Abstract
Plants and animals deploy intracellular immune receptors that perceive specific pathogen effector proteins and microbial products delivered into the host cell. We demonstrate that the ADR1 family of Arabidopsis nucleotide-binding leucine-rich repeat (NB-LRR) receptors regulates accumulation of the defense hormone salicylic acid during three different types of immune response: (i) ADRs are required as “helper NB-LRRs” to transduce signals downstream of specific NB-LRR receptor activation during effector-triggered immunity; (ii) ADRs are required for basal defense against virulent pathogens; and (iii) ADRs regulate microbial-associated molecular pattern-dependent salicylic acid accumulation induced by infection with a disarmed pathogen. Remarkably, these functions do not require an intact P-loop motif for at least one ADR1 family member. Our results suggest that some NB-LRR proteins can serve additional functions beyond canonical, P-loop–dependent activation by specific virulence effectors, extending analogies between intracellular innate immune receptor function from plants and animals.
Keywords: nucleotide-binding domain and leucine-rich repeat-containing protein receptors, plant immune system, effector-triggered immunity, microbial-associated molecular pattern-triggered immunity
Plants respond to attempted microbial infection with a two-tiered immune system. In the first tier, extracellular pattern recognition receptors (PRRs) bind conserved microbial-associated molecular pattern (MAMP) ligands, activating a complex host response that results in MAMP-triggered immunity (MTI). Successful pathogens deploy suites of virulence effectors that delay or suppress MTI, allowing infection. In the second tier, plant intracellular immune receptors of the nucleotide-binding leucine-rich repeat (NB-LRR) protein family can be activated either by direct binding of effectors or, alternatively, by effector action on an associated target protein that generates a “modified-self” molecule (1, 2). Effector-mediated NB-LRR activation results in effector-triggered immunity (ETI), a rapid and high-amplitude output significantly overlapping with MTI (1). ETI is typically accompanied by the hypersensitive cell death response (HR), limited to the site of pathogen attack. Both MTI and some cases of NB-LRR–mediated ETI require the salicylic acid (SA)-signaling molecule as a downstream mediator of transcriptional output responses (3, 4).
Plant NB-LRR proteins belong to the STAND (signal transduction ATPases with numerous domains) superfamily, which includes the animal apoptotic proteins Apaf-1/CED4 and innate immune receptors of the nucleotide-binding domain and leucine-rich repeat-containing proteins (NLR) family (5). Animal NLRs are activated by MAMPs and by modified-self molecules in the form of danger-associated molecular patterns (6) and regulate inflammasome activation, autophagy, and cell death (7). STAND protein functions require an intact P-loop motif (GxxxxGKT/S) that coordinates ATP binding. STAND proteins are molecular switches that toggle from an ADP-bound “off” position to an ATP-bound “on” state that activates downstream signaling. Activation of both animal NLR and plant NB-LRR receptors results in intra- and intermolecular conformational changes driven by nucleotide binding and hydrolysis (8, 9).
Plant NB-LRR proteins studied to date function in ETI, although gain-of-function mutations or ectopic over-expression can lead to additional phenotypes (10). Epistatic interactions involving NB-LRR genes can result in autoimmune-like responses (11). In some cases, ETI responses require a pair of NB-LRR proteins, although it is unclear whether these proteins interact directly. In these cases, one NB-LRR acts as the genetically defined “effector-sensor” and the other is required for its function, but is not implicated in effector perception per se (12). We suggest the term “helper NB-LRRs” for the latter category. Recent phylogenetic evidence suggests that a small, but ancient, subclade of plant coiled-coil (CCR)-NB-LRR proteins has evolved to fulfill “helper NB-LRR” function during ETI (13). Here, we demonstrate that the three members of the Arabidopsis CCR-NB-LRR ADR1 protein family are helper NB-LRRs in ETI and function in basal defense and in response to a disarmed pathogen via regulation of SA accumulation and subsequent activation of SA-dependent responses. We further show, surprisingly, that the P-loop is dispensable for these functions, suggesting that CCR-NB-LRR proteins can use an activation mechanism that differs from that of all other NB-LRR proteins studied to date in these contexts.
