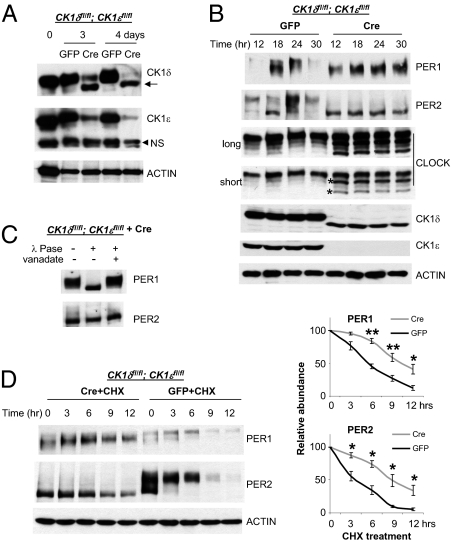
Fig. 1.
PER1/2 do not oscillate, and their phosphorylation is severely compromised in CK1 double-mutant (CK1δΔ2/Δ2:CK1ε−/−) fibroblasts. (A) CK1δ and ε were successfully deleted by introducing adenoviral cre into the double-floxed mutant cells. Cells were harvested 3 and 4 d after adenoviral cre or gfp infection, and then subjected to immunoblotting for CK1δ/ε and ACTIN. Note that a small band—indicated by the arrow and representing a deletion mutant, inactive form of CK1δ—is present, as reported by Etchegaray et al. (27). The arrowhead indicates a nonspecific band (NS). (B) PER does not oscillate in CK1δ/ε double-mutant cells. Cells were infected with adenoviral cre or gfp and harvested 5–6 d later at the indicated times after a 2-h serum shock. It is shown that two phosphorylated CLOCK isoforms are derived from two nonphosphorylated isoforms (indicated by asterisks) in fibroblasts as previously reported in liver (13). (C) PER1 is still phosphorylated, but PER2 is not apparently phosphorylated in the double-mutant cells. Immunoprecipitated PER1/2 from the double-mutant cells were subjected to λ-phosphatase (λ-Pase) reaction to reveal any mobility shift due to phosphorylation, as done previously (12). Vanadate, a phosphatase inhibitor, prevents the change in PER1 mobility. (D) PER1/2 are more stable in CK1δ/ε-deficient cells. The control and mutant cells were treated with CHX and harvested at the indicated times for immunoblotting assays. (Right) Results quantified by densitometric scanning. Values shown are mean ± SEM from four independent experiments. P values were calculated by Student t test between two groups. *P < 0.05; **P < 0.01.
When the double-floxed mutant cells were infected with adenoviral gfp, PER phosphorylation remained rhythmic and was followed by degradation (Fig. 1B, Left four lanes), similar to WT cells (13, 21). However, when CK1δ/ε were disrupted by adenoviral cre, PER rhythms in abundance and phosphorylation were completely abolished (Fig. 1B, Right four lanes). Furthermore, phosphorylation levels were dramatically reduced, with PER2 showing no mobility shift, whereas PER1 exhibited only half the mobility shift of PER1 in control cells (Fig. 1 B and C; see Fig. 2B for side-by-side comparison). PER1 protein levels in CK1δ/ε-null cells were similar to peak levels in control cells, whereas PER2 levels were somewhat lower than peak levels. Considering the low Per mRNA levels in the double-mutant cells (Fig. S3), these protein levels suggest that PER1 and PER2 are more stable in the mutant cells.
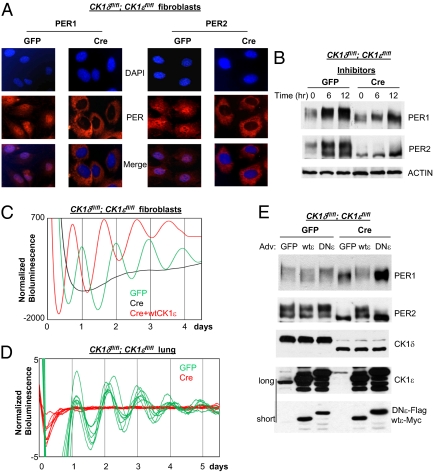
Fig. 2.
