Abstract
We previously described four small molecules that reduced the growth of lung adenocarcinoma cell lines with either epidermal growth factor receptor (EGFR) or KRAS mutations in a high-throughout chemical screen. By combining affinity proteomics and gene expression analysis, we now propose superoxide dismutase 1 (SOD1) as the most likely target of one of these small molecules, referred to as lung cancer screen 1 (LCS-1). siRNAs against SOD1 slowed the growth of LCS-1 sensitive cell lines; conversely, expression of a SOD1 cDNA increased proliferation of H358 cells and reduced sensitivity of these cells to LCS-1. In addition, SOD1 enzymatic activity was inhibited in vitro by LCS-1 and two closely related analogs. These results suggest that SOD1 is an LCS-1–binding protein that may act in concert with mutant proteins, such as EGFR and KRAS, to promote cell growth, providing a therapeutic target for compounds like LCS-1.
Keywords: chemical biology, target discovery and validation, gene interaction network
Lung cancer is the leading cause of cancer-related deaths in the United States and worldwide (1). Efforts to characterize the four histological forms of lung cancers by genetic and biochemical methods have increased our understanding of this disease and offered opportunities for development of targeted therapies. For example, many lung adenocarcinomas carry somatic mutations affecting components of known signaling pathways (2, 3), and these mutations make the tumors more vulnerable to treatment with small molecules or antibodies that interfere with the mutated target proteins.
Other potentially fruitful approaches, including high throughput screens for responses of cancer cells to small molecules or to inhibitory RNAs (4) may identify unsuspected vulnerabilities in cancer cells that depend on known or unrecognized mutations, changes in gene expression, or alterations in signaling pathways that create novel dependencies, a phenomenon called “synthetic lethality” (5).
In a previous report (6), we selected four compounds (referred to as lung cancer screen 1–4, LCS-1–4), which showed selectivity toward some or all of lung adenocarcinoma cell lines, were not similar to traditional kinase inhibitors, and offered other favorable properties for further study (6). In the current study, we have sought to identify the molecular target of LCS-1. By integrating data from affinity chromatography with two LCS-1 derivatives and from gene expression arrays, we propose that superoxide dismutase 1 (SOD1) is a target of LCS-1 (4,5-dichloro-2-m-tolylpyridazin-3(2H)-one).
Results
Derivatives of LCS-1 Inhibit the Growth of Lung Adenocarcinoma Cell Lines.
In our previous screen of 189,290 compounds, we uncovered several that inhibited the growth of one or more of four human lung adenocarcinoma cell lines with epidermal growth factor receptor (EGFR) or KRAS mutations more potently than the growth of nontumor cells (6). The four selected for further analysis showed strong structure–activity relationships, had chemical structures that can be readily modified to make affinity reagents, were not structurally similar to traditional kinase inhibitors, and were more potent against tumor cell lines than against nontumorigenic cells.
To identify the molecular target of one of these candidate compounds, LCS-1, we began by testing its effect on the growth of a wider panel of human lung adenocarcinoma cell lines that included lines with mutations in EGFR, ERBB2, KRAS, NRAS, or MAP2K4 or amplification of MET or TITF (NKX2-1). Two types of human noncancerous cells [normal human bronchial epithelial (NHBE) cells and WI-38 human lung fibroblasts] were used as controls (Table S1). The structures of several LCS-1 analogs (active and inactive) are displayed in Fig. S1. There was a gradient of responses to LCS-1, with the two nontumor cells being the least sensitive. A total of 10/27 adenocarcinoma cell lines were ∼10-fold more sensitive (median IC50 = 0.20 μM) to the growth inhibitory effect of LCS-1 than NHBE cells (IC50 = 2.66 μM), and these were considered LCS-1 sensitive. There was no apparent correlation between growth inhibition and known mutations in the cell lines, suggesting that it was unlikely that LCS-1 was targeting any of the proteins encoded by the known mutant genes.
Identification of Superoxide Dismutase 1 (SOD1) as an LCS-1–Binding Protein.