Results
PHOENIX21 Is a Positive Regulator of lsd1 Runaway Cell Death and Is a Member of the ADR1 Family of CCR-NB-LRR Proteins.
Mutants exist that cannot limit the spread of HR after NB-LRR activation. In the Arabidopsis lsd1 (lesions simulating disease 1) mutant, HR occurs normally, but the oxidative burst generated by pathogen recognition triggers a superoxide-dependent signal leading to “runaway cell death” that spreads beyond infection sites (14). This phenotype requires accumulation of SA and additional components of ETI/MTI signaling (15). In a screen for lsd1 suppressors, we isolated mutations in the PHOENIX (PHX) loci. One recessive complementation group of two alleles (phx21-1 and phx21-2) allowed neither the initiation nor the propagation of SA-induced lsd1 runaway cell death in the Wassilewskija (Ws) ecotype (14) (Fig. S1 A and B). We isolated PHX21 (At5g04720; hereafter, ADR1-L2) by map-based cloning (SI Materials and Methods). ADR1-L2 encodes a CCR-NB-LRR protein belonging to a small clade that includes ADR1 (ACTIVATED DISEASE RESISTANCE 1; At1g33560) and ADR1-L1 (At4g33300) (16) (Fig. S2A), previously characterized by the gain-of-function, over-expression phenotype of adr1 (17, 18). Over-expression of ADR1 results in the constitutive activation of the defense responses as well as in drought tolerance; both of these phenotypes are SA-dependent (17, 18). Loss of function for adr1-L1 results in modest suppression of ETI (19). We investigated ADR1 family functions in the Columbia (Col-0) ecotype after observing that an isogenic Col-0 allele (adr1-L2-4) also suppressed lsd1 runaway cell death (Fig. S1 C and D).
ADR1 Proteins Function as Helper NB-LRRs for Some, but Not All, ETI Responses and Are Required for Basal Defense to Virulent Pathogens.
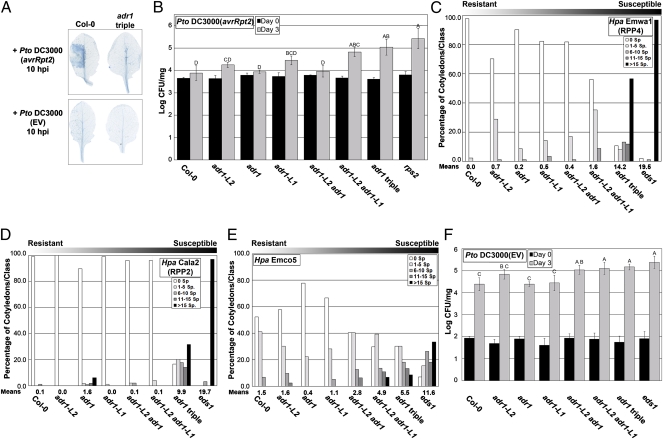
We assessed whether the ADR1 proteins can function as helper NB-LRRs for well-defined NB-LRR–mediated ETI responses. We challenged adr1, adr1-L1, and adr1-L2 single knock-out mutants (Fig. S2B), combinatorial double adr1 mutants, and the adr1 adr1-L1 adr1-L2 triple mutant (hereafter “adr1 triple”) with the bacterial pathogen Pseudomonas syringae pv. tomato (Pto) DC3000 expressing either the AvrRpm1 or AvrRpt2 effectors or with two isolates of the obligate biotrophic oomycete Hyaloperonospora arabidopsidis (Hpa isolates Emwa1 and Cala2). In Col-0, these effectors are recognized by either CC-NB-LRR proteins [RPM1 (20) and RPS2 (21)] or by the Toll/interleukin-1 receptor (TIR)-NB-LRR proteins [RPP4 (22) and RPP2 (23)], respectively. RPS2-mediated HR (Fig. 1A and Fig. S3A) and ETI (Fig. 1B) were significantly compromised in the adr1 triple mutant. RIN4 cleavage by the cysteine protease effector AvrRpt2 (24), which initiates RPS2-mediated ETI, was maintained (Fig. S3B). Hence, the ADR1 genes function downstream of this event in RPS2 signaling. Both RPP4- and RPP2-mediated