Abstract
EF4 (LepA), a strongly conserved protein, is important for bacterial growth and functional protein biosynthesis under certain conditions and is quite similar structurally to the translocase EF-G. The elongation cycle in protein synthesis is characterized by ribosome oscillation between pretranslocation (PRE) and posttranslocation (POST) complexes. Here, using ensemble single turnover and equilibrium experiments, as well as single molecule FRET measurements, we demonstrate that EF4 can compete with EF-G for binding to the PRE complex. Such EF4 binding results in formation of a complex, denoted X3, that effectively sequesters a catalytically active ribosome, leading to a transient inhibition of elongation that provides a mechanism for optimization of functional protein synthesis. Earlier [Liu H, et al. (2010) J Mol Biol 396:1043–1052] we demonstrated that EF4 also reacts with POST complex, leading to the formation of a complex, I3, that appears to be identical with X3. Our present results strongly suggest that PRE complex is the principal target of EF4 action on translation, rather than POST complex as had been previously supposed.
Keywords: pretranslocation complex, posttranslocation complex, hybrid state, fluorescent tRNA
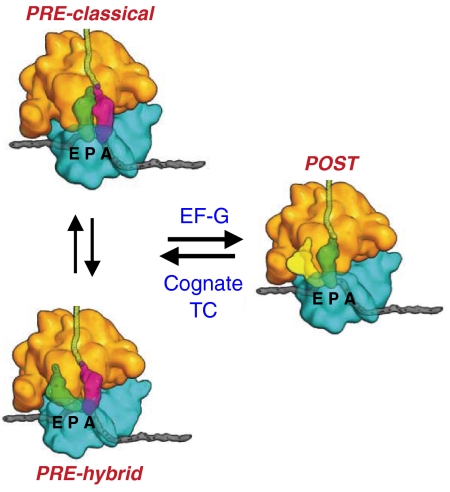
The elongation cycle in protein synthesis is characterized by ribosome oscillation between pretranslocation (PRE) and posttranslocation (POST) complexes [Fig. 1 (1)]. In the PRE complex, peptidyl-tRNA and deacylated tRNA fluctuate between classical (A/A and P/P sites, respectively) and hybrid (A/P and P/E sites, respectively) states (2, 3). Binding of the translocase EF-G•GTP induces translocation of peptidyl tRNA and deacylated tRNA into the P and E sites, respectively, with movement of the mRNA by one codon and GTP hydrolysis. Dissociation of EF-G•GDP and Pi leads to formation of the POST complex. Aminoacyl tRNA, in the form of a TC with EF-Tu and GTP and cognate to the codon in the A site, binds to the POST complex, either prior to or following deacylated tRNA dissociation. Such binding is followed by GTP hydrolysis, peptide bond formation leading to elongation of the nascent peptide chain, and dissociation of EF-Tu•GDP and Pi, leading to PRE complex formation and completing the elongation cycle. Interestingly, POST complexes can spontaneously back translocate to PRE complexes, but at rates too slow to be important as an in vivo process (4, 5).
Fig. 1.
Ribosome oscillation between PRE and POST complexes during elongation. [Reprinted by permission from Macmillan Publishers Ltd: Ref. 1, copyright 2009.] The ribosome has three binding sites for tRNA, A, P, and E. tRNAs can move independently with respect to the 50 S and 30 S subunits during the elongation cycle, leading to the formation of hybrid tRNA binding sites A/P and P/E. EF-G•GTP converts PRE complexes into POST complex. Binding of cognate ternary complex (TC) and dissociation of deacylated tRNA from the E site converts POST complex into PRE complex.
EF4 (LepA), which has recently been shown to interact with POST complex and act as a partial back translocase (6, 7) and has been identified as a bona fide translation factor (8), is a highly conserved protein present in bacteria, mitochondria, and chloroplasts but not in archaea or in the cytoplasm of eukaryotes. EF4 and EF-G share strong structural similarities, with four out of the five EF-G domains—I, II, III, and V—resembling corresponding domains in EF4. EF4 also has a unique C-terminal domain with an unusual fold, but lacks domain IV and the G′ subdomain of domain I that are present in EF-G (9). The lack of domain IV in EF4 is noteworthy because this domain in EF-G binds to the ribosome in a position overlapping that of A-site tRNA, thereby preventing back translocation (10).