We attempted to identify the targets of LCS-1 by integrating the data obtained from two approaches: affinity chromatography combined with mass spectrometry (affinity proteomics) and gene expression arrays combined with a comparison of pathway signatures (expression analysis). The affinity proteomics approach allowed us to isolate binding partners of the compound by using the compound as an affinity reagent, as has been done before (7). Next, to be able to form a ranked list of candidate small molecule targets that can be systematically validated, we performed transcription profiling of cells that were treated with compounds for a short time to identify pathways that were modulated by the small molecules. This approach was based on the idea that pathways regulated by the compound target would also be modulated by the compound and thus reflected in the compound's transcriptional signature. Finally, to identify which small molecule-binding proteins were also linked to the expression signature of the compound, we used a human gene interaction database that is based on known pathways and protein–protein interactions obtained from curated sources; these proteins were considered candidate small molecule targets for further analysis.
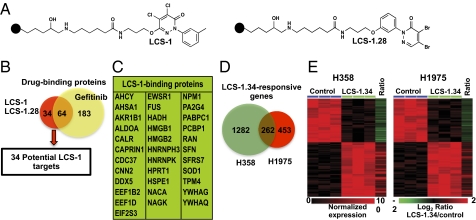
We first validated the experimental strategy with gefitinib, a known inhibitor of EGFR with higher activity against some mutant forms of the protein (8) (SI Results and SI Materials and Methods, Fig. S2, Table S2, and Dataset S1) and then applied the same methods to try to identify possible target(s) of LCS-1. A focused library of 41 LCS-1 analogs was synthesized to identify derivatives that could be modified to make affinity reagents that still retain biological activity (6). We generated two LCS-1 affinity reagents using LCS-1 and an analog that had almost identical activity profile against human lung adenocarcinoma cell lines {LCS-1.28 [4,5-dibromo-2-m-tolylpyridazin-3(2H)-one]} (Fig. 1A). LCS-1– and LCS-1.28–interacting proteins from extracts of two compound-responsive cell lines (H358 and H1975) were eluted from the affinity columns with unbound compounds (0.5 μM). Eluted proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We expected valid LCS-1 targets to bind to both affinity columns; proteins identified in at least three of the four conditions were considered possible LCS-1 targets. A list of the candidate LCS-1 targets is presented in Table S3 and summarized in Fig. 1B. A total of 98 proteins were eluted from the two LCS-1 columns. Sixty-four of these proteins were also eluted from the gefitinib column and therefore were not considered likely LCS-1 targets. The remaining 34 proteins that were eluted from both LCS-1 affinity columns but did not bind to gefiitnib were considered candidate LCS-1 targets for further validation and are given in Fig. 1C.
Fig. 1.
Identification of superoxide dismutase 1 (SOD1) as a likely target of LCS-1. (A) Two biologically active 2-phenylpyridazin-3(2H)-one analogs (LCS-1 and LCS-1.28) were immobilized to sepharose 6B beads (closed circles) and used to isolate binding partners in cell extracts of H358 and H1975 cells. (B) The numbers of proteins eluted with unbound compounds (0.5 mM) from the LCS-1 and LCS-1.28 columns and identified by LC-MS/MS. Affinity chromatography and MS identification were performed at least three times for each cell line. (C) The 34 proteins that were eluted from the LCS-1 and LCS-1.28 affinity columns, but not from the gefitinib column were considered candidate targets of LCS-1. (D) H358 and H1975 cells were treated with 0.25 μM LCS-1.34 for 3 h to generate expression signatures and the number of LCS-1.34–regulated genes is shown for each cell line. The 262 genes that were regulated in both H358 and H1975 comprise the LCS-1 gene signature. (E) “Heatmaps” of the altered expression of the 262 LCS-1 gene signature in H358 (Left) and H1975 (Right) cells. The ratio (LCS-1/control) of the average expression of each gene in three replicates is shown in the last column of the heatmaps.
