Querying the past is hard. Speciation and extinction processes are on a scale of thousands to millions of years. Thus, they are most often studied by reconstructing the evolutionary past. This past is reconstructed using phylogenetic methods either on the basis of data from living species or by directly examining the fossil record. Robust methods for inferring the evolutionary past purely on the basis of living species would allow us to understand speciation and extinction processes for the large number of groups without a good fossil record.
Generally, studies using living species infer lower extinction rates than the rates suggested by the fossil record (1, 2). A new study in PNAS (3) suggests that this mismatch is due to our use of oversimplified models of speciation and extinction.
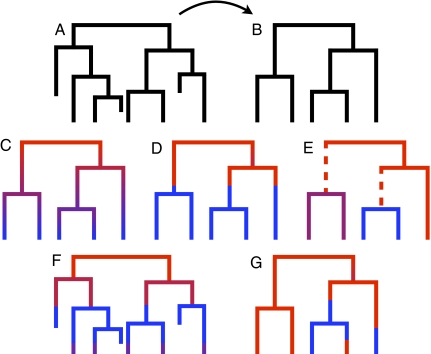
Fifteen years ago, Nee et al. (4) presented the first method to infer speciation and extinction rates on the basis of “reconstructed” phylogenies, i.e., phylogenies inferred on only extant species (Fig. 1 A and B). This first likelihood method relied on the idea that lineages in a reconstructed phylogeny accumulate through time with rate λ − μ (where λ is the speciation rate and μ is the extinction rate) and accumulate in the very recent past with rate λ. The change in rate of lineage accumulation from λ − μ to λ, called the “pull-of-the-present” (5), allows us to estimate both λ and μ given only data from living species.
Fig. 1.
(A and B) Complete phylogeny (A) with associated reconstructed phylogeny (B), which is obtained by suppressing all extinct lineages. (C–G) Models for speciation and extinction. Red denotes a fast rate, purple an intermediate rate, and blue a slow rate of speciation. C–E indicate the three models accounting for rate heterogeneity through time and across subclades: (C) Morlon et al. (3) (continuous change over time), (D) Stadler (8) (change at one time point), and (E) Alfaro et al. (6) (change in the dashed edges above subclades). (F) Density-dependent model (the complete phylogeny instead of the reconstructed phylogeny is plotted as the speciation rate depends on the total number of lineages). (G) Trait-dependent model by Maddison et al. (14) (changes between the two rates at arbitrary times).
The method of Nee et al. has been widely used for estimating speciation and extinction rates. Unfortunately, it often produces estimates of μ near zero (1, 2). Morlon et al. (3) suggest that the low extinction rate estimates might be due to the assumption that λ and μ are constant, which is, in particular for large groups, most likely wrong. For large clades, we expect both rate heterogeneity through time due to environmental factors and rate heterogeneity across subclades due to subclade-specific traits influencing speciation and extinction rates.
Several likelihood-based approaches now exist that infer speciation and extinction rates under each of these two scenarios of rate heterogeneity, based on reconstructed phylogenies. The basic idea is the same for the different approaches: Speciation and extinction rates are determined that maximize the likelihood of the reconstructed tree. The challenge has been to derive an analytic formula for the likelihood of the tree under the complex dynamics. Three recent PNAS studies provide more general analytic likelihood functions:
Alfaro et al. (6) provide a likelihood approach in which the speciation and extinction rates may vary across subclades, but each subclade has a constant rate (Fig. 1E). The original method derivation is provided in ref. 7 and found in the package MEDUSA. Such a model allows for detecting subclade-specific speciation and extinction processes.
Stadler (8) relaxes the assumption of constant rates by allowing for rates changing at specific points in time (Fig. 1D). Such a model allows for detecting rapid changes in speciation and extinction rates due to environmental effects like at the Cretaceous–Tertiary boundary at 65 Ma.
Morlon et al. (3) extend the two methods such that rates may change continuously through time (instead of discretely as in ref. 8), and subclades may have different speciation and extinction rates (as in ref. 6) (Fig. 1C).
