Abstract
Cell death is a complex process that plays a vital role in development, homeostasis, and disease. Our understanding of and ability to control cell death is impeded by an incomplete characterization of the full range of cell death processes that occur in mammalian systems, especially in response to exogenous perturbations. We present here a general approach to address this problem, which we call modulatory profiling. Modulatory profiles are composed of the changes in potency and efficacy of lethal compounds produced by a second cell death-modulating agent in human cell lines. We show that compounds with the same characterized mechanism of action have similar modulatory profiles. Furthermore, clustering of modulatory profiles revealed relationships not evident when clustering lethal compounds based on gene expression profiles alone. Finally, modulatory profiling of compounds correctly predicted three previously uncharacterized compounds to be microtubule-destabilizing agents, classified numerous compounds that act nonspecifically, and identified compounds that cause cell death through a mechanism that is morphologically and biochemically distinct from previously established ones.
Keywords: apoptosis, chemical biology, small molecules
Cell death has historically been viewed as a binary phenomenon. Cells were described to die in one of two ways—through a controlled and ordered process (apoptosis) or an unregulated and chaotic process (necrosis) (1, 2). Not only were these often considered the only two possible mechanisms, but they were also frequently viewed as morphologically and biochemically uniform (3, 4). A great deal of research in recent decades has not only shown the complexity and heterogeneity of apoptotic and necrotic signaling, but also that cells can die in physiological and nonphysiological contexts through processes morphologically and biochemically distinct from both apoptosis and necrosis.
Activation of caspases, a family of cysteine proteases, is essential for producing the full morphological characteristics of apoptosis. There are at least three distinct pathways that can lead to the activation of effector caspases—the extrinsic death receptor pathway, the intrinsic mitochondrial pathway, and the Granzyme B pathway (5)—but numerous mechanisms feed into these three pathways. There is substantial evidence that necrosis, long considered to be a disorganized and unregulated process, can proceed through an evolutionarily conserved pathway in a highly orchestrated fashion. Necrosis-like morphology has been observed after death receptor stimulation (6–12), after treatment with DNA-damaging agents (13–15), and in Caenorhabditis elegans in response to a variety of stresses (16, 17). These examples illustrate the heterogeneity of cell death processes resembling necrosis. A widely debated nonapoptotic, nonnecrotic cell death mechanism is autophagic cell death, which has been implicated in vivo in the involution of the salivary gland in Drosophila (18) and in death because of the hypersensitivity response in Arabidopsis (19) as well as a number of cell culture systems (20, 21). Additional forms of cell death have been described and reviewed elsewhere (22–24). Many of these alternative death programs have been observed only in specialized cells or under unusual conditions, and they are often limited to morphological rather than molecular-level descriptions. Rigorous functional understanding of these processes and their mechanistic relationship to each other is lacking.
New lethal reagents are routinely generated in anticancer drug discovery, chemical biology screens, and cell death research. However, despite our improved understanding of cell death, the investigation and characterization of such lethal reagents typically proceeds in an ad hoc way. There is no standardized process to compare lethal compounds or identify those compounds acting through specific mechanisms. Hence, rigorous characterization is only performed on lethal compounds that are active in more selective and inherently interesting secondary assays. The remaining uncharacterized orphan lethal compounds represent an untapped resource. Investigation into the mechanism of action of such compounds could reveal information about the scope and detail of death pathways that can be activated in cells.
A major use of specific secondary assays is for the elimination of compounds that kill cells through nonspecific mechanisms. Such compounds are not useful in investigating the signaling pathways that govern cell death, and they are not attractive leads for drug development. For instance, compounds can act nonspecifically on cells through chemical reactivity (25), by forming small molecule aggregates (26), or disrupting membranes (27). These properties of small molecules are not easy to predict a priori. Although reactivity can often be predicted from chemical structure, some reactive compounds kill cells specifically, whereas others are not lethal at all (28). Small molecule aggregates are well-established as nonspecific inhibitors in in vitro biochemical assays (29, 30), but only limited studies have been performed on their persistence in the presence of high protein concentrations (31) and cell culture (26). Most drug-like small molecules are lipophilic (32), and determining those molecules that act primarily through membrane disruption is not obvious.