PER1/2 are predominantly cytoplasmic, and circadian rhythms are completely disrupted in CK1δ/ε-deficient cells. (A) PER1/2 fail to accumulate in the nucleus in the double-mutant cells. The double-floxed mutant cells were infected with adenoviral cre or gfp, given a 2-h serum shock 5 d later, and immunostained for PER1/2 at 24 h after the serum shock. More cells are shown in Fig. S4 A and B. (B) Lack of hyperphosphorylated species of PER is not due to reduced stability of these species in the mutant cells. PER levels are increased in both control (GFP) and mutant (Cre) cells by proteasome inhibitor mixture treatment. Cells were harvested at the indicated times after the drug treatment. Blots are representative of several experiments. (C) Circadian rhythms are disrupted in the mutant cells and tissue but can be readily rescued by exogenous expression of wtCK1ε. Bioluminescence rhythms were measured by infecting the adenoviral Per2Luc reporter into control and mutant cells. To restore wtCK1ε expression in the mutant cells, adenoviral wtCK1ε was infected into the mutant cells at the same time as the reporter construct. GFP (n = 5): period (hr) = 23.3 ± 0.21; amplitude = 361 ± 65. CRE + wtCK1ε (n = 5): period = 22.6 ± 0.12; amplitude = 512 ± 65. Values presented are mean ± SEM from three experiments. (D) Floxed CK1δ/ε genes were deleted in lung from the double-floxed mutant mice by adenoviral cre, and bioluminescence rhythms were measured using the reporter virus. Adenovirus can effectively infect lung tissue as shown in Fig. S8. (E) PER phosphorylation is also rescued in the mutant cells by exogenous expression of wtCK1ε, but not by a dominant negative mutant CK1ε (DNCK1ε). Adenoviral gfp, wtCK1ε, or DNCK1ε was infected into control and mutant cells. Two exposures (long and short) are shown for the CK1ε immunoblot.
Our findings are further supported when degradation rates were measured after the cells were treated with cycloheximide. Both PER1 and PER2 were significantly more stable in the mutant cells compared with control cells (Fig. 1D). Nonphosphorylated CLOCK was also more pronounced in the mutant cells (Fig. 1B), which suggests that CLOCK is also a substrate of CK1δ/ε, likely via CK1δ/ε:PER complexes. This finding is reminiscent of what has been found in Neurospora and Drosophila (33–36).
Deletion of CK1δ/ε also caused PER1 and PER2 to accumulate predominantly in the cytoplasm, whereas they were mostly nuclear in control cells (Fig. 2A and Fig. S4 A and B). These results suggest that PER phosphorylation is required for nuclear translocation or accumulation, as has been reported in Drosophila (37). Our findings are consistent with data obtained from liver tissue, where hypophosphorylated PER species are cytoplasmic and not complexed with CK1δ/ε (12).
Because previous research suggested that hyperphosphorylation targets PER for proteasomal degradation (22, 28, 38), we considered the possibility that the lack of hyperphosphorylated, nuclear PER1/2 may have been due to rapid degradation as opposed to reduced phosphorylation and lack of nuclear transport. To test this possibility, we treated the cells with a mixture of proteasome inhibitors (MG132 + PSI + lactacystin). In control cells, inhibition of proteasomal degradation increased levels of hyperphosphorylated PER1 and both hypo- and hyperphosphorylated PER2 levels, consistent with previous work (Fig. 2B) (21). The same treatment in the CK1δ/ε-deficient cells did not result in the detection of hyperphosphorylated forms, demonstrating that elimination of CK1δ/ε does prevent a substantial amount of PER phosphorylation. However, the treatment induced accumulation of hypophosphorylated PER in the CK1δ/ε-deficient cells, albeit at a slower rate than in control cells (Fig. 2B), consistent with the data in Fig. 1D. Thus, though CK1δ/ε are essential for PER nuclear transport and regulate PER degradation, PER can still be targeted for proteasomal degradation in the absence of CK1δ/ε.