EF4 has ribosome-dependent GTPase activity and occupies the characteristic G-protein position on the ribosome (11) in the PRE-like complex, denoted PRE(L), that is formed on prolonged incubation of POST complex with EF4 (12). Although ΔEF4 cells grown in rich medium have no phenotype (13), recent results have demonstrated that EF4 can improve the yield of functional protein synthesis (6) and that, under certain stress conditions, including high salt, low pH, and low temperatures, a ΔEF4 strain is overgrown by wild-type bacterial cells (14). In addition, bacterial ΔEF4 strains have been shown to be hypersensitive to potassium tellurite and to penicillin [Escherichia coli (8)] and to have increased levels of the calcium-dependent antibiotic nonribosomal peptide synthetases [Streptomyces coelicor (15)].
These somewhat disparate results raise the question of what role EF4 plays as a translation factor. The strong structural similarity of EF4 and EF-G raises the possibility that EF4 might, in addition to its interaction with POST complex, also interact with PRE complex, the substrate for EF-G. Indeed, we demonstrate below that EF4 reacts with PRE complex as rapidly as does EF-G and in a competitive fashion with EF-G, leading to the formation of a complex, X3, having properties intermediate between those of PRE and POST complexes. For example, EF4 binding to a PRE complex shifts the distribution between classical and hybrid states strongly in favor of the hybrid state. Formation of X3 transiently sequesters a catalytically active ribosome, suggesting a specific role for EF4 in optimizing functional protein synthesis, as discussed below.
Earlier we demonstrated that back translocation in the presence of EF4 proceeds from POST complex to PRE complex via at least three intermediates: i.e., POST → I1 → I2 → I3 → PRE(L) (7). I3, which is distinct from the intermediate species, INT, formed during EF-G catalysis of PRE to POST conversion (16), accumulates transiently in the presence of EF4, which does not catalyze I3 to PRE(L) conversion. Our present results demonstrate that X3 is similar, if not identical, to I3 and suggest that X3/I3 formation during protein synthesis is more likely to occur from EF4 reaction with PRE complex than with POST complex.
Results
EF4 Interaction with PRE Complex.
Reversible increase in Flu-mRNA014 fluorescence.
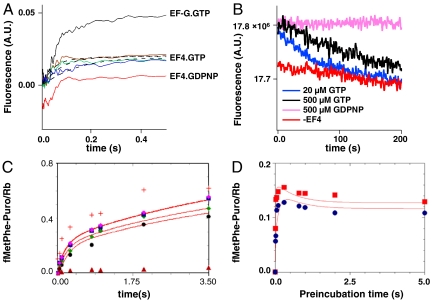
Changes in the fluorescence of 3′-labeled mRNA have been used to monitor the movement of mRNA during EF-G-facilitated translocation of PRE complex (17, 18) and EF4-facilitated partial back translocation of POST complex (7). The time-dependent change in fluorescence of a PRE complex programmed with Flu-mRNA014 on rapid mixing EF4•GTP (5 μM) is very similar to that seen on rapid mixing with EF-G•GTP (5 μM) (Fig. 2A) with a lag period followed by an increase, although the magnitude of the increase seen with EF4 is only about half of that seen with EF-G. From previous work (16, 17), 5 μM EF-G is close to saturating with respect to the rate of translocation of a PRE complex. The results presented in Fig. 2A demonstrate that 5 μM is also saturating with respect to both the rate and magnitude of fluorescence change induced by added EF4. Furthermore, such change does not depend on GTP hydrolysis, because an essentially identical change is observed when the nonhydrolyzable analogue GDPNP is added in place of GTP. Added viomycin (1 mM) or spectinomycin (3 mM) have no effect on the rate or magnitude of EF4•GTP-induced fluorescence change (Fig. S1), contrasting sharply with the ability of these antibiotics to inhibit EF-G dependent translocation (16, 19).
Fig. 2.
EF4 interaction with PRE complex. (A) PRE complexes programmed with Flu-mRNA014 (0.1 μM) were rapidly mixed in a stopped-flow spectrophotometer with 5 μM EF-G•GTP (black trace) or EF4•GTP (1 μM, blue trace; 2.5 μM, green trace; 5 μM, brown trace) or EF4•GDPNP (5 μM, red trace). The latter trace is offset on the Y axis for clarity. The magnitudes of the increases seen with 5 μM EF4•GTP and 5 μM EF4•GDPNP were indistinguishable. Dotted lines are fits to Scheme 1. (B) Reversibility of X3 formation. PRE complex (0.1 μM) containing Flu-MFK014 was mixed with 3 μM EF4 and 20 μM GTP (blue trace) or 500 μM GTP (black trace) or 500 μM GDPNP (pink trace) or PRE complex was mixed without the addition of EF4 (red trace). Excitation was at 460 nm and emission was monitored at 527 nm. (C) Isolated PRE complexes (0.1 μM) programmed with mRNA MFK were rapidly mixed with 3 mM puromycin and the indicated concentrations of EF4•GTP (0.5 μM, black circles; 1 μM, green diamonds; 2 μM, blue squares; 5 μM, pink diamonds), or 3 μM EF-G•GTP (red cross) or buffer (brown triangles), quenched at the indicated times, and fMetPhe-puromycin formation was quantified. Red lines are fits to Scheme 1. (D) PRE complexes (0.1 μM) as in C were preincubated with 1 μM EF4•GTP (blue circles) or 2 μM EF4•GTP (red squares) for various times, then rapidly mixed with 3 mM puromycin and allowed to react for 0.3 s prior to quenching and quantification of fMetPhe-puromycin formation. Red lines are fits to Scheme 1.
