Abstract
The exquisite specificity of proteins is a key feature driving their application to anticancer therapies. The therapeutic potential of another fundamental property of proteins, their ability to be regulated by molecular cues in their environment, is unknown. Here, we describe a synthetic biology strategy for designing protein therapeutics that autonomously activate a therapeutic function in response to a specific cancer marker of choice. We demonstrate this approach by creating a prodrug-activating enzyme that selectively kills human cancer cells that accumulate the marker hypoxia-inducible factor 1α. This property arises primarily through increased cellular accumulation of the enzyme in the presence of the marker. Our strategy offers a platform for the development of inherently selective protein therapeutics for cancer and other diseases.
Keywords: directed evolution, protein engineering, protein switch, enzyme/prodrug therapy
The attractiveness of proteins as therapeutics stems in part from the exquisite specificity by which they execute diverse functions—e.g., they catalyze exactly the right reaction or inhibit exactly the right cell receptor. Proteins possess another fundamental property central to life whose therapeutic potential has not been exploited—the ability to be regulated at the protein level by molecular signals. Prevailing approaches to cancer protein therapeutics focus on cancer marker recognition/modulation and downstream effects of that modulation (1). Such an approach is limiting because the therapeutic mechanism is restricted to those that naturally arise from modulation of the cancer marker. Furthermore, this approach precludes the use of cancer markers for which modulators cannot be found or for which modulation does not produce a therapeutic effect (2). In addition, many potential protein therapies lack the desired selective cancer cell targeting. The ability to link recognition of any cancer marker with activation of any desired therapeutic function would enormously expand the number of possible protein therapeutics. Here is where the therapeutic potential of regulation can be realized.
One approach to establishing unique regulatory relationships is to build proteins that function as switches—proteins whose cellular level of activity is modulated through interactions with an input signal such as a protein or small molecule. Our switch design strategy views all proteins as an extensive parts list from which switches can be built using domains with the prerequisite input and output functions of the desired switch. The design challenge for this approach is how to fuse the input (i.e., signal recognizing) and output domains (i.e., the function to be modulated) such that the input domain regulates the output domain’s function. We have explored a directed evolution approach to this general design problem in which libraries of random insertions of one domain into the other are subjected to selections and screens designed to identify library members with switching behavior (3–5). Our approach extensively explores the geometric space of how two protein domains can be fused through insertion of one domain into another (3, 4). For example, we have identified switch proteins with maltose-activated β-lactamase activity from libraries of circular permuted β-lactamase genes inserted into the gene encoding maltose-binding protein (MBP) (4, 5). Many of these switches function as allosteric enzymes with maltose binding to the MBP domain inducing conformational changes in the β-lactamase domain that affect its catalytic activity (4, 6). For other switches, maltose binding does not affect the specific activity of the protein but instead increases the accumulation of the switch protein in the cell (7).
Here, we propose a synthetic biology strategy for designing protein therapeutics that link activation of a chosen therapeutic function to a specific cancer marker of choice. We demonstrate this strategy by creating a protein switch that renders human colon and breast cancer cells susceptible to the prodrug 5-fluorocytosine (5FC) in response to the cancer marker hypoxia-inducible factor 1α (HIF-1a).
Results and Discussion
A Protein Therapeutic Modality Based on Regulation.
Our approach greatly expands the number of possible therapeutic strategies by providing a platform for how a cancer marker of choice can be used to trigger any therapeutic function of choice (Fig. 1). Central to this strategy is the development of a protein switch that couples recognition of the cancer marker to a therapeutic function. Such coupling could arise via newly established allosteric interactions (Fig. 1B) or through increased cellular accumulation in the presence of the marker (Fig. 1C). We sought to demonstrate this strategy by linking the unrelated functions of recognition of tumor-marker HIF-1a and the enzymatic production of the chemotherapeutic 5-fluorouracil (5FU) from the prodrug 5FC by cytosine deaminase (Fig. 2A). HIF-1a accumulates to a high level in many solid tumors, including breast, prostate, colorectal, and pancreatic cancers (8–10) but is virtually undetectable in normal, well-oxygenated tissues (11–13). Cancer cells that contain high levels of HIF-1a are able to survive under extreme conditions, are resistant to therapy, and have a greater potential for metastasis (10, 14–18). Thus, the linking of HIF-1a levels to 5FU production could in theory attack the most aggressive tumors, while having limited or no effect on well-oxygenated, normal cells. Such a strategy, like any strategy that utilizes an intracellular target, faces the challenge of efficient delivery of the therapeutic gene or protein to the cancer cell.