ETI were also significantly compromised in the adr1 triple mutant (Fig. 1 C and D) and weakly compromised in single and combinatorial mutants. The adr1 triple mutant was nearly as susceptible to infection as the TIR-NB-LRR/basal defense signaling mutant enhanced disease susceptibility 1 (eds1) (25). These results extend evidence that ADR1 and the NRG1 (N requirement gene 1) CCR-NB-LRR proteins are helper NB-LRRs (13). RPM1-mediated ETI was not altered (Fig. S4). Thus, some, but not all, effector-mediated ETI responses tested required ADR1 proteins.
Fig. 1.
ADR1 family members function in ETI and basal defense. (A) Pto DC3000(avrRpt2) or Pto DC3000(EV) (empty vector) was hand-infiltrated into leaves of 4-wk-old plants. Leaves were collected and stained with trypan blue to visualize cell death. (B) Twenty-day-old seedlings were dip-inoculated with Pto DC3000(avrRpt2), and bacterial growth was assessed at 0 and 3 d post inoculation (dpi). (C) Ten-day-old seedlings were inoculated with Hpa Emwa1. Sporangiophores per cotyledon were counted at 5 dpi (average of 100 cotyledons per genotype). Cotyledons were classified as supporting no sporulation (0 sporangiophores/cotyledon), light sporulation (1–5 and 6–10), medium sporulation (11–15), or heavy sporulation (>15). Means of sporangiophores/cotyledon for each genotype are noted below. (D) Ten-day-old seedlings were inoculated with Hpa Cala2. Sporangiophores/cotyledon were counted at 6 dpi as described above. (E) Ten-day-old seedlings were inoculated with Hpa Emco5. Sporangiophores/cotyledon were counted at 4 dpi as described above. (F) Twenty-day-old seedlings were dip-inoculated with Pto DC3000(EV). Bacterial growth was assessed at 0 and 3 dpi. Values in B and F are mean cfu/mg ± 2 × SE (n = 4). Letters indicate a significant difference following post-ANOVA Tukey's test (α = 0.05).
NB-LRR protein function has been implicated in basal defense (26), which is activated by virulent pathogens on susceptible hosts and can limit pathogen growth (4). We thus tested the ability of the adr1 family mutants to restrict the growth of virulent pathogens. The adr1 triple mutant was more susceptible to Hpa Emco5 and to Pto DC3000 compared with wild-type or single adr1 mutants (Fig. 1 E and F). Hence, ADR1 proteins act as redundant regulators of basal defense.
ADR1 Proteins Regulate SA Accumulation Following an Oxidative Burst.
SA is a key downstream mediator of plant defense against biotrophic pathogens like those used here (27). Systemic acquired resistance (SAR) (28), basal defense (4), MTI (3), and some, but not all, NB-LRR–mediated ETI responses (29) require SA accumulation, which in turn controls transcriptional reprogramming through the BTB/POZ/ankyrin coactivator NPR1 (Nonexpresser of PR genes 1) (28). For example, RPS2 function is partially compromised in mutants that do not accumulate SA during ETI, but RPM1 is not.