Here, we present a methodology for systematically characterizing lethal compounds based on functional profiles. This method, called modulatory profiling, systematically analyzes the changes in the lethality of a compound when used in combination with each member of a panel of cell death modulators. These modulators were selected to modulate established cell death processes. Applying this method to both characterized and uncharacterized compounds has allowed us to identify previously unidentified microtubule-destabilizing agents, segregate compounds that act nonspecifically, and identify compounds that induce death through morphologically and biochemically distinct mechanisms.
Results
Creating and Clustering Modulatory Profiles.
Our initial tasks were to (i) assemble a collection of reagents that can modulate cell death-related processes (cell death modulators), (ii) identify a test pool of characterized lethal compounds, and (iii) choose cell lines for testing. First, we chose chemical modulators (SI Appendix, Table S1) based on literature precedent. These compounds include reactive oxygen species scavengers and inhibitors of calcium signaling, protein synthesis, and proteases, among others. We also included four genetic modulators of cell death (SI Appendix, Fig. S1).
For the lethal compounds, we chose a set of 28 well-characterized compounds (SI Appendix, Table S2), including inhibitors of topoisomerases, microtubule assembly, proteasomes, histone deacetylases (HDACs), kinases, and various stages of mitochondrial respiration. We chose two cell lines, a fibrosarcoma cell line (HT-1080) and an engineered tumorigenic line derived from human foreskin fibroblasts (BJ-TERT/LT/ST/RASV12). Both of these cell lines grow rapidly and uniformly in 384-well assay plates. We reasoned that it would be advantageous to explore both an engineered tumorigenic line lacking mutations in uncharacterized death pathways that may be found in cancers as well as a cancer cell line with WT p53 that could be easily infected with lentivirus.
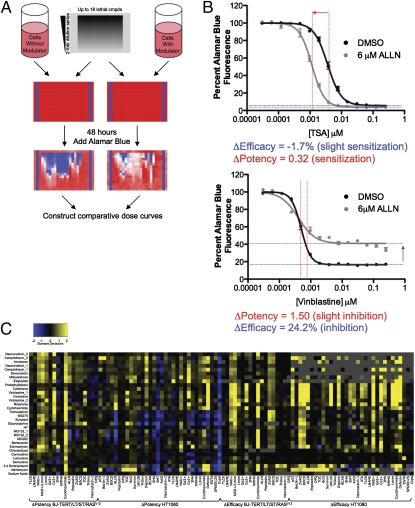
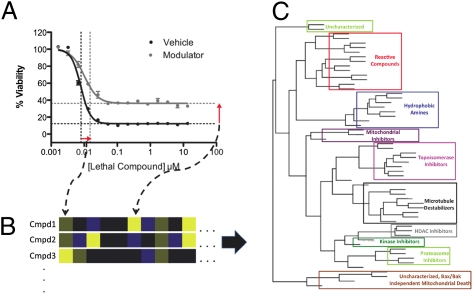
We then tested each lethal compound in combination with each cell death modulator (Fig. 1A). We chose a single concentration of each modulator (based on literature precedent and our optimization experiments) and used a 14-point, twofold dilution series of each lethal compound. Comparing survival in the presence of each modulator to survival without a modulator allowed us to construct comparative concentration response curves (Fig. 1B and SI Appendix, Fig. S2). We extracted two parameters from each graph—the change in potency (Fig. 1B Upper) and the change in efficacy (Fig. 1B Lower) caused by each modulator. The modulatory profile of a lethal compound was defined as the dimensional vector of its activity changes (i.e., changes in potency and efficacy) induced by each modulator across distinct cell lines. The modulatory profiles of known lethal compounds are graphically depicted in Fig. 1C and numerically depicted in SI Appendix, Table S3. We tested the data requirements of the profiles by calculating the parameters based on a subset of the data (SI Appendix, SI Text and SI Appendix, Fig. S3A). Experiments to determine the reproducibility of the measurements between batches, the similarities and differences in the parameters between cell lines, the dependence of the measured parameters on the detection reagent, and the effects of the modulators on the detection reagent and the cells are shown in SI Appendix, Fig. S3 B–F and discussed in SI Appendix, SI Text.