As loss of CK1δ/ε disrupts rhythms in PER abundance and nuclear transport, it also disrupts bioluminescence rhythms measured using a Per2 promoter-driven luciferase reporter (Fig. 2C and Fig. S4 C and D). Similar results were obtained with lung tissue explants after CK1δ/ε genes were deleted by adenoviral cre (Fig. 2D and Fig. S4E). The defects in PER phosphorylation and rhythmicity were readily rescued by exogenous expression of CK1ε in the mutant cells, indicating that the circadian defects were not due to general damage to cell physiology in the mutant cells caused by kinase deficiency or by the adenoviral cre vector (Fig. 2 C and E and Fig. S4F). The period was significantly shorter in CK1δ/ε mutant cells expressing transgenic CK1ε, probably due to overexpression of the transgenic CK1ε relative to endogenous CK1δ/ε (Fig. 2E) (21). This dose-dependent effect of CK1δ/ε on period length is further supported by a dramatic period lengthening observed in CK1δΔ2/Δ2:CK1ε−/+ cells where only one allele of CK1ε is preserved (Fig. S4G). The period was lengthened by ∼2.5 h relative to WT cells, or ∼1 h relative to CK1δΔ2/Δ2:CK1ε+/+ cells (27). The single-copy CK1ε cells were generated by expressing Cre in CK1δfl/fl:CK1ε fl/+ cells. PER1/PER2 phosphorylation was completely rescued by exogenous expression of wtCK1ε but not by a dominant negative CK1ε (DNCK1ε; Fig. 2E). Interestingly, expression of DNCK1ε increased levels of PER1/2 in the mutant cells, suggesting that DNCK1ε may interfere with targeted degradation of PER even in the absence of endogenous CK1δ/ε.
Inhibition of Protein Phosphatases by Calyculin A Induces Rapid Hyperphosphorylation of PER and Shortened Circadian Periods.
The slow and progressive PER phosphorylation seen in vivo, in contrast to the rapid PER phosphorylation seen in an in vitro kinase assay (21), may be attributed to either down-regulation of CK1δ/ε enzyme activity or a counterbalancing of PER phosphorylation by phosphatases. Although the enzyme activity of CK1δ/ε can be down-regulated by autophosphorylation in vitro, we have not observed such autophosphorylated isoforms for CK1δ/ε in fibroblasts or tissues in vivo (12, 21). Moreover, because protein phosphatases PP1 and PP2A have been implicated as clock components in various model organisms (39–43), we focused on the hypothesis that one or both of these two phosphatases are important for regulating progressive PER phosphorylation in mammals.
To test our hypothesis, we evaluated how phosphatase inhibitors, okadaic acid (OA) and calyculin A (CA), affect circadian rhythms in Per2Luc fibroblasts and lung explants (4). Because both OA and CA have cytotoxic effects, we could measure bioluminescence rhythms for only a couple of days after the drug treatment. When Per2Luc cells were treated with OA, bioluminescence rhythms and PER phosphorylation were not significantly altered (Fig. 3A and Fig. S5A). However, the rhythms were immediately abolished by treatment with CA (Fig. 3A), which is a much more potent PP1 inhibitor than OA (44). At lower doses, CA treatment induced shortened periods in a dose-dependent manner (Fig. 3B and Fig. S5B). All CA treatment conditions (continuous incubation or short incubation followed by wash-off) that significantly shortened the circadian period also caused cell death and allowed only one cycle after the CA treatment. Despite numerous efforts, we could not find conditions that induced short periods without causing cell death, suggesting that cell death is induced at lower doses than those affecting circadian rhythms. Immunoblots revealed striking effects of CA on endogenous PER. PER1 and PER2 from CA-treated cells migrated much more slowly compared with those from control and OA-treated cells, and these isoforms were hyperphosphorylated (Fig. 3C and Fig. S5C). The unusually hyperphosphorylated PER isoforms seem to be very unstable, as they have not been seen previously in vivo and they disappeared rapidly. Indeed, they were stabilized when the CA cells were treated with the proteasome inhibitor mixture (Fig. S5D).