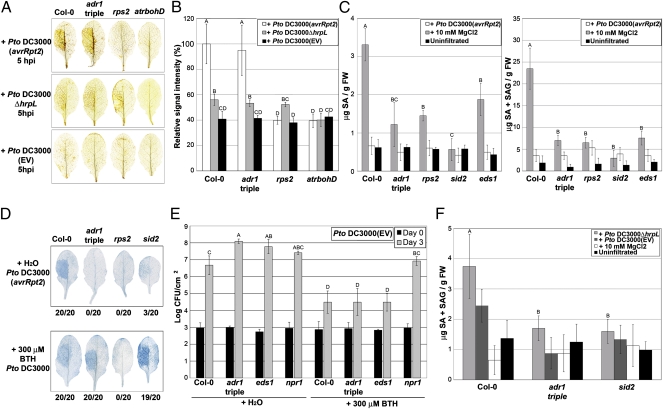
An extracellular burst of superoxide derived from the NADPH oxidase AtrbohD and subsequent hydrogen peroxide (H2O2) production are also hallmarks of early MTI and ETI responses (30). In wild-type plants, this oxidative burst signals cells surrounding an infection site to up-regulate defense and anti-oxidant gene transcription and to down-regulate cell death (14, 31). Reactive oxygen and SA gradients surrounding an infection site are also part of a signal amplification system (32) that sets a cell death threshold controlled by LSD1 (30). We thus speculated that the ADR1 proteins might regulate SA homeostasis following an oxidative burst. Infection with Pto DC3000(avrRpt2) triggered an RPS2-dependent oxidative burst in the triple adr1 triple mutant (Fig. 2 A and B). However, both free and total SA levels were poorly induced in this experiment, to levels as low as in either rps2 or eds1, but slightly more than in the SA biosynthentic mutant sid2 (Fig. 2C).
Fig. 2.
ADR1 proteins are required for effector-independent SA accumulation following a superoxide burst. (A) Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000(avrRpt2), Pto DC3000ΔhrpL, or Pto DC3000(EV). H2O2 accumulation was monitored by 3′,3′-diaminobenzidine (DAB) staining at 5 h post inoculation (hpi). Leaves are representative of 10 individuals. (B) DAB staining shown in A was quantified (mean ± 2 × SE, n = 5). Letters indicate a significant difference following post-ANOVA Student's t test (α = 0.05). (C) Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000(avrRpt2) or with MgCl2. Free (Left) and total SA (Right) were measured at 24 hpi (mean ± 2 × SE, n = 4). (D) Pto DC3000(avrRpt2) was hand-infiltrated into leaves from 4-wk-old plants pretreated with either H2O (Upper) or BTH (Lower) 24 h before bacterial infiltration. Leaves were collected 10 hpi and stained with trypan blue. Leaves are representative of 20 individuals. Numbers indicate how many leaves showed HR out of the total number of leaves analyzed. (E) Four-week-old plants were sprayed with either H2O or BTH. Leaves were hand-infiltrated with Pto DC3000(EV) 2 d post application (dpa). Bacterial growth was monitored at 0 and 3 dpi, mean ± 2 × SE (n = 4). (F) Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000ΔhrpL, Pto DC3000(EV), or MgCl2. Total SA was measured at 9 hpi (mean ± 2 × SE, n = 4) and compared with SA levels from uninfiltrated plants. Letters indicate a significant difference among genotypes infiltrated with Pto DC3000ΔhrpL following post-ANOVA Student's t test (α = 0.05). The experiments in A–F were repeated three times with similar results.
Provision of an SA analog (benzothiadiazole, or BTH) (33) rescued the defective ETI and basal defense responses of the adr1 triple mutant. Pretreatment with BTH restored RPS2-dependent HR in the adr1 triple mutant and the SA biosynthetic mutant sid2 (Fig. 2D). BTH also rescued the enhanced susceptibility to Pto DC3000(EV) detected in both the adr1 triple mutant and the eds1 controls pretreated with water (Fig. 2E). This phenotype required NPR1. We conclude that these deficient ETI and basal defense responses are the consequences of the adr1 triple mutant's inability to accumulate SA. Our results suggest that ADR1 proteins are required for SA accumulation following an intact oxidative burst upon effector and MAMP recognition.