The models above have in common that the speciation and extinction rates within subclades are a function of time only, meaning that the rates are changing only due to external factors (i.e., the environment). In particular in ref. 6, the rates within subclades are constant.
Are the Speciation and Extinction Rates Governed by Environmental Effects?
For cetaceans (whales, dophins, and porpoises), Morlon et al. (3) show that incorporating rate heterogeneity produces speciation and extinction rate estimates in good agreement with the fossil record. In particular, the authors infer an exponentially decreasing speciation rate and a constant extinction rate for the cetacean subclade Balaenopteridae.
An immediate follow-up question is: Why is the speciation rate declining in this group? Is the speciation rate change really caused by the environment (which is implicitly assumed by the underlying model)?
An alternative explanation for a decreasing speciation rate is density-dependent speciation (e.g., ref. 9), meaning that the speciation rate depends on the number of species instead of time (Fig. 1F): A clade begins to radiate quickly into non-occupied niches, and the process slows down as the niches fill. The initial adaptive radiation is typically caused by a key innovation or a move to a new habitat of the ancestral species. There is no closed-form likelihood expression for density-dependent speciation models available yet. A first step is taken in ref. 10 where a likelihood function is provided for scenarios when the extinction rate is zero.
Density-dependent speciation yields a declining speciation rate through time; analog, density-dependent extinction yields an increasing extinction rate through time. A likelihood approach will allow us to formally contrast density-dependent against time-dependent models for clades such as the baleen whales.
Unfortunately, neither time-dependent nor density-dependent models can explain the speciation process completely, as the resulting trees induced by these models are too balanced: A reconstructed phylogeny is fully characterized by the times of lineage accumulation (continuous part) and the ranked tree shape (11) (discrete part). Time-dependent and density-dependent models assume exchangeable species, meaning that at each point in time each species undergoes the same speciation and extinction mechanism. Aldous (12) showed that all exchangeable species models induce the same distribution on ranked tree shape (but can induce a wide variety of lineage accumulation patterns). Because empirical trees are less balanced than trees under exchangeable species models (13), the exchangeable species models lack an important feature.
Imbalance can be explained by trait-dependent speciation and extinction. Two PNAS studies (3, 6) take a first step to account for trait-dependent speciation by assuming heterogeneity across subclades. However, the change in trait is not explicitly modeled; a trait change (and thus a change in speciation and extinction rates) is added if the likelihood increases sufficiently. Maddison et al. (14) introduced a likelihood approach assuming discrete traits evolving under a Markov model (Fig. 1G); Fitzjohn (15) generalized the approach by allowing for continuous traits. However, these approaches lack the rate heterogeneity through time.
Are Models with Environmental-, Density-, and Trait-Dependent Speciation and Extinction Rates Sufficient?
Morlon et al. (3) obtain speciation and extinction rate estimates for whales that are in agreement with the fossil record. In particular, the extinction rates are not underestimated. This result suggests that we might be able to reconcile molecular phylogenies with the fossil record in general.
However, the bias of underestimating extinction rates in reconstructed phylogenies will not completely vanish with the methods accounting for environmental, density-dependent, and trait-dependent effects: Underestimating extinction is primarily due to the fact that very few reconstructed phylogenies have a pronounced pull-of-the-present effect; i.e., often the lineages do not accumulate faster in the recent past. In fact, often the most recent lineage accumulation is slower instead of faster [very pronounced, e.g., for mammals (16) and birds (17)].
It is well recognized that this apparent slowdown can be due to simply not sampling all of the species, and approaches dealing with this missing data problem have recently become available (18, 19).
However, we observe the slowdown even in complete phylogenies like mammals. The reason for this slowdown may be that very recent speciation events have not yet been identified (20). The delay of recognizing species after two populations started diverging needs to be recognized by our methods to avoid biases in speciation and extinction rate estimates.