The increase seen in Fig. 2A is slowly reversible when EF4•GTP but not EF4•GDPNP is added to PRE complex (Fig. 2B). When the initial GTP concentration is 500 μM, a lag period of approximately 30 s is seen prior to the onset of the decrease in fluorescence, which proceeds with a rate constant of 0.007 ± 0.001 s-1. Decreasing GTP concentration to 20 μM strongly decreases the lag period to approximately 2 s, whereas the rate constant for fluorescence decrease is similar (0.009 ± 0.001 s-1). These results are most simply explained by the assumptions that hydrolysis of EF4•GTP to EF4•GDP on the ribosome (6, 7) weakens the strong binding displayed by EF4•GTP (and mimicked by EF4•GDPNP) and that rapid exchange of GTP for GDP maintains high affinity EF4 binding, preserving the fluorescence increase seen in Fig. 2A. When all the GTP in solution is hydrolyzed, EF4•GDP dissociates from the ribosome, leading to slow reformation of PRE complex, and a concomitant fluorescence decrease. This explanation accounts for the dramatic decrease in the length of the lag period as GTP concentration is lowered from 500 to 20 μM.
Increase in peptidyl-tRNA reactivity toward puromycin.
fMetPhe-tRNAPhe reacts with puromycin to form fMetPhe-puromycin at a rate that is 103–104-fold slower when it is bound in a PRE complex than when it is bound in a POST complex (7, 16, 19–21), permitting fMetPhe-tRNAPhe reactivity toward puromycin to be used as a measure of peptidyl-tRNA positioning in the ribosome. Rapid mixing of PRE complex with EF4•GTP and puromycin led to a biphasic increase in fMetPhe-puromycin formation, with an initial rapid rate of increase giving way eventually to a slower rate (Fig. 2C). As is the case with the mRNA fluorescence increase, both phases are clearly saturated at 5 μM EF4•GTP.
The increase in puromycin reactivity that follows EF4•GTP addition to PRE complex was also examined using a double incubation experiment, in which PRE complex was preincubated for various times with EF4•GTP before being rapidly mixed for a fixed amount of time with puromycin (Fig. 2D). This experiment, showing that fMetPhe-puromycin formation goes through a maximum at short preincubation times, indicates transient formation of a species with higher puromycin reactivity than that which is formed on more prolonged preincubation.
A quantitative kinetic scheme for EF4•GTP interaction with PRE complex.
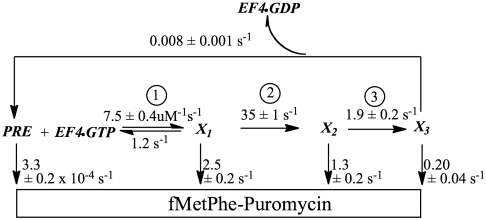
The results in Fig. 2 A–D can be quantitatively fit to the kinetic scheme (Scheme 1), in which EF4•GTP converts PRE complex into a complex denoted as X3, which is formed via complexes X1 and X2 in a three-step mechanism. Evidence for the first two of these steps comes from the stopped-flow fluorescence results (Fig. 2A), which show a lag phase followed by an increase in fluorescence intensity that is complete within 0.1 s. Evidence for the third step is provided by the much slower decline in the rate of fMetPhe-puromycin formation (Fig. 2D), which is only complete after 1.5 s.
Scheme 1.
A quantitative kinetic scheme for EF4 interaction with PRE complex. Experimental conditions were as described in the legend to Fig. 2.