The gene expression profiles of H358 and H1975 cells before and after compound treatment were compared using the Illumina Human HT-12 expression. For these experiments, the cells were treated for 3 h with LCS-1.34 (an active LCS-1 derivative with increased stability) (6). At the P < 0.01 level, the expression of 1,544 and 715 genes was significantly altered by LCS-1.34 treatment in H358 and H1975 cells, respectively (Fig. 1 D and E), with an overlap of 262 LCS-1.34–regulated genes (P = 1.6811e-11, Fisher's exact test). As observed with gefitinib treatment (Fig. S2), the directionality of changes in expression of the overlapping genes was consistent between the two cell lines (Fig. 1E). We used the human gene interaction network (SI Results and SI Materials and Methods) to screen for proteins that showed a significant link to the LCS-1.34 transcriptional signature. Of the 34 potential LCS-1 candidates from affinity proteomics, only one, superoxide dismutase 1 (SOD1) had a statistically significant connection to the LCS-1.34 gene signature (P = 0.007, Fisher's exact test, Dataset S1).
siRNAs to SOD1 Inhibit the Growth of LCS-1–Sensitive Cell Lines.
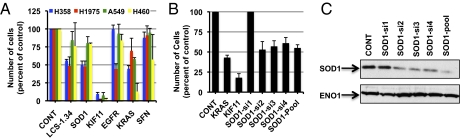
To determine directly whether inhibition of the SOD1 gene can impair cell growth, we reduced production of SOD1 with siRNAs. A pool of four SOD1 siRNAs was introduced into two cell lines with high sensitivity to LCS-1 (H358 and H1975) and two with low sensitivity (A549 and H460). Cells were exposed to siRNAs for 96 h, and the number of cells was counted by microscopy. Several control siRNAs were also used (nontargeting siRNAs and siRNAs specific for EGFR, KRAS, and KIF11). siRNAs to KIF11 inhibited growth of all cell lines, as expected, because this gene is an integral part of the cell cycle machinery (9) (Fig. 2A). EGFR siRNAs reduced the growth of the EGFR-mutant cell line H1975 by ∼70% (33 ± 5% of control, P < 0.05), without causing a significant reduction in the growth of cell lines with oncogenic KRAS (Fig. 2A). In contrast, siRNAs to KRAS reduced the growth of the mutant KRAS cell lines H358 (to 45 ± 4%), A549 (to 58 ± 2%), and H460 (to 18 ± 11%) more than it did the growth of H1975 (to 70 ± 18%). The growth of H358 and H1975 was reduced to 50 ± 5% and 48 ± 8% of control values by SOD1 siRNAs, respectively, whereas growth of A549 (80 ± 9%) and H460 (79 ± 1%) was not as markedly affected by these siRNAs. As an additional control, we also reduced production of SFN, one of the proteins that bound to LCS-1 but was considered unlikely to be a target of LCS-1 due to a lack of connection to the LCS-1 gene expression signature. SFN siRNAs had no statistically significant effect on the growth of any of the cell lines (P < 0.05, Fig. 2A). As a precaution, we also tested siRNAs to 29/34 of the genes whose product showed specific affinity for LCS-1 (Fig. 1C). None of these siRNAs inhibited growth in a pattern that mirrored the activity of LCS-1 (Fig. S3).
Fig. 2.
SiRNAs to SOD1 reduce the growth of cell lines relatively sensitive to LCS-1. (A) Cell lines that were relatively sensitive (H358 and H1975) or relatively insensitive (A549 and H460) to LCS-1 were transfected with pools of four of the indicated siRNAs. Twenty-four hours after transfection, wells with nontargeting control siRNAs (CONT) were also treated with 0.5 μM LCS-1.34. (B) H358 cells were transfected with the indicated individual siRNAs and 96 h later images of Hoechst-stained cells were taken with an IN Cell Analyzer 2000 wide-field epifluorescence microscope and the total number of Hoechst-labeled nuclei in nine fields per well counted using the object recognition function of Developer 1.7 software. Results represent the average ± SD of two to five experiments in which each condition was assayed in triplicate. In A, the results obtained with each siRNA are expressed relative to the control transfection (CONT) for the respective cell line. (C) H358 cells were transfected with the same siRNAs as in B for 48 h, then extracts prepared and immunoblotted for SOD1 or ENO1 (loading control).
To ensure that the reduction in cell growth we observed in the presence of SOD1 siRNAs was not likely to be due to any off-target effects of the pool of siRNAs, we tested five individual SOD1 siRNAs on cell growth. Four of the single SOD1 siRNAs reduced growth of H358 cells, similar to the SOD1 siRNA pool (Fig. 2B). We also observed a good correlation between inhibition of growth and reduction in SOD1 protein levels by siRNAs; the only siRNA that did not block cell growth (SOD1-si1) had little effect on SOD1 expression (Fig. 2C).