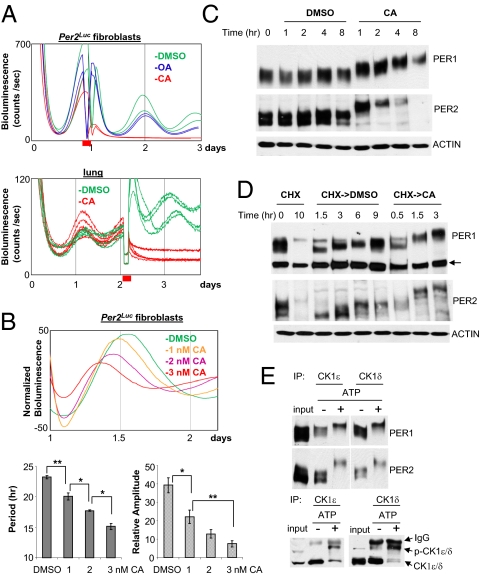
Fig. 3.
Phosphatase inhibition by calyculin A (CA) causes rapid hyperphosphorylation and degradation of PER, and disruption of circadian rhythms in fibroblasts and lung explants. (A) Circadian rhythms are abolished by CA treatment at a high dose. Per2Luc fibroblasts were treated with DMSO, OA, or CA for 2 h at the first peak of the bioluminescence rhythm, then washed and returned to the Lumicycle. Per2Luc lung was treated with DMSO or CA for 2 h near the second peak, then washed and returned to the Lumicycle. Red bar indicates the timing of the treatment. (B) Circadian rhythms are shortened by CA treatment at low doses in a dose-responsive manner. The cells were treated with CA for 6 h at the first peak and then measured for bioluminescence rhythms. Because only one complete cycle was observed after the CA treatment, period and amplitude of the one cycle were compared. (Upper) Comparison of representative traces. (Lower) Period and relative amplitude values presented as mean ± SEM from a representative of several experiments. Sample number: DMSO = 5; 1 nM = 5; 2 nM = 3; 3 nM = 5. P values were calculated by Student t test between two groups. *P < 0.05; **P < 0.01. (C) Hyperphosphorylation and rapid degradation of PER1/2 by CA treatment. Desynchronized Per2Luc fibroblasts were treated with DMSO or CA and harvested at the indicated times. (D) Accelerated PER phosphorylation by CA treatment. Fibroblasts were treated with cycloheximide for 10 h to remove existing PER, washed, and treated with DMSO or CA. The arrow indicates a nonspecific band. (E) In vitro kinase assay of in vivo PER:CK1δ/ε complexes. In vivo PER:CK1δ or ε complexes were purified by immunoprecipitation with antibody to CK1δ or CK1ε, and subjected to in vitro kinase reaction for 30 min. The resulting samples were subjected to immunoblotting for PER1/2 and CK1δ/ε. Note that CK1δ/ε were also phosphorylated by the kinase reaction (p-CK1δ/ε).
For a more quantitative assessment, we assessed PER phosphorylation rate from de novo synthesized proteins after existing PER proteins were depleted by cycloheximide treatment. Under normal conditions, PER1 and PER2 reached their maximum phosphorylation state (lowest mobility form) between 3 and 6 h after cycloheximide removal (Fig. 3D). However, PER achieved an even slower mobility form (extrahyperphosphorylated state) in <1.5 h in CA-treated cells. PER was similarly extrahyperphosphorylated within 30 min when PER:CK1δ/ε complexes were purified from cell extracts and incubated in vitro with ATP, suggesting that CK1δ/ε and/or other copurified kinases can phosphorylate PER rapidly in vitro to a level similar to that seen in CA-treated cells (Fig. 3E). Consistent with previous findings (45), CK1δ/ε were also phosphorylated under these conditions. CA treatment also allowed the accumulation of hyperphosphorylated CK1δ/ε, suggesting that CK1δ/ε, like PER, are dephosphorylated by phosphatases in vivo, and that phosphorylation promotes degradation (Fig. S6) (45).
PP1 Is a Major Phosphatase That Counteracts PER Kinases to Ensure That PER Phosphorylation Occurs Gradually During the Circadian Cycle.