In Scheme 1, the increase in Flu-mRNA014 fluorescence results from conversion of X1 to X2. X1 has a puromycin reactivity comparable to that of POST complex, whereas X2 and X3 have 2-fold and 12-fold lower reactivities, respectively. The high puromycin reactivity of X1 implies that initial binding of EF4 leads to full accommodation of peptidyl tRNA into the acceptor position of the peptidyl transferase center. The lower puromycin reactivities of X2 and X3 may result from direct EF4 interaction with the 3′ end of peptidyl tRNA (12), which could follow EF4•GTP hydrolysis. Dissociation of EF4•GDP from X3, which may be quite rapid, allows the slow reformation of PRE complex.
The puromycin reactivities and fluorescence intensities of X1, X2, and X3 clearly demonstrate that movement of the highly flexible 3′ end of peptidyl tRNA is largely decoupled from mRNA movement. Thus, the two largest changes in puromycin reactivity are seen during X1 and X3 formation, both of which occur with no change in mRNA fluorescence, whereas X2 formation is accompanied by a relatively large change in mRNA fluorescence and only a modest change in puromycin reactivity. We previously observed similar decoupling during EF4 facilitated formation of I3 from POST complex (7).
EF4 Competes with EF-G for Interaction with PRE Complex.
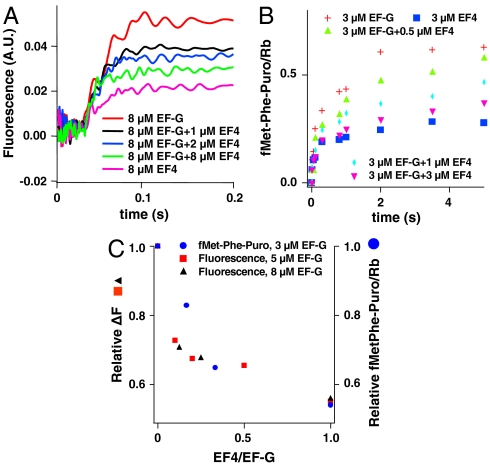
Measuring either the fluorescence change of Flu-mRNA014 or the reactivity of fMetPhe-tRNAPhe toward puromycin when PRE complex is rapidly mixed with fixed concentrations of EF-G•GTP and varying concentrations of EF4•GTP provides evidence that EF4 competes with EF-G for binding to PRE complex (Fig. 3 A and B). In both cases, the response seen in the presence of EF-G•GTP alone (POST complex formation) is converted monotonically into the response seen with added EF4•GTP alone (X3 complex formation) as the EF4/EF-G ratio is increased (Fig. 3C). Evidence that the change is a result of EF4 competition with EF-G is provided by the similarity in ratio of EF4/EF-G needed to produce a given decrease in fluorescence change as EF-G concentration is increased from 5 to 8 μM. As determined from both fluorescence change and puromycin reactivity measurements, an EF4/EF-G ratio of 0.1–0.2 suffices to achieve half conversion of PRE complex to X3, suggesting an apparent affinity of EF4 for PRE complex that is some 5–10 times higher than that of EF-G.
Fig. 3.
EF4 competition with EF-G for PRE complex. (A) PRE complexes programmed with Flu-mRNA014 (0.1 μM) were rapidly mixed in a stopped-flow spectrophotometer with EF-G•GTP, EF4•GTP or mixtures of EF-G•GTP and EF4•GTP at the indicated concentrations. (B) PRE complexes (0.1 μM) programmed with mRNA MFK were rapidly mixed with 3 mM puromycin and the indicated concentrations of EF-G•GTP, EF4•GTP or mixtures of EF-G•GTP and EF4•GTP, quenched at the indicated times, and fMetPhe-puromycin formation was quantified. (C) Relative changes in Flu-mRNA014 fluorescence (after 1 s; see A) and fMetPhe-puromycin formation (after 3.5 s; see B) as a function of the EF4/EF-G concentration ratio.
EF4•GTP Reaction with PRE Complex Favors Hybrid State Formation.