HrpL-deficient Pto DC3000ΔhrpL is a potent MTI trigger because it cannot deliver MTI-suppressing effectors to the host (34). Pto DC3000ΔhrpL induced a weak, but detectable, oxidative burst (Fig. 2 A and B) sufficient to trigger SA accumulation in Col-0 (3), but not in the adr1 triple mutant (Fig. 2F). Hence, recognition of one or more MAMPs expressed by Pto DC3000ΔhrpL activates MTI responses that result in SA accumulation regulated by the ADR1 proteins.
The two best-characterized MTI responses follow specific recognition of peptides derived from either bacterial flagellin (flg22) or elongation factor (elf18) by the PRRs FLS2 and EFR, respectively (35, 36). The accumulation of a functional epitope-tagged ADR1-L2 protein expressed from its native promoter was up-regulated by both flg22 and elf18 peptide treatments, as well as upon BTH application (Fig. S5), yet the adr1 triple mutant exhibited normal oxidative burst, MAPK activation, and callose deposition following treatment with either peptide (Fig. S6). Collectively, these findings indicate that the ADR1 proteins act in MTI downstream or independently of early events subsequent to EFR or FLS2 activation but upstream of SA accumulation.
An Intact P-Loop Domain Is Dispensable for Any of the ADR1-L2 Functions.
STAND proteins bind ATP and most act as ATPases. These include plant NB-LRR proteins (8), a variety of animal NLRs and cell death control proteins (5), and functionally diverse bacterial proteins (9). To date, there is no crystal structure of a full-length NB-LRR or NLR immune receptor. However, structures for both Apaf1 [a functional ATPase (37)] and CED4, which binds ATP (38) but does not hydrolyze it (39), are available (39, 40). Homology modeling of the CCR-NB domain (residues 40–671) of ADR1-L2 with Apaf-1 (residues 1–586) confirmed the location of conserved functionally relevant glycine and lysine residues (GK212/213) analogous to Apaf-1 (GK159/160) in the ATP-binding pocket of the P-loop (Fig. S7 A and B). The P-loop directly interacts with the β-phosphate of ADP (41). An invariant GK residue pair is crucial for this interaction, and mutation of these residues abrogates nucleotide binding and/or ATPase activity and functions across kingdoms (41, 42) (Fig. S7G).
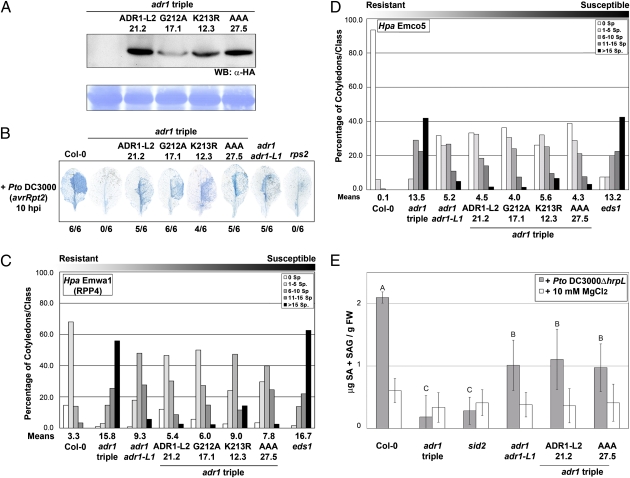
We constructed transgenic adr1 triple-mutant transgenic plants expressing wild-type ADR1-L2, ADR1-L2G212A, ADR1-L2K213R, or ADR1-L2AAA (GKT212/213/214AAA) with C-terminal HA-epitope tags under the control of the native promoter (Fig. S7 C–E). Surprisingly, homozygous transgenic adr1 triple-mutant lines expressing these alleles (Fig. 3A) complemented RPS2-mediated HR (Fig. 3B), RPP4-dependent ETI (Fig. 3C), basal defense (Fig. 3D), and Pto DC3000ΔhrpL-induced SA accumulation (Fig. 3E) to levels comparable to those observed in the adr1 adr1-L1 double mutant. Thus, an intact P-loop motif is dispensable for ADR1-L2 functions in ETI, basal defense, and MTI. Additionally, an allele of ADR1-L2 that lacks a functional P-loop can function in the absence of the other two ADR1 family members.