Fig. 1.
Creating modulatory profiles. (A) Cells with or without modulator were seeded in 384-well plates. Lethal compounds in dilution series were then added, and the plates were incubated for 48 h before the addition of the cell viability dye Alamar blue. Readout of fluorescence after a 14-h incubation with Alamar blue allowed the construction of comparative concentration response curves. (B) Two examples of comparative concentration response curves and an illustration of the two parameters—the change in efficacy and change in potency—extracted from each pairwise combination of modulators and lethal compounds. Both examples use HT-1080 cells. (C) Heat map depicting 32 modulatory profiles of characterized lethal compounds (28 distinct compounds and 4 repeat compounds). Lethal compounds are listed on the y axis, and modulators, cell lines, and parameter types are on the x axis. Missing values are depicted in gray.
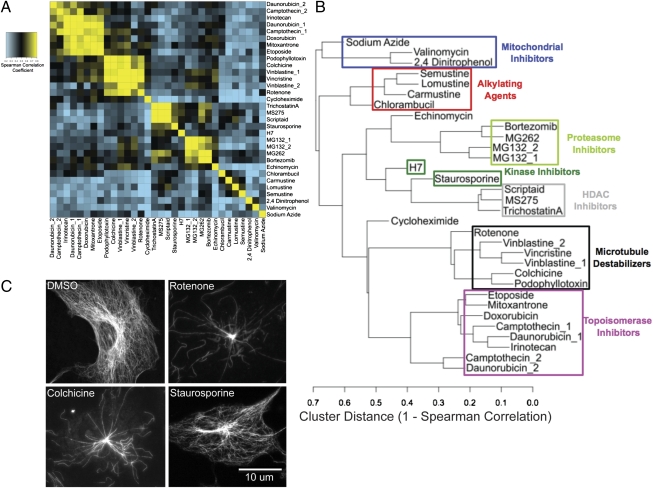
Classifying the lethal compounds based on the effects of a single modulator was ineffective (SI Appendix, SI Text and SI Appendix, Fig. S4), necessitating the use of the full modulatory profiles. We compared modulatory profiles of lethal compounds using Spearman correlations between pairs of compounds (the similarity matrix is shown in Fig. 2A). Hierarchically clustering the axes of the matrix caused compounds with well-correlated modulatory profiles to appear clearly along the axis. The clustering was also visualized with a dendrogram (Fig. 2B). Independent replicates of the same compound from separate batches were clustered closely, showing reproducibility. Compounds with the same well-characterized mechanisms of action were found to cluster together. Correct clustering, based on an established mechanism, was observed for topoisomerase inhibitors, proteasome inhibitors, microtubule-destabilizing agents, HDAC inhibitors, alkylating agents, and mitochondrial inhibitors (aside from rotenone, which is discussed below). Interestingly, compounds that did not cluster together were mechanistically distinct from each other. Cycloheximide is a translational inhibitor, which is quite different from echinomycin, a bis-DNA intercalator and transcriptional inhibitor. Two broad-spectrum kinase inhibitors, although clustered relatively close, are not in an exclusive cluster, likely because these inhibitors target different kinases to varying extents.
Fig. 2.