SOD1 Expression Correlates Inversely with Sensitivity to LCS-1.
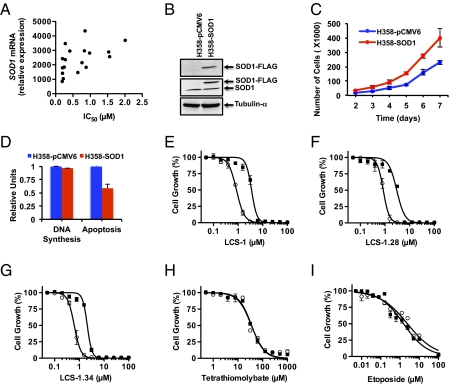
To gain insight into any relationship between sensitivity to LCS-1 and SOD1 expression, we tested whether there was a correlation between these two parameters. We analyzed 18 lung adenocarcinoma cell lines for which we had data for both SOD1 expression and sensitivity to LCS-1 (Table S1). Tumor cell lines that we considered “sensitive” had significantly lower levels of SOD1 RNA compared with the lines we categorized as “insensitive” (mean SOD1 RNA in sensitive lines was 1,563 ± 236 relative units, whereas the mean SOD1 RNA in insensitive lines was 2,793 ± 232 relative units, P = 0.0199, two-tailed t test). We observed an inverse correlation between SOD1 RNA level and sensitivity to LCS-1 (Fig. 3A, Spearman's r = 0.6270, P = 0.0054, two tailed). A similar inverse correlation was also observed between SOD1 protein level and sensitivity to LCS-1 (Fig. S4).
Fig. 3.
Expression of SOD1 correlates inversely with sensitivity to LCS-1 and promotes growth. (A) The IC50 values shown in Table S1 for inhibition of growth of 18 human adenocarcinoma cell lines by LCS-1 are plotted as a function of SOD1 mRNA levels. Spearman's correlation test determined that there was a significant correlation between SOD1 levels and sensitivity to LCS-1 (Spearman's r = 0.6270, P = 0.0054). (B) H358 cells were transfected with a FLAG-tagged SOD1 CDNA or pCMV6 empty vector and G418-resistant populations were selected. Cell extracts were immunoblotted with anti-FLAG (Top), anti-SOD1 (middle) or antitubulin-α (Bottom). (C) Equal number of H358 cells stably expressing SOD1 (H358-SOD1) or control vector (H358-pCMV6) were plated and cell numbers determined daily, starting at day 2. Results represent the mean ± SD of two experiments (performed several months apart) in which each condition was assayed in duplicate. (D) 3H-thymidine incorporation and caspase-3/7 activity were determined 24 h after plating equal numbers of the indicated cell lines as described in SI Materials and Methods. Results represent the mean ± SD of two experiments in which each condition was assayed in triplicate. (E–I) H358-SOD1 (open circle) or H358-pCMV6 (solid square) cells were plated in medium containing increasing concentrations of the indicated compounds; then the numbers of viable cells remaining after 72 h were assessed using AlamarBlue viability dye. Results represent the mean ± SD of at least two experiments in which each condition was assayed in duplicate.
Expression of a SOD1 cDNA Increases Cell Number, Decreases Apoptosis, and Reduces Sensitivity to LCS-1 Analogs.
To determine whether SOD1 directly modulates growth and response of lung cancer cell lines to LCS-1, we generated two stable H358 cell lines, one expressing a SOD1 cDNA encoding FLAG and MYC epitope tags at the N terminus (H358-SOD1) and one with the same pCMV expression vector lacking the SOD1 insert (H358-pCMV). Western blots with anti-FLAG and anti-SOD1 antisera confirmed the expression of FLAG-tagged SOD1 (Fig. 3B), which migrated more slowly in polyacrylamide gel electrophoresis because of the two epitope tags. In two independent experiments conducted several months apart, we observed that H358-SOD1 cells grew at a faster rate than the control H358-pCMV6 cells (Fig. 3C), but there was no appreciable difference in the rate of DNA synthesis (0.96 ± 0.03-fold over control, Fig. 3D). However, caspase-3/7 activity in H358-SOD1 cells was 0.58 ± 0.08-fold of the activity in H358-pCMV6 cells (Fig. 3D), suggesting that SOD1 expression may increase the number of viable cells by inhibiting apoptotic pathways.