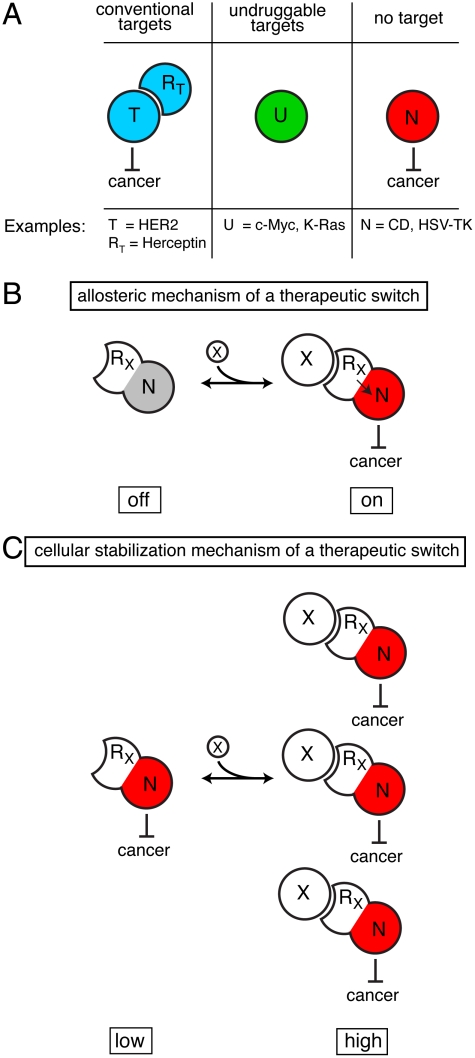
Fig. 1.
A therapeutic modality based on regulation expands potential strategies. (A) Cancer marker T is a conventional protein target in which recognition and modulation of T by protein RT causes a therapeutic effect. Cancer marker U is considered undruggable: Either a protein capable of recognizing and modulating U cannot be found or a therapeutic benefit for modulation of U does not exist. N is a protein with inherent therapeutic activity but is not targeted to cancer cells. (B) A therapeutic switch functioning by an allosteric mechanism couples RX’s recognition of cancer marker X to a therapeutic effect mediated by protein N. The switch is a fusion protein of RX and N such that binding of X regulates N’s therapeutic effect. (C) A therapeutic switch functioning by a cellular stabilization mechanism couples RX’s recognition of cancer marker X to cellular accumulation. The switch’s therapeutic activity is higher in the presence of X, not because of change in specific activity of the switch, but because the switch accumulates to a higher level in the cell. These two switch mechanisms are not mutually exclusive and either can cause an otherwise nonspecific therapeutic effect to depend on the presence of any cancer maker, whether conventional or undruggable.
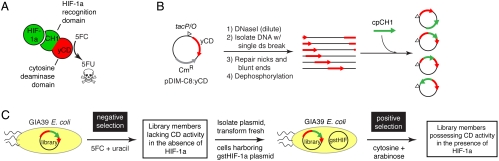
Fig. 2.
Protein switch concept, creation, and isolation. (A) The desired switch is a fusion protein between a HIF-1a binding domain (CH1) and yeast cytosine deaminase (yCD) that increases the cellular conversion of 5FC to 5FU in the presence of HIF-1a. (B) Library creation. A plasmid containing the yCD gene is randomly linearized and ligated to a collection of circularly permuted CH1 genes (cpCH1; see Fig. S1), resulting in a library of random insertions of cpCH1 into the plasmid. (C) Two-tiered genetic selection for yCD-CH1 hybrids that possess cytosine deaminase activity only in the presence of HIF-1a.
We constructed our switch using the CH1 domain from the human p300 protein as the HIF-1a recognition input domain and yeast cytosine deaminase (yCD) as the prodrug activation output domain (Fig. 2A). The p300/CBP protein complex binds HIF-1a in the cytoplasm and translocates it to the nucleus (13, 19–21). CH1 is a small 100 amino acid domain of the p300 protein that retains full HIF-1a affinity (19, 21). CH1 interacts with other cancer markers besides HIF-1a such as p53 (20) and HIF-2a (10)—offering other cancer-specific signals that could cause prodrug activation by our desired switch. yCD is an enzyme that converts 5FC to 5FU. We utilized the triple mutant of yCD (A23L/V108I/I140L) shown to have increased thermostability and full enzyme efficiency (22). 5FC is a nontoxic compound (LD50 → 15 g/kg in rats), and 5FU is a well-established, highly active chemotherapeutic used to treat multiple types of cancer, including breast, colorectal, and pancreatic cancers. Cytosine deaminase has been previously shown to activate the prodrug 5FC into the toxin 5FU in animal models (23) and the 5FC/CD combination has been used in gene-directed enzyme prodrug therapy (GDEPT) treatments in clinical trials (24).