The difference in the effects of the two phosphatase inhibitors on PER phosphorylation state and bioluminescence rhythms is informative. OA inhibits PP2A with >100-fold greater potency than PP1, and our dose was >2 orders of magnitude higher than the IC50 for PP2A; these results suggest that inhibition of PP2A does not affect the circadian period. In contrast, CA inhibits PP1 and PP2A with almost equal potency (44). Thus, our results suggest that PP1 plays a critical role in the mammalian clock mechanism. To test by a nonpharmacological approach whether PP2A plays a significant role in PER dephosphorylation and in circadian clock function, we used mouse embryonic fibroblasts (MEFs) derived from the α4 mutant mouse in which PP2A activity is severely compromised (>70%). α4 is a regulatory protein that binds to and regulates the activity and/or stability of the major phosphatases, PP2A, PP4, and PP6 (46–49). Although α4 deletion usually leads to immediate cell death, the p53−/− background renders the mutant cells viable (48). We measured PER phosphorylation in α4flp53−/− cre-ER MEFs where α4 is conditionally deleted by 4-hydroxy-tamoxifen treatment. As reported by Kong et al. (48), levels of α4 and PP2Ac were dramatically reduced in the cells 6 d after 4-hydroxy-tamoxifen treatment (Fig. 4A). PER levels were slightly reduced, but phosphorylation levels were not significantly altered, indicating that PP2A, PP4, and PP6 do not play a major role in dephosphorylating PER.
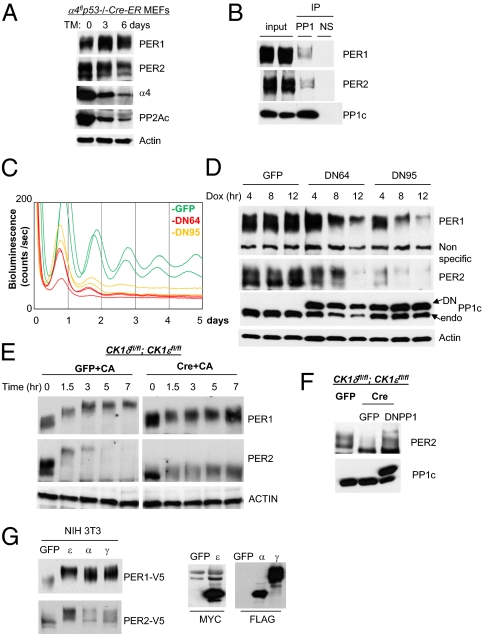
Fig. 4.
PP1 is a major PER phosphatase and essential for rhythm generation. (A) PP2Ac levels are dramatically reduced in α4 mutant fibroblasts, but PER phosphorylation is intact. Cre (used for conditional deletion of floxed α4) was activated by addition of 4-hydroxytamoxifen (TM) to the medium. (B) Coimmunoprecipitation of PER with PP1 catalytic subunit PP1c. (C) Shortened and compromised bioluminescence rhythms in dominant negative PP1 (DN64 or DN95)-expressing cells. Adenoviral DN64, DN95, or gfp was infected into Per2Luc fibroblasts, and expression of DN was initiated by doxycycline. The period (measured from the first two peaks) of the bioluminescence rhythms was 3–4 h shorter in DN-expressing cells. GFP: n = 6; period (hr) = 23.9 ± 0.2; relative amplitude = 53.0 ± 3.3. DN64: n = 4; period = 20.0 ± 0.1; amplitude = 15.4 ± 2.9. DN95: n = 4; period = 21.1 ± 0.1; amplitude = 26.5 ± 3.6. Values presented are mean ± SEM from three experiments. (D) Hyperphosphorylation and rapid degradation of PER induced by DN expression. Desynchronized cells were infected with adenovirus, treated with doxycycline, and harvested at the indicated times. (E) PER2 phosphorylation after CA treatment in CK1δ/ε-deficient cells. The mutant cells were harvested at the indicated times after CA treatment. (F) PER2 phosphorylation in the mutant cells with and without expression of DN64 (DNPP1). (G) PER phosphorylation by CK1α/γ in vitro. The kinases and Per were cotransfected into NIH 3T3 cells and harvested 48 h later. ε, CK1ε; α, CK1α; γ, CK1γ.