Fig. 3.
An intact P-loop catalytic domain is dispensable for ADR1-L2 to function in ETI, basal defense, and MTI. (A) Protein extracts were sampled from stable homozygous transgenic adr1 triple plants expressing HA-tagged ADR1-L2, ADR1-L2G212A, ADR1-L2K213R, or ADR1-L2AAA (ADR1-L2, G212A, K213R, AAA, respectively). Numbers indicate the identity of the transgenic lines used. An anti-HA antibody was used to detect ADR1-L2 protein accumulation. Equal loading was verified by Coomassie staining (Lower). (B) Pto DC3000(avrRpt2) was hand-infiltrated into leaves from 4-wk-old plants and stained with trypan blue. Numbers indicate how many leaves showed HR out of the total number of leaves analyzed. (C) Ten-day-old seedlings were inoculated with Hpa Emwa1. Sporangiophores/cotyledon were counted at 5 dpi, and cotyledons were classified as in Fig. 1. (D) Ten-day-old seedlings were inoculated with Hpa Emco5. Sporangiophores were counted at 4 dpi as above. (E) Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000ΔhrpL or MgCl2. Total SA was measured at 9 hpi (mean ± 2 × SE, n = 4) and compared with SA levels from mock-treated plants. Letters indicate a significant difference among genotypes infiltrated with Pto DC3000ΔhrpL following post-ANOVA Student's t test (α = 0.05). The assays in B–D were repeated three times with similar results.
Discussion
We demonstrate that the CCR-NB-LRR ADR1 proteins are functionally distinct innate plant immune receptors. They regulate SA-dependent defense in three contexts: MTI responses against a disarmed pathogen, basal defense against virulent pathogens, and some, but not all, ETI responses. Moreover, at least ADR1-L2 function in these contexts is P-loop–independent. Hence, ADR1-L2 is not activated via the canonical mechanism used by NB-LRR and NLR receptors as microbial sensors, at least for the phenotypes that we describe.
NB-LRR pairs acting in ETI have been reported (12). The lack of physical interaction between the pairs analyzed to date supports a scenario in which the helper NB-LRRs might constitute convergence points in defense responses downstream of recognition and oxidative burst mediated by either PRRs or by NB-LRR sensors activated via effector-driven, P-loop–dependent conformational changes. Our analysis of the CCR-NB-LRR ADR1 proteins is consistent with their evolutionary history and divergence from other CC-NB-LRR family members (13).
We speculate that the CCR-NB-LRR ADR1 proteins might function as signaling scaffolds and regulators of signal transduction processes leading to SA accumulation and consequent defense outputs. This is reminiscent of the function of NLRC5 and NLRP12, NLRs that do not function as microbial sensors per se but rather regulate intracellular signaling pathways (43). These proinflammatory NLRs might function in conjunction with additional NLRs, as suggested by the physical interaction of NLRP12 with NOD2 (44). By analogy, the plant CCR-NB-LRR ADR1 proteins might mediate signal transduction in response to common upstream stimuli. Consequently, the microbial sensor function of immune receptors might be the result of the coordination between effector-mediated sensor activation and a more general signaling function provided by CCR-NB-LRR ADR1 proteins.