Library Creation and Selection.
We constructed a library in which DNA encoding the CH1 domain was randomly inserted into a plasmid encoding yCD (Fig. 2B, Fig. S1, and SI Text). Our yCD-CH1 library contained approximately 10 million members of which 25% had the DNA encoding the CH1 domain inserted somewhere in the yCD gene. We devised a two-tier genetic selection to isolate yCD-CH1 hybrids that exhibit low cellular activity in the absence of HIF-1a, but high cellular activity in the presence of HIF-1a (Fig. 2C). The library was transformed into GIA39 Escherichia coli—a uracil auxotroph that lacks a functional E. coli cytosine deaminase (25, 26). In the negative selection tier, library members deficient in 5FC to 5FU conversion in the absence of HIF-1a were selected. In the subsequent positive selection tier, library plasmids were transformed into GIA39 cells that harbored a plasmid encoding a fusion protein of GST and the C-TAD domain of HIF-1a under the control of the arabinose promoter. Freedman et al. demonstrated that this fusion (referred to here as “gstHIF-1a”) and the CH1 domain maintain the high affinity of the full-length proteins and could be copurified when they are expressed within the same E. coli cell (20). Our selections resulted in the identification of two recombinant genes that conferred to GIA39 cells a gstHIF-1a-dependent sensitivity to 5FC. These nearly identical proteins were named “Haps3” and “Haps59” for HIF-1a activated protein switch (Fig. 3A).
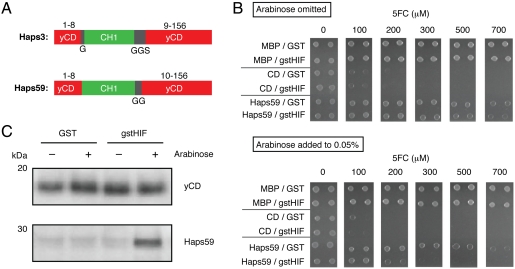
Fig. 3.
Characterization of Haps59 in E. coli. (A) Switches contain a complete, noncircularly permuted CH1 domain (green) flanked by short linker sequences of the indicated sequences (black) inserted after amino acid 8 of yCD (red). The black numbers indicate the corresponding amino acid numbers in the wild-type yCD protein. (B) Growth of GIA39 cells expressing MBP, yCD, or Haps59 and either GST or gstHIF-1a on minimal media agar plates as a function of 5FC concentration. (Top) In the absence of arabinose and (Bottom) in the presence of arabinose to induce GST and gstHIF-1a expression. (C) Accumulation of yCD and Haps59 in GIA39 E. coli in the absence and presence of coexpressed GST or gstHIF-1a as detected by Western blot with anti-yCD antibodies.
Characterization of Haps Proteins in Bacteria.
Initial experiments suggested that haps59 conferred to E. coli the greatest HIF-1a dependence on 5FC sensitivity. To quantify the effect, the plasmid encoding Haps59 was isolated and retransformed into fresh GIA39 cells harboring either the gstHIF-1a plasmid or an analogous negative control plasmid encoding only GST. These cells and appropriate control cells were challenged to grow on agar plates containing 5FC (Fig. 3B and Fig. S2). Only the combination of the presence of the gstHIF-1a gene and the addition of arabinose, which induces gstHIF-1a expression, increased the 5FC sensitivity of cells expressing Haps59. Qualitatively similar results were obtained in liquid media (Fig. S3). The results indicate that HIF-1a significantly increases the 5FC deaminase activity of cells expressing Haps59.
Copurification experiments indicated that Haps59 interacts with gstHIF-1a in E. coli (Fig. S4). Presumably this interaction must allosterically activate the cytosine deaminase activity of the switch and/or cause the increased accumulation of the switch in vivo. We observed a substantial increase in the accumulation of Haps59 when gstHIF-1a was coexpressed in vivo (Fig. 3C). Purified Haps59 exhibited a small increase in the 5FC deaminase activity in the presence of gstHIF-1a, but poor Haps59 stability complicated quantification of the effect (see SI Text). We conclude that increased accumulation of Haps59 in the presence of gstHIF-1a is likely to be the major mechanism by which haps59 confers HIF-1a-dependent sensitivity to 5FC (i.e., the mechanism of Fig. 1C). The CH1 domain has a molten globule state in the absence of HIF-1a and transforms into a stable, structurally well-defined domain upon binding to HIF-1a (27). HIF-1a-induced stabilization of the CH1 domain of Haps59 might stabilize the entire fusion resulting in increased cellular accumulation. However, why Haps59 and not other yCD-CH1 fusions would have this property is a challenging problem. This result highlights both the difficulty in designing proteins that function in the complex cellular environment and the power of directed evolution to identify proteins that provide the desired cellular phenotype when the appropriate genetic selection is applied.