Furthermore, we found that PP1 coimmunoprecipitates with PER in vivo, as CK1δ and CK1ε do, although the PP1/PER interaction seems to be significantly weaker than that between PER and CK1δ/ε (Fig. 4B). Interestingly, coimmunoprecipitated PER was mid- to hyperphosphorylated, suggesting that these species have higher affinity for PP1 than non- or hypophosphorylated species. We then specifically disrupted PP1 activity using dominant negative mutants of PP1. Like PP2A, PP1 cannot be constitutively disrupted due to its essential role in cell physiology (50, 51). We generated inducible adenoviral vectors to express two different dominant negative PP1 mutants (DN64 and DN95) (52) in a doxycycline (Dox)-dependent manner. These DN mutants induced cell death in rapidly growing cells, consistent with significant interference with endogenous PP1 activity. However, quiescent cells in serum-free medium could tolerate the DN mutants, and we normally measure circadian rhythms under these conditions. When expression of either PP1 mutant was induced by Dox, bioluminescence rhythms were severely disrupted and basal levels of bioluminescence were lowered (Fig. 4C). Further, consistent with accelerated PER phosphorylation, the period (measured between the first two peaks) was dramatically shortened (3–4 h) compared with that of control cells. However, a normal period was readily restored when Dox was washed off, indicating that period shortening was caused by the reversible inhibition of PP1 activity, probably on PER rather than cytotoxic effects of PP1 inhibition (Fig. S7A). In desynchronized cells following Dox treatment, PER1 and PER2 were progressively hyperphosphorylated and disappeared (Fig. 4D), indicating that PP1 is a major phosphatase counteracting the activity of PER kinases CK1δ/ε in vivo. The extrahyperphosphorylated species of PER seen in CA-treated cells were not observed in DN PP1 cells, probably because PP1 activity was not disrupted as severely as by CA treatment. Taken together, our data reveal that temporal phosphorylation of PER is determined by the balance between the opposing actions of CK1δ/ε and PP1.
Kinases other than CK1δ/ε may also play a role (28–32). When we inhibited PP1/2A with CA in the CK1δ/ε mutant cells, PER1 did not reach the same hyperphosphorylated state as seen in WT cells treated with CA (cf. Figs. 4E and 3C). However, hypophosphorylated forms of PER1 seemed to become a little more phosphorylated over time (Fig. 4E). PER2 was hyperphosphorylated in the mutant cells treated with CA to a level similar to that in control cells before CA treatment (Fig. 4E). This result suggests that PER1 and PER2 are phosphorylated by kinases other than CK1δ/ε, but the phosphorylation is rapidly reversed by phosphatases. Phosphorylation by these kinases may target PER for proteasomal degradation, as we had found that PER is degraded by the proteasome even in the absence of CK1δ/ε (Fig. 2B). Expression of DN64PP1 in the mutant cells also induced hyperphosphorylation of PER2, confirming that PP1 is a major phosphatase for PER2 and opposes the non-CK1δ/ε kinases (Fig. 4F). Based on previous in vitro studies, other CK1 members, such as CK1α and CK1γ, may be responsible for this mobility-shifting PER phosphorylation in the CK1δ/ε mutant cells (28, 29). In our in vitro assay using transiently expressed proteins in cultured cells, CK1α and γ can induce mobility-shifting PER phosphorylation, though not as efficiently as CK1ε (Fig. 4G); their lower activity is consistent with the finding that their interaction with PER is significantly weaker than interaction between PER and CKIδ/ε (28, 29, 53). It is also possible that PER phosphorylation under these conditions may be attributed to other kinases, such as CK2, GSK3β, and ERK2, previously implicated as PER kinases by in vitro studies (29–32).