To test the effect of SOD1 overexpression on sensitivity to LCS-1 analogs, we determined the IC50 for inhibition of the growth of H358-SOD1 and H358-pCMV cells by LCS-1, LCS-1.28, and LCS-1.34. Growth of H358-pCMV6 cells was inhibited by LCS-1, LCS-1.28, and LCS-1.34 with IC50 values of 0.8 μM, 0.8 μM, and 0.6 μM, respectively (Fig. 3 E–G). Cells overexpressing SOD1 showed about a threefold reduction in sensitivity to LCS-1 (IC50 = 3.5 μM), LCS-1.28 (IC50 = 3.0 μM), and LCS-1.34 (IC50 = 2.2 μM). This ability of high levels of SOD1 to reduce sensitivity to LCS-1 analogs was not seen when cells were challenged with the copper chelators ammonium tetrathiomolybate (Fig. 3H) and triethylenetetramine tetrahydrochloride (TETC), both of which inhibit SOD1 indirectly by chelating copper or by etoposide (Fig. 3I). These results suggest that sensitivity to LCS-1 is directly determined by SOD1 levels, with higher SOD1 levels correlating with reduced sensitivity to LCS-1.
Only Biologically Active LCS-1 Analogs Inhibit SOD1 Activity in Vitro.
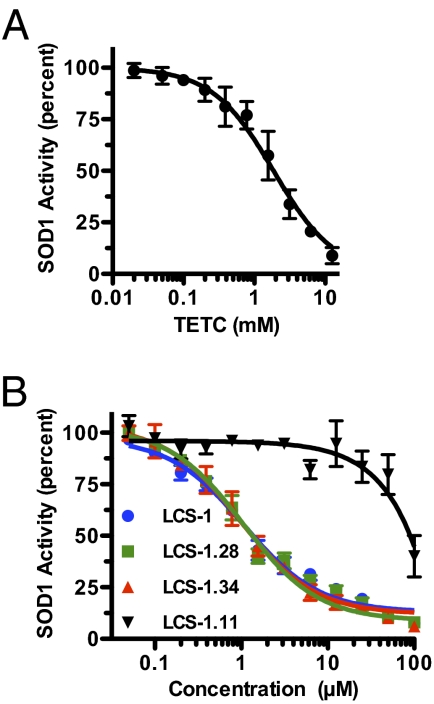
To further explore the relationship between SOD1 and LCS-1, we tested whether LCS-1 can inhibit SOD1 activity directly, using an in vitro enzymatic assay. We tested three active LCS-1 analogs that inhibited the growth of lung cancer cell lines (LCS-1, LCS-1.28, and LCS-1.34, Fig. S1), one analog that had little effect on cell growth (LCS-1.11; Fig. S1) (6), and TETC. As expected, TETC inhibited SOD1 activity in a dose-dependent manner (Fig. 4A, IC50 = 1.86 mM). The three active LCS-1 analogs also inhibited SOD1 in a dose-dependent manner with similar IC50 values (LCS-1, 1.07 μM; LCS-1.28, 0.95 μM; and LCS-1.34, 1.05 μM; Fig. 4B). In contrast, the relatively inactive LCS-1.11 inhibited SOD1 activity only at high concentrations (>10 μM). These results support the proposal that SOD1 is a direct target of LCS-1.
Fig. 4.
Active LCS-1 analogs inhibit SOD1 enzymatic activity in vitro. (A and B) SOD1 enzymatic activity was assayed in vitro in the presence of increasing concentrations of (A) triethylenetetramine tetrahydrochloride or (B) LCS-1 analogs. Note that LCS-1.11 does not inhibit the growth of lung cancer cells. Results represent the mean ± SE of at least three experiments in which each condition was assayed in duplicate.
Most Human Lung Adenocarcinomas Express SOD1 at Higher than Normal Levels.