Haps59 Increases the 5FC Sensitivity of Cancer Cells Expressing HIF-1a.
We next examined whether Haps59 would function in human cancer cells and confer sensitivity to 5FC selectively under conditions that cause HIF-1a to accumulate. Colorectal solid tumors accumulate high levels of HIF-1a (8–10). RKO is a colorectal cancer cell line that is known to accumulate high levels of HIF-1a in hypoxia (28) and is sensitive to 5FU in culture (29). 5FC was highly toxic to stable cell lines of RKO expressing yCD (LC50 = ∼ 15 μM) but nontoxic up to approximately 2 mM to stable cell lines constructed with the empty vector control (Fig. S5). We chose the CMV promoter to control expression of Haps59 and yCD because hypoxic conditions do not change the level of expression from this promoter (30). Initial experiments used the addition of Co2+ to the media to cause HIF-1a accumulation. Co2+ disrupts the degradation pathway of HIF-1a, allowing the protein to accumulate in the cytoplasm of the cell (31) (Fig. 4A). In the presence of Co2+, Haps59-expressing RKO cell lines were 10-fold more sensitive to 5FC (Fig. 4B). Hypoxic growth conditions (1% O2) also caused a marked increase in the 5FC sensitivity of these cells (Fig. 4B). Neither hypoxic growth conditions nor the addition of Co2+ caused 5FC sensitivity to the empty vector control cells or altered the 5FC sensitivity of cells expressing yCD (Fig. 4B and Fig. S5). Additionally, neither the presence of Co2+ nor hypoxic conditions were inherently toxic to Haps59-expressing RKO cells (Fig. S6A).
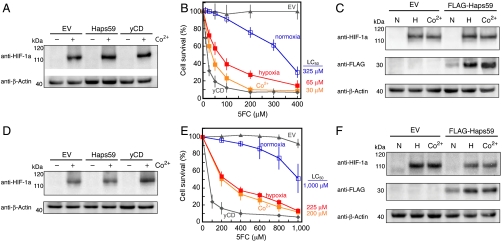
Fig. 4.
Characterization of Haps59 in human cancer cells. (A–C) RKO colorectal cancer cells; (D–F) MCF7 breast cancer cells. (A and D) Co2+ causes accumulation of HIF-1a in cells expressing Haps59, yCD, or the empty vector control as detected by Western blot with anti-HIF-1a antibodies. (B and E) Both Co2+ (orange solid squares) and exposure to hypoxic growth conditions (red solid squares) increase the 5FC sensitivity of Haps59-expressing cells compared to Haps59-expressing cells not exposed to those conditions (open blue squares). The 5FC sensitivities of cells expressing yCD (solid diamonds) and EV control (solid triangle) are shown for comparison (see Fig. S5 for additional controls). Each point represents the mean from three different clones of each cell line, and each clone was tested in three separate experiments (error bars, SD, N = 9). (C and F) Haps59 accumulates at higher levels in cells when HIF-1a is present. In these experiments a FLAG epitope was appended to the N terminus of Haps59 (FLAG-Haps59) for switch detection by Western blot. Cells were cultured under normoxic (N), hypoxic (H), or normoxic conditions with the addition of 100 μM cobalt (Co2+). Detection of β-actin served as a loading control.
Analogous experiments with MCF7 breast cancer cells illustrated the generality of the approach (Fig. 4 D and E). Mirroring results with E. coli cells, Haps59 accumulated at a higher level in the presence of HIF-1a in both RKO and MCF7 cells (Fig. 4 C and F), whereas yCD did not (Fig. S7A). This finding further supports the hypothesis that the increase in 5FC sensitivity results from a Haps59-HIF-1a interaction that causes increased accumulation of Haps59. This interaction was substantiated in RKO cells by coimmunoprecipitation experiments (Fig. S7B) and is the designed input signal to activate 5FU production. We confirmed the production of 5FU in Haps59-expressing RKO cells and found the production to be 2.5-fold higher in cells exposed to Co2+ (Fig. S6B). These results suggest that Haps59 functions as designed—increasing 5FU production in cancer cells in response to HIF-1a.
Haps59’s Potential as a Cancer Therapeutic.
HIF-1a is a molecular signature of cancer cells that can survive under extreme conditions, are resistant to therapy, and have a greater potential for metastasis (14–18). A treatment method that could specifically target these cells could significantly improve the effectiveness of cancer treatments when used in combination with traditional therapies. Haps59 is designed to establish a direct relationship between HIF-1a levels and the intracellular production of a chemotherapeutic drug. Haps59’s therapeutic potential is derived from this unique regulatory property. Our establishment of this regulation results in a complex protein that autonomously “decides” whether its drug-producing capability should be activated.