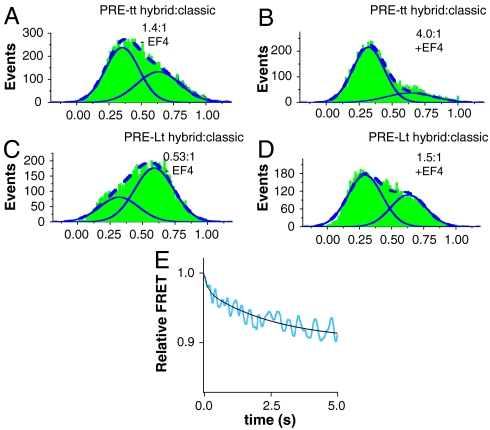
Single molecule FRET (smFRET) studies have shown that PRE complexes are distributed between hybrid and classical states (2, 3) and that a fraction of these complexes fluctuate between these states (2, 22). Following Chen et al. (22), we used smFRET to examine PRE complexes formed using two different sets of fluorescently labeled materials. PRE-Lt complex contains L11 labeled with Cy3 at position 87 (L11Cy3) and fMetPhe-tRNAPhe (Cy5) in the A site and unlabeled tRNAfMet in the P site. PRE-tt complex contains tRNAArg (Cy3) in the P site and ArgPhe-tRNAPhe (Cy5) in the A site. We found that classical and hybrid states of PRE-Lt complex had FRET efficiencies of 0.60 and 0.33, respectively, and that the corresponding efficiencies for the PRE-tt complex were 0.63 and 0.36, respectively. Addition of EF4•GTP to fluctuating PRE-Lt (Fig. S2) and PRE-tt complexes dramatically shifts the ratio of hybrid:classical state from 0.53∶1 to 1.5∶1 (Lt complexes) and from 1.4∶1 to 4∶1 (tt complexes) (Fig. 4 A–D). Shifts in distribution favoring hybrid states, albeit less dramatic, are also seen for nonfluctuating PRE complexes (Fig. S3).
Fig. 4.
The hybrid state is favored in X3. (A–D) Single molecule FRET determinations of the influence of added EF4•GTP on the distribution of fluctuating ribosome complexes (Fig. S2) between high FRET (classical) and low FRET (hybrid) states. The sum of the two states is displayed as a dashed line. X axes show FRET values. (A) PRE-tt complexes containing P-site tRNAArg (Cy3) and A-site ArgPhe-tRNAPhe (Cy5). (B) Same as A, but following addition of EF4•GTP (2 μM). (C) PRE-Lt complexes containing L1187-Cy3 ribosomes and A-site fMetPhe-tRNAPhe (Cy5). (D) Same as C, but following addition of EF4•GTP (2 μM). EF4 addition also favors hybrid state for nonfluctuating complexes (Fig. S3). (E) Ensemble stopped-flow FRET measurements following addition of EF4•GTP (3 μM) to a PRE-Lt complex (0.16 μM) in buffer A at 25 °C, with excitation at 530 nm and monitoring at 680 ± 10 nm. Shown is the corrected relative FRET (see Fig. S4). The solid line is fit to the rate constants shown in Scheme 1, with relative FRET values for the PRE, X1, X2, and X3 complexes of 1.00, 1.00, 0.97, and 0.91, respectively.
Because EF4•GTP addition to PRE-Lt complex led to increased hybrid state formation having lower FRET efficiency, we reasoned that the ensemble kinetics of this process could be monitored by measuring the time dependence of the expected decrease in overall FRET efficiency. Indeed, rapid mixing of PRE-Lt complex with EF4•GTP leads to a biphasic decrease in relative FRET efficiency, on time scales consistent with X2 and X3 formation (Scheme 1), with no FRET efficiency change occurring on X1 formation (Fig. 4E and Fig. S4). These results suggest that increased stabilization of the hybrid state occurs as a result of conformational changes that follow EF4•GTP binding, rather than from EF4•GTP binding itself.
X3 and I3 Are Similar and Possibly Identical.
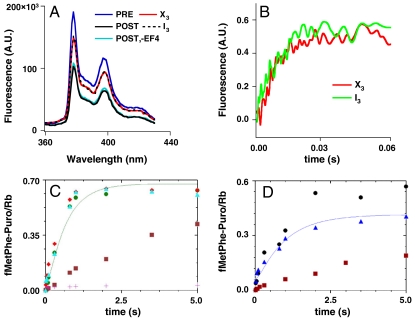
By four experimental criteria, described below, the complex X3 formed on reaction of PRE complex with EF4 (Scheme 1) is very similar, if not identical, to the previously identified complex I3 formed on EF4 interaction with POST complex (7). First, I3 and X3 have identical puromycin reactivities toward peptidyl-tRNA (apparent rate constants of 0.13 ± 0.02 s-1 and 0.13 ± 0.03 s-1 for I3 and X3, respectively, at 2 mM puromycin) and quite different from the higher and much lower reactivities of POST and PRE complexes, respectively (Scheme 1). Second, the fluorescence change of Flu-mRNA014 when PRE complex is converted to X3 falls in between the change seen on conversion of PRE to POST complex (Figs. 2A and 3A). This parallels the partial change observed when POST complex is converted to I3 as compared with the change seen on POST to PRE conversion (7). A more quantitative illustration of this point comes from comparison of the emission spectra of PRE, POST, I3, and X3 complexes for ribosomes programmed with a fluorescent mRNA, pyrene-mRNA09 (18). Such ribosomes display a larger fluorescence change on conversion of PRE to POST complex than ribosomes programmed with Flu-mRNA014. Translocation of pyrene-mRNA09 by one codon results in a 46% decrease in fluorescence intensity as PRE complex is converted to POST, and both I3 and X3 show an identical decrease, 21%, as compared with PRE complex (Fig. 5A).