Future Challenges
In large clades, speciation and extinction rates were most likely not constant throughout evolutionary time. Rates change due to (i) a changing environment, (ii) density-dependent speciation and extinction, and (iii) trait-dependent speciation and extinction. The study in PNAS (3) improves our understanding of scenario i, but we require more work on scenario ii to test scenario i against ii. Scenarios i and ii will help account for the mode of lineage accumulation. With an improved understanding of scenario iii, we may also be able to account for tree shape. As lineage accumulation and tree shape fully describe a reconstructed phylogeny, a combination of models i–iii may have the power to describe the process of speciation and extinction.
However, in addition, we need to take into account the mode of data collection to avoid biases: We have to be aware that we may not be able to recognize very young species and we may not sample all extant species of a clade.
It is a puzzle for the future to combine the time-dependent, density-dependent, and trait-dependent models with the data collection biases to produce one unified approach that can be used in a likelihood framework. Morlon et al. (3) give us an elegant version of the time-dependent piece to fit in.
Footnotes
The author declares no conflict of interest.
See companion article on page 16327.
References
- 1.Nee S. Birth-death models in macroevolution. Annu Rev Ecol Evol Syst. 2006;37:1–17. [Google Scholar]
- 2.Purvis A. Phylogenetic approaches to the study of extinction. Annu Rev Ecol Evol Syst. 2008;39:301–319. [Google Scholar]
- 3.Morlon H, Parsons TL, Plotkin JB. Reconciling molecular phylogenies with the fossil record. Proc Natl Acad Sci USA. 2011;108:16327–16332. doi: 10.1073/pnas.1102543108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nee S, May RM, Harvey PH. The reconstructed evolutionary process. Philos Trans R Soc Lond B Biol Sci. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. [DOI] [PubMed] [Google Scholar]
- 5.Nee S, Holmes EC, May RM, Harvey PH. Extinction rates can be estimated from molecular phylogenies. Philos Trans R Soc Lond B Biol Sci. 1994;344:77–82. doi: 10.1098/rstb.1994.0054. [DOI] [PubMed] [Google Scholar]
- 6.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106:13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabosky DL, Donnellan SC, Talaba AL, Lovette IJ. Exceptional among-lineage variation in diversification rates during the radiation of Australia's most diverse vertebrate clade. Proc Biol Sci. 2007;274:2915–2923. doi: 10.1098/rspb.2007.0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci USA. 2011;108:6187–6192. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillimore AB, Price TD. Density-dependent cladogenesis in birds. PLoS Biol. 2008;6:e71. doi: 10.1371/journal.pbio.0060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabosky DL, Lovette IJ. Density-dependent diversification in North American wood warblers. Proc Biol Sci. 2008;275:2363–2371. doi: 10.1098/rspb.2008.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gernhard T. The conditioned reconstructed process. J Theor Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Aldous DJ. Stochastic models and descriptive statistics for phylogenetic trees, from Yule to today. Stat Sci. 2001;16:23–34. [Google Scholar]
- 13.Blum MG, François O. Which random processes describe the tree of life? A large-scale study of phylogenetic tree imbalance. Syst Biol. 2006;55:685–691. doi: 10.1080/10635150600889625. [DOI] [PubMed] [Google Scholar]
- 14.Maddison WP, Midford PE, Otto SP. Estimating a binary character's effect on speciation and extinction. Syst Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 15.FitzJohn RG. Quantitative traits and diversification. Syst Biol. 2010;59:619–633. doi: 10.1093/sysbio/syq053. [DOI] [PubMed] [Google Scholar]
- 16.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 17.Klicka J, Zink RM. The importance of recent ice ages in speciation: A failed paradigm. Science. 1997;277:1666–1669. [Google Scholar]
- 18.Cusimano N, Renner SS. Slowdowns in diversification rates from real phylogenies may not be real. Syst Biol. 2010;59:458–464. doi: 10.1093/sysbio/syq032. [DOI] [PubMed] [Google Scholar]
- 19.Höhna S, Stadler T, Ronquist F, Britton T. Inferring speciation and extinction rates under different species sampling schemes. Mol Biol Evol. 2011;28:2577–2589. doi: 10.1093/molbev/msr095. [DOI] [PubMed] [Google Scholar]
- 20.Avise JC, Walker D. Pleistocene phylogeographic effects on avian populations and the speciation process. Proc Biol Sci. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]