Discussion
The importance of PER kinases has long been recognized, and previous work has shown that CK1δ/ε are important (14–18, 21, 23–25, 27, 54, 55), but our generation of CK1δ/ε-deficient cells enabled us to study the roles of these kinases with unprecedented rigor. Although the previous studies suggested that CK1δ and CK1ε are essential kinases for PER phosphorylation and a functioning clock, our present study clearly establishes that CK1δ/ε are redundant for PER phosphorylation. Additional kinases such as CK1α/γ may also regulate PER phosphorylation and stability, but the high affinity of CK1δ/ε for PER (and possibly their abundance relative to other CK1s) would ensure that their contribution to phosphorylation would outcompete that of CK1α/γ. CK2 and GSK3β may also phosphorylate PER on important residues in vivo. However, we show that CK1δ/ε are irreplaceable for promoting nuclear entry/accumulation of PER, which is a key event in the negative feedback loop.
We also reveal an interesting difference in regulation between PER1 and PER2. In the absence of CK1δ/ε, PER1 (unlike PER2) is stably phosphorylated to an intermediate level. If further phosphorylation by CK1δ/ε requires this initial phosphorylation, then the priming kinase(s) may also be essential for the circadian clock. The PER1 rhythm would be compromised without the priming phosphorylation, and this could lead to a complete disruption of the molecular oscillator, as seen in cells with constitutively expressed PER1 (11).
Our data suggest that the gradual, progressive phosphorylation of PER proteins occurs because the rate of PER dephosphorylation is slower than that of phosphorylation. PER in mammals is mainly dephosphorylated by PP1, whereas PP2A has little effect. Our data are consistent with previous in vitro studies showing that PP1 antagonizes CK1δ/ε activity (43, 56). Our findings are also consistent with the presence in mammalian PERs of PP1-binding motifs (R/K-V/I-x-F; Fig. S7B) (41, 57, 58). This motif, commonly found in PP1-binding regulatory subunits, suggests that direct binding between the PP1 catalytic subunit and PER1/2 may occur. Our findings contrast with studies in Drosophila, where PP2A (rather than PP1) is the clock-relevant PER phosphatase (41, 42).
Because CK1δ/ε and PP1 activities do not oscillate (12, 56), overall speed of PER phosphorylation—resulting from the relative activity between CK1 and PP1—would remain constant over the circadian cycle. However, the relative activity can be modulated by disrupting CK1δ/ε or PP1 activities pharmacologically or genetically. If PP1 activity is disrupted (as by CA or DN-PP1), then the excess CK1δ/ε activity enhances the speed of PER phosphorylation, and the clock runs faster. However, if CK1δ/ε activity is disrupted, PER phosphorylation slows down, resulting in a slower oscillator (27, 54, 55). While our work was under review, Schmutz et al. (39) showed that inhibition of PP1 activity slightly lengthened the period of bioluminescence and behavioral rhythms. The difference in the effect of PP1 inhibition may be due to different levels of inhibition and/or different methods of inhibition. Because rhythms in PER phosphorylation determine phase and period of the molecular oscillator and circadian rhythms, our present study provides clear insights into how the clock can be regulated by posttranslational regulation of the critical clock component PER, and also provides well-defined targets for therapeutic intervention to treat circadian clock disorders.
Materials and Methods
Animals, Cells, and Antibodies.
All animals were maintained and used according to the Florida State University Animal Care and Use Committee's guidelines. The CK1δfl/fl;CK1εfl/fl double-floxed mutant fibroblasts were isolated from the tails of double-floxed mutant mice described previously (27) and immortalized as described previously (11). Cre was expressed in these cells by an adenoviral vector (SI Materials and Methods). Deletion of both CK1δ/ε genes was tolerated but induced slow growth in the immortalized cells. The α4fl p53−/−Cre-estrogen receptor (ER) mouse embryonic fibroblasts and anti-α4 antibody were kindly provided by Mei Kong and Craig Thompson (University of Pennsylvania, Philadelphia). In these cells, Cre was activated by the addition of 200 nM 4-hydroxytamoxifen (TM) to the medium. Per2Luc fibroblasts were described previously (4).
Generation of CK1δ/ε Double-Mutant Cells for Monitoring Bioluminescence Rhythms and Immunoblotting.