The expanded CCR-NB-LRR functions that we describe for at least ADR1-L2 require neither the canonical P-loop motif nor the other two full-length CCR-NB-LRR ADR1 family members. We note the presence of ADR1-L3 in the Col-0 genome (At5g47280), although it lacks approximately the first 190 amino acids at its N terminus and has no reported phenotype. Nevertheless, this protein could play a unique and equally noncanonical role in the phenotypes that we describe. We suggest that the unique signaling functions for at least ADR1-L2 are a consequence of its association with one or more yet-to-be-defined defense machines. The functions that we define for CCR-NB-LRR ADR1 proteins do not preclude an additional, undiscovered, P-loop–dependent function as an effector sensor that, in this context, may associate with and guard the hypothetical signaling machine. Interestingly, a rice protein was recently described that lacks an intact ATP-binding domain and functions in disease resistance against rice blast (45), suggesting a mode of action similar to the CCR-NB-LRR ADR1 proteins. We speculate that additional immune receptors of the NB-LRR and NLR classes might have the following mechanistically separable functions: (i) canonical P-loop–dependent activation via microbial perturbation of the inter- and intramolecular interactions that repress nucleotide exchange and/or hydrolysis and (ii) recognition-independent, P-loop–independent scaffold functioning as part of stress response machinery.
Materials and Methods
ADR1 CCR-NB-LRR Protein Family Nomenclature.
ADR1 (At1g33560) was identified via its ectopic over-expression phenotype, which resulted in constitutive SA-dependent defense gene activation and consequent induction of disease resistance, a common NB-LRR protein gain-of-function phenotype. ADR1-L1 (At4g33300) and ADR1-L2 (At5g04720) were identified by homology (16).
Plant Lines and Pathogens Strains.
T-DNA insertion lines in the Arabidopsis thaliana Col-0 accession were from public collections and were identified by searching the SiGNAL database (http://signal.salk.edu). Details of the mutants, the pathogen strains, and their growth quantification used in this study are provided in SI Materials and Methods.
DNA Manipulations.
Standard techniques of DNA manipulation were used.
Cell Death Assays.
Leaves were harvested and cell death was assessed by Trypan blue staining to visualize dead cells or by conductivity measurements as described in SI Materials and Methods.
Immunoblot Analysis.
Leaves from 4-wk-old plants were harvested, and total proteins were extracted by grinding frozen tissue in a buffer containing 20 mM Tris·HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.1% SDS, 10 mM DTT, and plant protein protease inhibitor mixture (Sigma-Aldrich). Samples were centrifuged at 14,000 × g for 15 min at 4 °C to pellet the debris. Proteins (75 μg) were separated on 7.5% or 12% SDS/PAGE for detection of ADR1-L2 or RIN4, respectively. Proteins were transferred to polyvinylidene difluoride membranes, and Western blots were performed using standard methods. Monoclonal anti-HA antibody (Santa Cruz Biotechnology) antibody was used at a 1:3,000 dilution, whereas anti-RIN4 serum was used at 1:2,000. Signals were detected by enhanced chemiluminescence using ECL Plus (Amersham Biosciences).
Detection of H2O2.
H2O2 was visualized in situ by 3,3′-diaminobenzidine (DAB) staining as described (30). Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000(avrRpt2) or Pto DC3000ΔhrpL at 107 cfu/mL and collected 10 h after infiltration. Leaves were vacuum-infiltrated with a solution containing 1 mg/mL DAB and placed in a dark plastic box under high humidity for an additional 8 h. Leaves were then destained in a solution of 3:1:1 ethanol/lactic acid/glycerol.
Free and Total SA Measurement.