Because SOD1 appears to increase growth of lung adenocarcinoma cells (Fig. 3C), we wanted to know whether expression of the gene was altered in human lung cancers. We compared SOD1 transcript levels observed in normal lung and in adenocarcinomas in two independent sample sets downloaded from the Gene Expression Omnibus. Dataset GSE10072 consisted of 58 lung adenocarcinomas and 49 normal lung tissue samples (31 pairs), and dataset GSE7670 contained 26 lung adenocarcinomas paired with adjacent normal lung samples. SOD1 RNA levels were slightly but significantly higher in tumors than in normal lung tissue in the GSE10072 dataset (Fig. S4A; mean expression tumors, 3,075 ± 120; mean expression normals, 2,454 ± 80; P = 0.0004, two-tailed Welch's T test). This observation was confirmed through the pairwise analysis of the samples in the GSE7670 dataset (Fig. S4B; mean expression tumor, 1,148 ± 80; mean expression normal, 956 ± 36; P = 0.0051, two-tailed paired T test). Taken together with the observation that overexpressing SOD1 in cells in culture increases growth, these findings support the notion that cells with increased SOD1 level may have a selective advantage.
Discussion
We used two approaches—affinity chromatography combined with mass spectrometry (affinity proteomics) and gene expression arrays combined with a comparison of pathway signatures (expression analysis)—to identify SOD1 as a target of LCS-1, after validating the approach with the EGFR inhibitor gefitinib. In both cases, combining the results of the two approaches pointed to only one target responsible for a biological action of the compounds, namely, growth inhibition of lung adenocarcinoma cell lines. Of course, LCS-1 may have targets other than SOD1, just as gefitinib can inhibit tyrosine kinases other than EGFR (10). It is possible that these additional LCS-1 targets may contribute to the effect of the compound on cell growth.
SOD1, also known as Cu/Zn SOD, is a copper-dependent enzyme that catalyzes the conversion of superoxide ion (O2−) into H2O2 and O2 to maintain low levels of reactive oxygen species (ROS) (11). There are two additional SOD genes (SOD2 and SOD3) that encode enzymes functionally similar to SOD1 but localized in different cellular compartments (12); therefore, they cannot substitute for each other in free radical detoxification. Our results imply SOD2 and SOD3 do not substitute for SOD1 when this enzyme was inhibited by LCS-1. This could be because LCS-1 inhibited all three SOD isoforms or that these proteins have additional unique functions. Given that we did not detect binding of SOD2 or SOD3 to LCS-1 and that the three proteins are very different chemically, it is unlikely that these two isoforms are targets for LCS-1. The SODs maintain tight regulation of ROS as these entities can either promote growth or have antiproliferative effects, depending on cellular concentration. The activities of a variety of transcription factors (e.g., AP1, p53, and NF-κB) have been shown to be regulated by the redox status of a cell (13). In addition, ROS can induce posttranslational modification of proteins involved in growth factor signaling pathways, leading to DNA damage and apoptosis (12). The three superoxide dismutase genes have been implicated in the pathophysiology of various diseases (12). For example, mutations in SOD1 have been identified in ∼20% of families with amyotrophic lateral sclerosis (14). In addition, the copper chelator ATN-224, which interferes with the activation of SOD1 (and other cellular processes), inhibits growth of tumor cells and blocks angiogenesis in animal models (15), suggesting a role for SOD1 in regulating tumorigenesis and angiogenesis. ATN-224 is currently being tested in clinical trials for the treatment of late stage solid tumors (16). However, there has been no clear evidence pointing to a specific role for SOD1 in tumorigenesis, and we are unaware of any previously reported SOD1 inhibitors known to be effective in cells, other than copper chelators.