Hypoxic regions within a solid tumor can vary from 10–80% (32), and aberrant HIF-1a accumulation as a result of both gain-of-function and loss-of-function mechanisms has been observed experimentally and clinically in normoxic conditions (10). Thus, a high percentage of solid tumor cells can potentially activate Haps59, and non-Haps59 expressing tumor cells would be susceptible to the strong 5FU bystander effect (33, 34). Haps59 exploits an intracellular cancer marker for this activation. Prevailing strategies for protein cancer therapeutics target more-readily accessible extracellular cancer markers such as membrane-bound cell receptors. However, its been estimated that < 10% of the human genome codes for cell surface proteins, limiting the available targets for therapy (2). There is a great need to develop effective targeting strategies, and this might be accomplished through intracellular cancer markers. Such strategies will face delivery challenges: Either the therapeutic protein or its corresponding gene must be efficiently delivered inside the cell. However, our placement of the selectivity at the protein level overcomes serious limitations of existing gene therapy approaches that require selective targeting of gene delivery to malignant cells. For example, the combination of specific delivery of a cytosine deaminase gene to cancer cells followed by 5FC administration is an example of GDEPT (33, 34). One major limitation to existing CD/5FC GDEPT strategies is the poor transmission efficiency of the CD gene using current viral vectors because of the need for transmission specifically to cancer cells (35). Our strategy overcomes this limitation by moving the specificity from the transductional level to the protein level, allowing efficient means of gene delivery to be used regardless of cell-type specificity. In addition, our approach is complementary to both transcriptional (36) and transductional targeting and might be combined with these approaches to afford a double or triple layer of specificity: at the gene delivery level, at the transcription level, and at the protein level. Recent advances in protein delivery offer a potential route toward achieving efficient delivery without gene therapy (37, 38).
Nature chose proteins as a molecule of choice to carry out a wide array of specific, intricately regulated functions. Our increasing ability to emulate nature and design proteins capable of complex, carefully regulated functions offers an attractive path to achieving the effective and selective therapeutics we seek. By connecting disparate functions, protein switches expand the scope of potential therapeutic strategies in a manner that inherently encompasses specificity.
Methods
Library Creation and Selection.
Creation of the yCD-CH1 hybrid library is described in detail in SI Text. For selections, yCD-CH1 library plasmid DNA was isolated from an aliquot of DH5α E. coli cells, and 25 ng of the purified plasmid was used to transform GIA39 E. coli cells (Coli Genetic Stock Center# 5594) by electroporation. The transformed cells were plated on LB agar containing 50 μg/mL chloramphenicol and then recovered using a sweep buffer (1× M9 salts containing 2% glucose and 15% glycerol) and stored in aliquots at -80 °C. These cells were first plated on 24.5 × 24.5 cm minimal media plates (1× nitrogen base, 1× yeast synthetic dropout without uracil, 2% glucose, and 20 g/L select agar) containing 75 μg/mL 5FC, 1 μg/mL uracil, 1 mM IPTG, and 50 μg/mL chloramphenicol. Cells were plated at a concentration of approximately 500,000 cfu/plate, with cfu defined on minimal media plates without 5FC but with 1 μg/mL uracil. Colonies on these plates were recovered into sweep buffer and replated at the same cfu density on the same type of plate media except the 5FC concentration was lowered to 50 μg/mL. Library members surviving this second negative selection plate were recovered into sweep buffer, aliquoted, and stored at -80 °C. Library plasmid DNA was isolated from these samples and used for the positive selection.
GIA39 cells harboring a gstHIF-1a plasmid were cotransformed with library plasmids from the negative selection and plated on LB media containing 100 μg/mL ampicillin and 50 μg/mL chloramphenicol. Cotransformed GIA39 cells were recovered from the plate using sweep buffer, aliquoted, and stored as described above. These cells were plated on 24.5 × 24.5 cm minimal media plates supplemented with 25 μg/mL cytosine, 1 mM IPTG, 0.15% arabinose, 50 μg/mL chloramphenicol, and 100 μg/mL ampicillin at a density of approximately 500,000 cfu/plate. A total of 99 colonies formed on this selection plate, all of which were screened by colony PCR for pDIM-C8 plasmids that contained the CH1 insert within the yCD gene. This was performed using primers that anneal outside the yCD gene followed by gel electrophoresis to observe the size of the PCR product. About 20% of these colonies harbored library members that contained a CH1 insert within the yCD gene, and these members were sequenced. Eight members of the 20 sequenced were in-frame, of which Haps3 appeared twice and Haps59 appeared three times. In-frame members were then individually replated on selection media to ensure their switching behavior in the presence of gstHIF-1a. Only Haps3 and Haps59 behaved as switches after replating.