Double-floxed (CK1δfl/fl; CK1εfl/fl) fibroblasts were grown in DMEM supplemented with 10% FBS. The fibroblasts were infected with adenoviral gfp or cre in 100-mm dishes when the cells had reached ∼70% confluency, and the cultures were then split twice over ∼5 d. At the third split, the cells were transferred into 35-mm dishes, infected with adenoviral-Per2-Luc reporter and/or other adenovirus for 1 d, and set up for measuring bioluminescence after a 2-h serum shock. For immunoblots, at the third split the cells were transferred into 60-mm dishes and infected with adenovirus for 1 d or treated with drugs before harvesting at specific times. Lung tissue was incubated with adenovirus cre for 5 d and then infected with the adenoviral Per2:Luc reporter. Period and amplitude in Figs. 2C, 3B, and 4C were calculated using the periodogram function in ClockLab software. First four, one, and two peaks after drug treatment or serum shock were considered for Figs. 2C, 3B, and 4C, respectively. P values were calculated by Student t test between two groups (*P < 0.05; **P < 0.01; ***P < 0.001).
See SI Materials and Methods for additional information.
Supplementary Material
Acknowledgments
We thank Qisen Li for excellent technical assistance during the project and Dennis Chang for assistance with manuscript revisions. This work was supported by National Institutes of Health Grants NS-053616 (to C.L.) and NS-056125 (to D.R.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107178108/-/DCSupplemental.
References
- 1.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 6.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005;6(Spec No):S9–S13. doi: 10.1038/sj.embor.7400424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuninkova L, Brown SA. Peripheral circadian oscillators: Interesting mechanisms and powerful tools. Ann N Y Acad Sci. 2008;1129:358–370. doi: 10.1196/annals.1417.005. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keesler GA, et al. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport. 2000;11:951–955. doi: 10.1097/00001756-200004070-00011. [DOI] [PubMed] [Google Scholar]
- 17.Camacho F, et al. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- 18.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 20.Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng QJ, et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 25.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 26.Vanselow K, et al. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etchegaray JP, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 29.Hirota T, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 31.Maier B, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuchiya Y, et al. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Q, et al. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci USA. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schafmeier T, et al. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 37.Price JL, et al. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 38.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 39.Schmutz I, et al. Protein phosphatase 1 (PP1) is a post-translational regulator of the mammalian circadian clock. PLoS ONE. 2011;6:e21325. doi: 10.1371/journal.pone.0021325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, et al. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev. 2004;18:255–260. doi: 10.1101/gad.1152604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- 43.Shanware NP, et al. Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J Biol Chem. 2011;286:12766–12774. doi: 10.1074/jbc.M111.224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swingle M, Ni L, Honkanen RE. Small-molecule inhibitors of ser/thr protein phosphatases: Specificity, use and common forms of abuse. Methods Mol Biol. 2007;365:23–38. doi: 10.1385/1-59745-267-X:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivers A, Gietzen KF, Vielhaber E, Virshup DM. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J Biol Chem. 1998;273:15980–15984. doi: 10.1074/jbc.273.26.15980. [DOI] [PubMed] [Google Scholar]
- 46.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 47.Murata K, Wu J, Brautigan DL. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci USA. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong M, Ditsworth D, Lindsten T, Thompson CB. Alpha4 is an essential regulator of PP2A phosphatase activity. Mol Cell. 2009;36:51–60. doi: 10.1016/j.molcel.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong M, et al. The PP2A-associated protein alpha4 is an essential inhibitor of apoptosis. Science. 2004;306:695–698. doi: 10.1126/science.1100537. [DOI] [PubMed] [Google Scholar]
- 50.Gallego M, Virshup DM. Protein serine/threonine phosphatases: Life, death, and sleeping. Curr Opin Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Zhang Z, Brew K, Lee EY. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 53.Dahlberg CL, Nguyen EZ, Goodlett D, Kimelman D. Interactions between Casein kinase Iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS ONE. 2009;4:e4766. doi: 10.1371/journal.pone.0004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng QJ, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci USA. 2010;107:15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walton KM, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 56.Gallego M, Kang H, Virshup DM. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem J. 2006;399:169–175. doi: 10.1042/BJ20060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Egloff MP, et al. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.