Fig. 5.
Similarities of X3 and I3. For all experiments, preformed PRE or POST complexes were added at a final concentration of 0.1 μM and incubations were performed at 25 °C. (A) Fluorescence spectra of ribosome complexes programmed with pyrene-labeled mRNA09. Samples were excited at 343 nm, and the emission spectrum from 360–430 nm was recorded. I3 was formed by incubating POST complex with 3 μM EF4•GDPNP and 0.15 μM tRNAfMet for 4 min. A control sample was prepared by incubating POST complex with only GDPNP and 0.15 μM tRNAfMet for 4 min. X3 was formed by preincubating PRE complex with 3 μM EF4•GDPNP and 0.1 μM tRNAfMet 30 s. (B–D) Rapid EF-G conversion of complexes X3 or I3 to POST complex as monitored by either Flu-mRNA014 fluorescence (B) or peptidyl-tRNA reactivity toward puromycin (C, X3), (D, I3). In B and C, X3 was formed by rapid mixing of PRE complex with 0.5 μM EF4•GTP and preincubation for 0.5 or 5 s, prior to a second rapid mixing step with either EF-G•GTP (3 μM, B) or EF-G•GTP (5 μM, C) and puromycin (2 mM, C) and further incubation for the indicated times. In B and D, I3 was formed by rapid mixing of POST complex with 1.25 μM EF4•GDPNP and preincubation for 2.5 min, prior to a second rapid mixing step with either EF-G•GTP (3 μM, B) or EF-G•GTP (5 μM, D) and puromycin (1.5 mM, D) and further incubation for the indicated times. Puromycin reactions in C and D were terminated by quenching. (B) The total fluorescence changes have been equalized for ease of comparison. (C) •, 0.5 s preincubation; ▴, 5 s preincubation. Control samples: +, PRE complex; ▪, as in •, but with EF-G•GTP omitted; □, PRE complex rapidly mixed directly with EF-G•GTP and puromycin. (D) ▴, double mixing experiment. Control samples: •, POST complex; ▪, as in ▴, but with EF-G•GTP omitted.
The third and fourth criteria concern the rates at which incubation of X3 or I3 with EF-G•GTP leads to formation of POST complex, as determined by measurement of either mRNA fluorescence or of puromycin reactivity toward fMetPhe-tRNA. The experiments examining this point again involved a double incubation protocol. X3 or I3 were prepared by a preincubation step (X3: PRE complex with EF4•GTP, for 0.5 s or 5 s; I3: POST complex with EF4•GDPNP for 2.5 min), which was followed by mixing with EF-G•GTP. EF-G•GTP addition to either X3 or I3 leads in both cases to very rapid increases in fluorescence intensity (Fig. 5B) corresponding to POST complex formation (Figs. 2A and 3A), with rates and magnitudes that were indistinguishable. Similarly, EF-G•GTP and puromycin addition to either X3 (Fig. 5C) or I3 (Fig. 5D) leads in both cases to rapid fMetPhe-puromycin formation, characteristic of POST complex. For X3 the apparent rate constant and stoichiometry of fMetPhe-puromycin formation are quite similar to those found on EF-G•GTP and puromycin addition to PRE complex. For I3, the apparent rate constant of fMetPhe-puromycin formation is similar to that found for puromycin addition to POST complex, but the stoichiometry is somewhat lower, due to the partial formation of PRE(L) complex, which is only slowly converted to POST complex on EF-G•GTP addition (Fig. S5).
Effect of High Mg2+ Concentration.