5FC Sensitivity Assays with GIA39 Cells on Solid Media.
Fresh GIA39 cell lines harboring a pDIM-C8 plasmid for expression of MBP (negative control), yCD (positive control), or Haps59 were cotransformed with either the gstHIF-1a plasmid or an analogous negative control plasmid encoding only GST were created. These six cell lines were cultured to midlog phase (0.3 OD) in minimal media and then serially diluted with minimal media 3.3-fold in 96-well format. Minimal media consisted of 1× nitrogen base, 1× yeast synthetic dropout without uracil, 2% glucose, 5 μg/mL uracil, 100 μg/mL of ampicillin, and 50 μg/mL chloramphenicol. One microliter of each cell line dilution was spotted on minimal media plates (OmniTray, 86 × 128 mm, Nunc) containing 1 mM IPTG, 20 g/L select agar, and different amounts of 5FC (0, 100, 200, 300, 500, and 700 μM). The media either omitted arabinose or contained 0.05% arabinose to induce GST and gstHIF-1a. The plates were incubated for 24–36 h at 37 °C, and the results of these experiments can be seen in Fig. 3 and Fig. S2.
Haps59 Accumulation Studies in E. coli.
Four GIA39 cell lines (expressing yCD + GST, yCD + gstHIF-1a, Haps59 + GST, or Haps59 + gstHIF-1a) were cultured in 25 mL of minimal media containing 1× nitrogen base, 1× yeast synthetic dropout without uracil, 2% glucose, 10 μg/mL uracil, 50 μg/mL chloramphenicol, and 100 μg/mL ampicillin overnight at 37 °C. These cultures were diluted into eight separate fresh minimal media flasks to compare the addition or omission of arabinose on yCD and Haps59 accumulation. These eight cultures were grown to an OD of 0.2, at which point IPTG (to 1 mM) was added to all cultures and 0.05% arabinose was added to half of the flasks to express GST or gstHIF-1a. The cultures were then incubated at 37 °C for 12–16 h. An equal amount of cells were aliquoted based on final OD measurements for lysis. The bacterial cells (680 million cells per sample) were lysed using BugBuster™ (Novagen) following the manufacturer’s protocol. Equal amounts of the cell lysates were separated on a 4–12% Bis-Tris NuPAGE gels (Invitrogen) and then transferred to PVDF membrane (BioRad). The Western protocol is described in SI Text.
5FC Sensitivity Experiments in RKO and MCF7 Cells Expressing yCD and Protein Switches.
RKO cells stably expressing an empty vector (one clone), yCD (three clones), or Haps59 (three clones) were used to seed 96-well plates, 1,500 cells per well, in 100 μL of MEM media supplemented with 10% fetal bovine serum and 1% antibiotic and antimycotic (ABAM) (all from Gibco). MCF7 cells stably expressing an empty vector (one clone), yCD (three clones), or Haps59 (three clones) were used to seed 96-well plates, 3,500 cells per well, in 100 μL of DMEM media supplemented with 10% fetal bovine serum and 1% ABAM (all from Gibco). After 24 h, 100 μL of MEM (RKO) or DMEM (MCF7) media containing 5FC (0–2 mM) was added to these cells. These experiments were carried out for 6 d with one media change after 3 d. After 6 d, the cells were lysed by rinsing twice with 100 μL of PBS and then incubated in 100 μL of 0.1 % SDS in water. The cells were incubated at 37 °C for 2 h in the SDS solution for complete lysis. To detect dsDNA, SYBR green (Invitrogen) was added to a final concentration of 0.075% to lysed cells and the fluorescence for each well was recorded at 520 nm after excitation at 485 nm. The percent survival was calculated using the equation (A)sample/(A)control × 100, where (A)sample is the fluorescence of sample wells with 5FC and (A)control is the fluorescence of the control well lacking 5FC. The results displayed in Fig. 4 represent the mean with experiments performed on three separate days (N = 6 for empty vector (EV), N = 9 for yCD, and N = 9 for Haps59). Error bars are the standard deviation from the mean.