Here we have demonstrated that overexpression of SOD1 in lung cancer cells promotes growth without affecting cell cycle progression. Instead, cells overexpressing SOD1 had a lower frequency of basal apoptosis, implying that SOD1 promotes growth by increasing survival. In agreement with this, we previously showed that the LCS-1 analog LCS-1.34 increased apoptosis in lung adenocarcinoma cell lines (6). It is unclear how SOD1 is able to influence survival pathways in lung cancer cell lines to permit growth. Recently, it was shown that the antiapoptotic protein XIAP (X-linked inhibitor of apoptosis) can activate SOD1 by ubiquitinating and activating the copper chaperone for superoxide dismutase (CCS), which delivers copper to SOD1, thereby enhancing its enzymatic function (17). It remains to be determined whether a similar process occurs in cancer cells, because the XIAP study was done in mammalian cell lines that were not derived from tumors. We have previously shown that LCS-1 can prevent serum-induced activation of the ERK and PI 3-kinase/AKT signaling pathways (6). In support of this, Juarez et al. reported that ATN-224–mediated inhibition of SOD1 activity prevents growth factor-induced activation of ERK (18). The authors speculated that, when SOD1 activity is low, there is an abundant supply of reactive oxygen species that prevents the oxidation of tyrosine phosphates (18). This causes the down-regulation of growth-promoting signals from tyrosine kinase receptors and this may be the mechanism by which ROS impair growth. In support of this antiproliferative role of ROS, two studies have identified two different ROS-inducing compounds from chemical screens that can inhibit the growth of KRAS-transformed 3T3 cells (19) and cancer cells (20). In addition, it was recently shown that the transcription factor Nrf2 (which controls a antioxidant pathway) was required for tumorigenesis in mice by oncogenes such as Kras (21). Taken together, these results suggest that modulating the redox status of a cell either by inhibiting SOD1 or by other mechanisms, could be exploited as an effective therapeutic strategy for treating cancer.
We report here that expression of SOD1 was significantly higher in most lung tumors from two independent datasets obtained from the Gene Expression Omnibus. Given our observation that overexpression of SOD1 leads to increased growth, it seems likely that cells with high levels of SOD1 have a selective advantage during tumorigenesis. Thus, SOD1 might be a valuable therapeutic target and LCS-1 compounds might be candidate lead compounds for development. Because tumor cell lines with higher levels of SOD1, like tumor cell lines engineered to express additional SOD1, appeared less sensitive to the growth inhibitory effects of LCS-1, SOD1 inhibitors (e.g., new LCS-1 analogs) with more potency and efficacy would be needed to overcome this inverse relationship between SOD1 expression and sensitivity to LCS-1. In addition, sequencing of SOD1 cDNAs derived from five lung cancer cell lines with low or high sensitivity to LCS-1 revealed no somatic mutations (data not shown), suggesting that aberrant expression of the wild-type protein itself may be involved in lung tumorigenesis.
Novel small molecules have not been widely used as probes to identify new therapeutic targets. As we have shown in this study, chemical biology approaches are valuable in identifying new therapeutic targets and biological mechanisms.
Materials and Methods
Preparation of Cell Extracts, Affinity Chromatography, and Mass Spectrometry.
Preparation of cell extracts and affinity chromatography was carried out at 4 °C as described (7) and bound proteins were eluted with 600 μL lysis buffer containing either 1 mM gefitinib, or 0.5 mM LCS-1, or LCS-1.28. Eluates were collected and concentrated using a vacuum dryer at room temperature. LC-MS/MS and protein identification was carried out as described in SI Materials and Methods.
SOD1 Enzymatic Assay.
SOD1 activity was determined in vitro using a Superoxide Dismutase Assay kit from Cayman Chemicals according to the manufacturer's instructions (22). See SI Materials and Methods for more information.
Gene Expression Profiling of Cells Exposed to Small Molecules.
Cells were plated at a density of 200,000 cells per well (six-well plate) for 24 h and then treated with fresh growth media containing either gefitinib (0.1 μM) or LCS-1.34 (0.25 μM) for 3 h. Cells were then lysed with 1 mL TRIzol per well, and RNA was extracted and profiled for expression analysis by the Memorial Sloan-Kettering Cancer Center (MSKCC) Genomics Core Facility using the Illumina Human HT-12 array platform according to the manufacturer's instructions. Three biological replicates were profiled for each condition with each experimental pair (control and compound treatment) prepared on different days. See SI Materials and Methods for more information on data analysis.
Pathway Analysis to Find Genes Linked to Expression Signatures.