Pech et al. (14) have shown that whereas added EF4 has little effect on the rate of in vitro poly(U)-directed poly(Phe) synthesis under optimal buffer conditions, such as those used in the present experiments reported in Figs. 2–5, it markedly relieves the inhibition observed when Mg2+ concentration is raised from 4.5 to 14 mM, a buffer change that is used to simulate an in vivo stress condition. We find that increasing Mg2+ concentration to 14 mM has no discernible effect on either EF-G catalyzed translocation, or on EF4 competition with EF-G for PRE complex, as measured by Flu-mRNA014 fluorescence change (Fig. S6) and has at most minor effects on EF4-catalyzed partial back translocation of POST complex to X3/I3 complex (7), as measured by fluorescence changes of either Flu-mRNA014 (Fig. S7A and B) or tRNAfMet (prf) (Fig. S7 C and D). However, raising Mg2+ does strongly inhibit the already slow conversion of X3/I3 to a PRE(L) complex (Fig. S7 C–F) (7). Such inhibition may reflect stabilization of EF4 interaction with the X3/I3 complex at high Mg2+ concentration.
Discussion
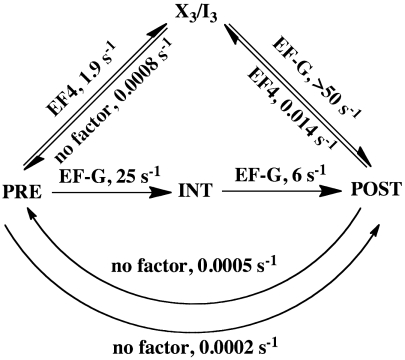
Direct in vitro and in vivo experiments (23–26), as well as inferences drawn from bioinformatic analyses of codon usage (27, 28), provide strong evidence that transient pausing of normal polypeptide elongation can increase the fraction of active protein produced by facilitating cotranslational protein folding (26). Qin et al. (6) have observed that added EF4 significantly increases the fraction of active protein made in a cell-free coupled transcription–translation system. The results presented here and in our earlier study (7) suggest that EF4 could effect transient pausing of polypeptide elongation via its ability to transiently sequester active ribosomes in the form of the X3/I3 complex, by its reaction with either the PRE or POST complex (Scheme 2). Although the X3/I3 complex formed by EF4•GTP interaction with PRE complex can be subsequently rapidly converted by EF-G into POST complex, conversion of PRE complex to POST complex via X3/I3 complex will proceed almost threefold slower than via INT complex (overall rate constants of approximately 1.8 s-1 and approximately 4.8 s-1, respectively) even in a single turnover reaction involving one EF4•GTP hydrolysis. Formation of POST complex would be slowed further when EF4 is present in sufficient concentration to effectively compete with EF-G for the X3/I3 complex, resulting in multiple rounds of EF4•GTP hydrolysis. Such inhibition of POST complex formation should be rather infrequent (< 1 elongation cycle in 5–10) under normal growth conditions, when EF4 is present in bacterial cytoplasm at approximately 0.02 copies/ribosome (14), some 50-fold less than EF-G, based on results showing that EF-G concentration equals that of the ribosome (29). However, EF4 concentration in the bacterial cytoplasm rises by almost an order of magnitude under conditions of growth stress, such as high Mg2+ concentration (14), significantly raising the probability of transient EF4 inhibition of PRE complex conversion to POST complex. Moreover, the results presented in Fig. S7 C–E suggest that high Mg2+ concentration also increases the lifetime of the X3/I3 complex.
Scheme 2.
EF4 modulates the elongation cycle. Kinetic scheme summarizing the modulating effects of EF4 on the elongation cycle. Based on results presented herein, X3 is assumed to be identical to I3. Rate constants are from work reported here, or by Pan et al. (16) (EF-G catalyzed translocation), by Liu et al. (7) (EF4-catalyzed POST to X3/I3 conversion; X3/I3 conversion to PRE in absence of EF4), or by Semenkov et al. (20) (translocation in absence of EF-G).
Formation of the X3/I3 complex via EF4•GTP interaction with POST complex (6, 7) presents another potential mechanism for transient inhibition of elongation. However, the sluggishness of EF4-facilitated X3/I3 complex formation from POST complex (Scheme 2) reduces the likelihood that this mechanism will, under normal conditions, lead to transient inhibition of elongation, although it might be pertinent under conditions of amino acid starvation. Indeed, there is evidence that EF4•GTP competes with the EF-Tu•GTP•Ala-tmRNA complex for binding to stalled ribosomes and inhibits A-site mRNA cleavage in such ribosomes (8).
As the above discussion makes clear, although EF4 was first described as an elongation factor because of its ability to react with POST complex and facilitate partial back-translocation (6, 7), our current results strongly suggest that its more important transient inhibitory effect on elongation derives from its very effective competition with EF-G for binding to PRE complex. Such inhibition would be expected to have salutary effects on the activity of the protein being synthesized during the decoding of only a limited number of mRNA codons within the full open reading frame, raising the question of how selectivity for such codons can be achieved. An intriguing speculation, which remains to be tested, is that the competition between EF-G and EF4 for PRE complex (or, less likely, between ternary complex and EF4 for POST complex) might be sensitive to the binding of specific mRNA sequences that result in alterations of ribosome conformation in the structurally flexible GTPase associated center (11).