To induce HIF-1a accumulation, MEM or DMEM media containing 75 μM CoCl2 was used. The presence of Co2+ had little effect on the growth of the RKO cells, regardless of whether or not the cells had been transfected with a gene (Fig. S6). Alternatively, a hypoxic environment was induced by incubation of the cancer cells in a MIC-101 hypoxia chamber (Billups–Rothenberg). The chamber was purged with three 3-min purges of 1% O2 gas containing 5% CO2 and 94% N2 over 2 h. A 3-min purge was then used every 24 h to maintain a hypoxic environment for the 6 d. Incubation at 1% O2 tension also had no effect on cell growth compared to the parental cell lines grown in the same conditions (Fig. S6). After 6 d (with one media change after 3 d), the cells were lysed and the percent survival was calculated using the same protocol as described above. Hypoxic environment experiments were repeated on separate days (N = 6 for all), and error was calculated by standard deviation from the mean.
HIF-1a Dependent Accumulation of Haps59 in RKO and MCF7 Cancer Cells.
yCD antibodies cross-reacted with many human proteins (Fig. S7A). To circumvent this problem, a FLAG™ tag (Sigma-Aldrich) was appended to the N terminus of Haps59 for protein switch accumulation experiments. RKO or MCF7 cells expressing an EV or Haps59 with the N-terminal FLAG™ tag (FLAG-Haps59) were incubated in normoxia (with and without 100 μM CoCl2), or hypoxia (1% O2) conditions for 12–18 h. After exposure, cell cultures were incubated at 4 °C for 30 min and then treated with radioimmunoprecipitation assay buffer (Sigma) containing protease inhibitor cocktail (Sigma). Cells were scrapped from the flask and placed in a 1.5-mL tube and incubated on ice for 30 min. Cell debris was removed by centrifugation (20,000 × g for 30 min, 4 °C) and the supernatant was transferred to a fresh 1.5-mL tube. Protein concentrations of the whole cell lysates were determined using the detergent compatible protein assay (BioRad). A total of 50 μg of each lysate were separated on 4–12% Bis-Tris NuPAGE gels (Invitrogen) and then transferred to PVDF membrane (BioRad). The membrane was blocked with 3% nonfat milk for 30 min.
Primary anti-FLAG™-HRP conjugated antibodies (Sigma) were diluted into the SignalBoost™ Immunoreaction Enhancer buffer (EMD Biosciences) according to the manufacturer’s protocol and incubated at room temperature for 1 h. The membrane was then washed and ECL was visualized using a Universal Hood II and QuanityOne software (BioRad); the results can be seen in Fig. 4 C and F. After detection of FLAG™, the membrane was stripped and then reprobed with beta-actin-HRP conjugated antibodies (Abcam) to verify protein-loading levels. For HIF-1a detection, 50 μg of the same samples were separated on 4–12% Bis-Tris NuPAGE gels and transferred to PVDF membrane. The membrane was blocked with 3% nonfat milk for 30 min. Primary antibodies for HIF-1a were diluted 1∶1,000 into blocking buffer and incubated at 4 °C overnight. The membrane was then washed, followed by the addition of mouse-HRP conjugated secondary antibodies, and ECL was visualized as described above.
Supplementary Material
Acknowledgments.
The authors thank Sharon Gerecht, Phillip Cole, and Denis Wirtz for comments on the manuscript and Gregg L. Semenza and Hirohiko Kamiyama for helpful discussions. This work was supported by National Institutes of Health Grants R01GM066972 (to M.O.) and R01CA130938 (to J.R.E.).
Footnotes
Conflict of interest statement: M.O. and C.M.W. declare a conflict of interest in the form of a patent application.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102803108/-/DCSupplemental.