A published human gene interaction network (Netbox) that is based on known pathways and protein–protein interaction data (23) was used to screen for genes that had a significant connection to the expression signature of compound treated cells. The network was created by combining interaction data from Human Protein Reference Database (24), Reactome (25), National Cancer Institute/Nature Pathway Interaction Database (26) and the MSKCC Cancer Cell Map (27), and contains 9,264 proteins. Fisher's exact test was used to assess whether a protein (e.g., a candidate target identified by MS) was linked to more transcriptionally altered genes than expected by chance, given its total number of neighbors in the network. For this we used the “linker gene” functionality of the Netbox software.
Supplementary Material
Acknowledgments
We thank L. Fabrizio for help with LC-MS/MS. We also extend our gratitude to Drs. G. Sukenick, S. Rusli, and H. Liu, and Miss H. Fang from Memorial Sloan-Kettering Cancer Center (MSKCC) Nuclear Magnetic Resonance Analytical Core for help with analytical data acquisition and interpretation. For helpful advice over the course of this study, we thank members of the H.E.V. laboratory at MSKCC and Mariana Yaneva. This work was supported by an National Institutes of Health Grant (PO1 CA129243-03 to Mark Kris) and a National Cancer Institute Cancer Center Support Grant (P30 CA08748). The High Throughput Screening Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, the William Randolph Hearst Fund in Experimental Therapeutics and the Lillian S. Wells Foundation. E.L. is supported by a Postdoctoral Fellowship from the Swedish Research Council. W.W.L. is a recipient of the Jean-Francois St-Denis Fellowship in Cancer Research from the Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113554108/-/DCSupplemental.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Lovly CM, Carbone DP. Lung cancer in 2010: One size does not fit all. Nat Rev Clin Oncol. 2011;8:68–70. doi: 10.1038/nrclinonc.2010.224. [DOI] [PubMed] [Google Scholar]
- 3.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber SL, et al. Cancer Target Discovery and Development Network. Towards patient-based cancer therapeutics. Nat Biotechnol. 2010;28:904–906. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011;21:34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Somwar R, Shum D, Djaballah H, Varmus H. Identification and preliminary characterization of novel small molecules that inhibit growth of human lung adenocarcinoma cells. J Biomol Screen. 2009;14:1176–1184. doi: 10.1177/1087057109350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godl K, et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rello-Varona S, et al. Preferential killing of tetraploid tumor cells by targeting the mitotic kinesin Eg5. Cell Cycle. 2009;8:1030–1035. doi: 10.4161/cc.8.7.7950. [DOI] [PubMed] [Google Scholar]
- 10.Brehmer D, et al. Cellular targets of gefitinib. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 11.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 12.Miao L, St Clair DK. Regulation of superoxide dismutase genes: Implications in disease. Free Radic Biol Med. 2009;47:344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 14.Ticozzi N, et al. Genetics of familial amyotrophic lateral sclerosis. Arch Ital Biol. 2011;149:65–82. doi: 10.4449/aib.v149i1.1262. [DOI] [PubMed] [Google Scholar]
- 15.Juarez JC, et al. Copper binding by tetrathiomolybdate attenuates angiogenesis and tumor cell proliferation through the inhibition of superoxide dismutase 1. Clin Cancer Res. 2006;12:4974–4982. doi: 10.1158/1078-0432.CCR-06-0171. [DOI] [PubMed] [Google Scholar]
- 16.Lowndes SA, et al. Phase I study of copper-binding agent ATN-224 in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7526–7534. doi: 10.1158/1078-0432.CCR-08-0315. [DOI] [PubMed] [Google Scholar]
- 17.Brady GF, et al. Regulation of the copper chaperone CCS by XIAP-mediated ubiquitination. Mol Cell Biol. 2010;30:1923–1936. doi: 10.1128/MCB.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juarez JC, et al. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci USA. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw AT, et al. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci USA. 2011;108:8773–8778. doi: 10.1073/pnas.1105941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj L, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chade AR, et al. Antioxidant intervention blunts renal injury in experimental renovascular disease. J Am Soc Nephrol. 2004;15:958–966. doi: 10.1097/01.asn.0000117774.83396.e9. [DOI] [PubMed] [Google Scholar]
- 23.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS ONE. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keshava Prasad TS, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database issue):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer CF, et al. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anonymous The Cancer Cell Map. 2006. Available at http://cancer.cellmap.org. Accessed (Memorial Sloan-Kettering Cancer Center, New York, NY). Accessed September 1, 2011.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.