Another open question concerns the structure of the EF4-bound X3/I3 complex, which can be formed from either the PRE or POST complex and has properties (mRNA fluorescence, puromycin reactivity, fraction present in the hybrid state, and rapidity of reaction with EF-G to form POST complex) intermediate between them. Recent time-resolved cryo-EM studies have provided evidence that PRE and POST complexes contain as many as five and three distinct structural states, respectively, that suggest trajectories coupling tRNA movement (PRE-classical via several intermediates to full PRE-hybrid and then to the various POST structures) with global and local conformational changes of the ribosome (30). It clearly would be of interest to determine whether overall ribosomal conformation and mRNA and tRNA placement within the EF4-bound X3/I3 complex resembles that found in any of the eight structures described in ref. 30, in particular those closest to the PRE:POST interface.
In summary, we demonstrate herein that EF4 competes effectively with EF-G for interaction with PRE complex, leading to rapid formation of a species, X3 (Scheme 1), that has properties intermediate between those of PRE and POST complexes, reacts very rapidly with EF-G to form POST complex, and appears to be identical to a species I3 that is formed more slowly by reaction of EF4 with POST complex. Our results strongly suggest that PRE complex is the principal target of EF4 action on translation, rather than POST complex as had been previously supposed.
Formation of the X3/I3 complex, principally from PRE complex, provides a plausible mechanism for transient inhibition of the ribosome elongation cycle that could optimize functional protein synthesis.
Materials and Methods
See also SI Materials and Methods. Most materials were obtained and complexes (initiation, ternary, PRE, and POST) were prepared as previously described (6, 7, 22, 31). Pyrene-mRNA09 with sequence AAG GAG GUA AAA AUG UUU GCU (initiator codon underlined) was a kind gift from Simpson Joseph (University of California, San Diego, La Jolla, CA). All rate measurements and associated incubation steps were carried out at 25 °C in buffer A [20 mM Hepes-KOH, pH 7.5 (at 0 °C), 150 mM NH4Ac, 4.5 mM MgAc2, 4 mM β-mercaptoethanol, 0.05 mM spermine, and 2 mM spermidine]. Measurements of steady-state and stopped-flow fluorescence, and of single molecule FRET, and rapid quench experiments were carried out as described (7, 22).
Supplementary Material
Acknowledgments.
We thank Professor Simpson Joseph (University of California, San Diego, La Jolla, CA) for a generous gift of pyrene-labeled mRNA09 and Professor Knud Nierhaus for sharing results with us prior to publication. This work was supported by National Institutes of Health Grants GM071014 and GM080376.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103820108/-/DCSupplemental.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konevega AL, et al. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 6.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: The EF4 (LepA) effect on back-translocation. J Mol Biol. 2010;396:1043–1052. doi: 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010;92:157–163. doi: 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci USA. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 13.Dibb NJ, Wolfe PB. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pech M, et al. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci USA. 2011;108:3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badu-Nkansah A, Sello JK. Deletion of the elongation factor 4 gene (lepA) in Streptomyces coelicolor enhances the production of the calcium-dependent antibiotic. FEMS Microbiol Lett. 2010;311:147–151. doi: 10.1111/j.1574-6968.2010.02083.x. [DOI] [PubMed] [Google Scholar]
- 16.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 18.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 19.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 20.Semenkov YP, Shapkina TG, Kirillov SV. Puromycin reaction of the A-site bound peptidyl-tRNA. Biochimie. 1992;74:411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 21.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, et al. Single molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 24.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 25.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 26.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 27.Purvis IJ, et al. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J Mol Biol. 1987;193:413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- 28.Makhoul CH, Trifonov EN. Distribution of rare triplets along mRNA and their relation to protein folding. J Biomol Struct Dyn. 2002;20:413–420. doi: 10.1080/07391102.2002.10506859. [DOI] [PubMed] [Google Scholar]
- 29.Gordon J. Regulation of the in vivo synthesis of the polypeptide chain elongation factors in Escherichia coli. Biochemistry. 1970;9:912–917. doi: 10.1021/bi00806a028. [DOI] [PubMed] [Google Scholar]
- 30.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 31.Pan D, Qin H, Cooperman BS. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. RNA. 2009;15:346–354. doi: 10.1261/rna.1257509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.