References
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: A summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: Lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 3.Guntas G, Ostermeier M. Creation of an allosteric enzyme by domain insertion. J Mol Biol. 2004;336:263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Guntas G, Mitchell SF, Ostermeier M. A molecular switch created by in vitro recombination of nonhomologous genes. Chem Biol. 2004;11:1483–1487. doi: 10.1016/j.chembiol.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci USA. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright CM, Majumdar A, Tolman JR, Ostermeier M. NMR characterization of an engineered domain fusion between maltose binding protein and TEM1 beta-lactamase provides insight into its structure and allosteric mechanism. Proteins. 2010;78:1423–1430. doi: 10.1002/prot.22657. [DOI] [PubMed] [Google Scholar]
- 7.Sohka T, et al. An externally tunable bacterial band-pass filter. Proc Natl Acad Sci USA. 2009;106:10135–10140. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong H, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 9.Mabjeesh NJ, Amir S. Hypoxia-inducible factor (HIF) in human tumorigenesis. Histol Histopathol. 2007;22:559–572. doi: 10.14670/HH-22.559. [DOI] [PubMed] [Google Scholar]
- 10.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu AY, et al. Temporal, spatial, and oxygen-regulated expression of hypoxia-inducible factor-1 in the lung. Am J Physiol. 1998;275:L818–826. doi: 10.1152/ajplung.1998.275.4.L818. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Hydroxylation of HIF-1: Oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 14.Sun HC, et al. Expression of hypoxia-inducible factor-1a and associated proteins in pancreatic ductal adenocarcinoma and their impact on prognosis. Int J Oncol. 2007;30:1359–1367. [PubMed] [Google Scholar]
- 15.Dales JP, et al. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: Retrospective study in a series of 745 patients. Int J Cancer. 2005;116:734–739. doi: 10.1002/ijc.20984. [DOI] [PubMed] [Google Scholar]
- 16.Akakura N, et al. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548–6554. [PubMed] [Google Scholar]
- 17.Khandrika L, et al. Hypoxia-associated p38 mitogen-activated protein kinase-mediated androgen receptor activation and increased HIF-1alpha levels contribute to emergence of an aggressive phenotype in prostate cancer. Oncogene. 2009;28:1248–1260. doi: 10.1038/onc.2008.476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–572. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 19.Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 20.Freedman SJ, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedman SJ, et al. Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 22.Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kievit E, et al. Superiority of yeast over bacterial cytosine deaminase for enzyme/prodrug gene therapy in colon cancer xenografts. Cancer Res. 1999;59:1417–1421. [PubMed] [Google Scholar]
- 24.Crystal RG, et al. Phase I study of direct administration of a replication deficient adenovirus vector containing the E. coli cytosine deaminase gene to metastatic colon carcinoma of the liver in association with the oral administration of the pro-drug 5-fluorocytosine. Hum Gene Ther. 1997;8:985–1001. doi: 10.1089/hum.1997.8.8-985. [DOI] [PubMed] [Google Scholar]
- 25.Mahan SD, Ireton GC, Stoddard BL, Black ME. Alanine-scanning mutagenesis reveals a cytosine deaminase mutant with altered substrate preference. Biochemistry. 2004;43:8957–8964. doi: 10.1021/bi049720z. [DOI] [PubMed] [Google Scholar]
- 26.Mahan SD, Ireton GC, Knoeber C, Stoddard BL, Black ME. Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy. Protein Eng Des Sel. 2004;17:625–633. doi: 10.1093/protein/gzh074. [DOI] [PubMed] [Google Scholar]
- 27.Dial R, Sun ZY, Freedman SJ. Three conformational states of the p300 CH1 domain define its functional properties. Biochemistry. 2003;42:9937–9945. doi: 10.1021/bi034989o. [DOI] [PubMed] [Google Scholar]
- 28.Dang DT, et al. Hypoxia-inducible factor-1alpha promotes nonhypoxia-mediated proliferation in colon cancer cells and xenografts. Cancer Res. 2006;66:1684–1936. doi: 10.1158/0008-5472.CAN-05-2887. [DOI] [PubMed] [Google Scholar]
- 29.Thant AA, et al. Role of caspases in 5-FU and selenium-induced growth inhibition of colorectal cancer cells. Anticancer Res. 2008;28:3579–3592. [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata T, Giaccia AJ, Brown JM. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 2000;7:493–498. doi: 10.1038/sj.gt.3301124. [DOI] [PubMed] [Google Scholar]
- 31.Epstein AC, et al. Celegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhao D, Ran S, Constantinescu A, Hahn EW, Mason RP. Tumor oxygen dynamics: Correlation of in vivo MRI with histological findings. Neoplasia. 2003;5:308–318. doi: 10.1016/S1476-5586(03)80024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greco O, Dachs GU. Gene directed enzyme/prodrug therapy of cancer: Historical appraisal and future prospectives. J Cell Physiol. 2001;187:22–36. doi: 10.1002/1097-4652(2001)9999:9999<::AID-JCP1060>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 34.Russell PJ, Khatri A. Novel gene-directed enzyme prodrug therapies against prostate cancer. Expert Opin Investig Drugs. 2006;15:947–961. doi: 10.1517/13543784.15.8.947. [DOI] [PubMed] [Google Scholar]
- 35.Schepelmann S, Springer CJ. Viral vectors for gene-directed enzyme prodrug therapy. Curr Gene Ther. 2006;6:647–670. doi: 10.2174/156652306779010679. [DOI] [PubMed] [Google Scholar]
- 36.Marignol L, et al. Hypoxia response element-driven cytosine deaminase/5-fluorocytosine gene therapy system: A highly effective approach to overcome the dynamics of tumour hypoxia and enhance the radiosensitivity of prostate cancer cells in vitro. J Gene Med. 2009;11:169–179. doi: 10.1002/jgm.1281. [DOI] [PubMed] [Google Scholar]
- 37.Yan M, et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 38.Cronican JJ, et al. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.