Leaves from 4-wk-old plants were hand-infiltrated with Pto DC3000(avrRpt2) at 107 cfu/mL or with Pto DC3000ΔhrpL at 5 × 107 cfu/mL free SA and glucose-conjugated SA (SAG) (SA + SAG) measurements were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Farid El Kasmi, Petra Epple, Marc Nishimura, and Nuria Sanchez-Coll for critical reading of the manuscript; Colleen Rice for technical support; Jane Parker for sharing eds1-2 seeds; Xiaoyou Zheng for assistance with the SA measurement; Christopher Willet and Darrel Stafford for sharing equipment; Brian Nalley and Susan Whitfield for graphics assistance; and John Craig and Derek Werner for greenhouse assistance. J.L.D. is a Howard Hughes Medical Institute-Gordon and Betty Moore Foundation Plant Science Investigator. This work was funded by the National Institutes of Health (Grants RO1GM066025 and R01GM057171 with an American Recovery & Reinvestment Act supplement to J.L.D.) and the National Science Foundation (Arabidopsis 2010 Program Grant IOS-0929410 to J.L.D.); V.B. was supported by the Human Frontier Science Program (Grant LT00905/2006-L); S.T. was supported by the National Natural Science Foundation of China (Grant 30671180).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113726108/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 3.Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- 4.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 5.Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 7.Philpott DJ, Girardin SE. Nod-like receptors: Sentinels at host membranes. Curr Opin Immunol. 2010;22:428–434. doi: 10.1016/j.coi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 9.Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: A mechanistic insight into signaling by STAND proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Tameling WI, Joosten MH. The diverse roles of NB-LRR proteins in plants. Physiol Mol Plant Pathol. 2007;71:126–134. [Google Scholar]
- 11.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eitas TK, Dangl JL. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011;24:918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 14.Jabs T, Dietrich RA, Dangl JL. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 15.Aviv DH, et al. Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 2002;29:381–391. doi: 10.1046/j.0960-7412.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- 16.Chini A, Loake GJ. Motifs specific for the ADR1 NBS-LRR protein family in Arabidopsis are conserved among NBS-LRR sequences from both dicotyledonous and monocotyledonous plants. Planta. 2005;221:597–601. doi: 10.1007/s00425-005-1499-3. [DOI] [PubMed] [Google Scholar]
- 17.Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 2004;38:810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- 18.Grant JJ, Chini A, Basu D, Loake GJ. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact. 2003;16:669–680. doi: 10.1094/MPMI.2003.16.8.669. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackey D, Holt BF., III Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 21.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 22.Holub EB, Beynon JL, Crute IR. Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol Plant Microbe Interact. 1994;7:223–239. [Google Scholar]
- 23.Holub EB, Beynon J. Symbiology of mouse-ear cress (Arabidopsis thaliana) and oomycetes. Adv Bot Res. 1997;24:228–273. [Google Scholar]
- 24.Kim HS, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt BF., III Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 27.Loake G, Grant M. Salicylic acid in plant defence: The players and protagonists. Curr Opin Plant Biol. 2007;10:466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 29.McDowell JM, et al. Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 2000;22:523–529. doi: 10.1046/j.1365-313x.2000.00771.x. [DOI] [PubMed] [Google Scholar]
- 30.Torres MA, Jones JD, Dangl JL. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- 31.Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL. LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol Plant Microbe Interact. 1999;12:1022–1026. doi: 10.1094/MPMI.1999.12.11.1022. [DOI] [PubMed] [Google Scholar]
- 32.Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawton KA, et al. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- 34.Block A, Alfano JR. Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr Opin Microbiol. 2011;14:39–46. doi: 10.1016/j.mib.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 36.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Benedict MA, Ding L, Núñez G. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J. 1999;18:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chinnaiyan AM, Chaudhary D, O'Rourke K, Koonin EV, Dixit VM. Role of CED-4 in the activation of CED-3. Nature. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 39.Yan N, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 40.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 41.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 42.Tameling WI, et al. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. [DOI] [PubMed] [Google Scholar]
- 44.Wagner RN, Proell M, Kufer TA, Schwarzenbacher R. Evaluation of Nod-like receptor (NLR) effector domain interactions. PLoS ONE. 2009;4:e4931. doi: 10.1371/journal.pone.0004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi N, et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.