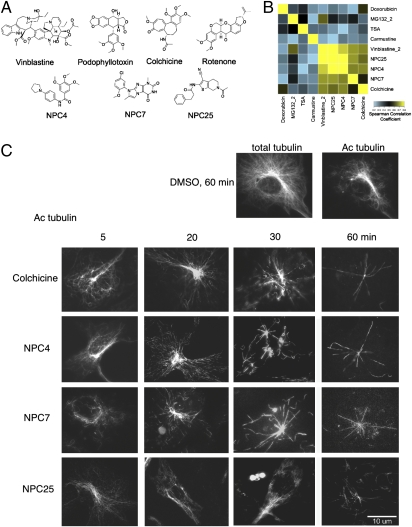
Clustering characterized lethal compounds based on modulatory profiles. (A) Heat map of the similarity matrix showing the Spearman correlation between modulatory profiles of lethal compounds. (B) Dendrogram derived from hierarchical clustering of the similarity matrix in A. (C) TC-7 cells stained for acetylated tubulin after 60-min treatment with vehicle (DMSO), 5 μM rotenone, 1 μM colchicine, or 1 μM staurosporine. Representative images were chosen for each treatment.
One of the mitochondrial inhibitors—rotenone, a complex I inhibitor—was placed into a cluster with microtubule-destabilizing agents. Rotenone has, in fact, been shown to destabilize microtubules both in vitro and in cells (33, 34). We confirmed by immunofluorescence that rotenone causes a rapid loss of microtubule polymers in HT-1080 cells (SI Appendix, Fig. S5) as well as in the flatter TC-7 cells (Fig. 2C) and that other inhibitors of mitochondrial respiration do not affect microtubules (SI Appendix, Fig. S6). The unbiased recapitulation of rotenone's antimicrotubule activity suggests that, when a lethal compound operates through the same mechanism of action as a characterized compound, the mechanism of action can be predicted based on modulatory profiling. Importantly, it also suggests that if a compound has multiple targets, modulatory profiling can identify which of these targets is relevant to cell death.
Clustering of Compounds Based on Gene Expression Profiles or Chemical Structure.
Gene expression profiling using cDNA microarrays has been applied widely in biology and medicine. Although there have been notable successes using gene expression profiling to investigate small molecule mechanisms of action (35–39), the use of straightforward statistical methods to compare small molecules based on their gene expression profiles in mammalian systems has largely proven disappointing.
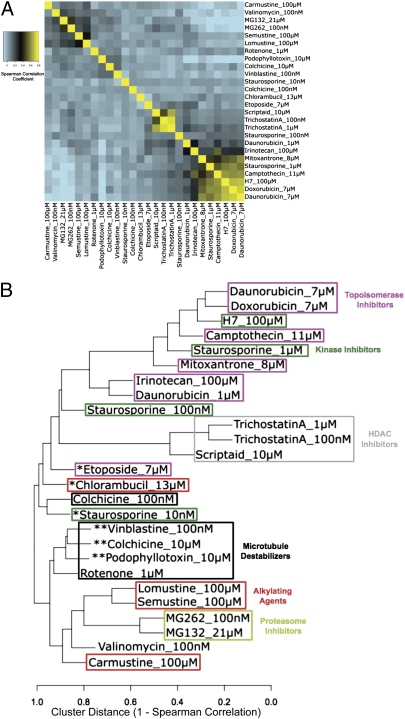
We confirmed this observation by clustering lethal compounds based on gene expression profiling using the same clustering algorithm that we applied to the modulatory profiles. We assembled gene expression profiling data from the Broad Institute's connectivity map (http://www.broadinstitute.org/cmap/) for lethal compounds that we also tested in the modulatory profiling system. These data were obtained relatively early after compound treatment (6 h) in an attempt to capture the primary mechanism of action. We used data from 281 separate microarray chips, including 86 drug treatments and 195 vehicle controls. The compounds, concentrations, and cell lines used are listed in SI Appendix, Table S4. We treated each concentration as a separate instance and calculated Spearman correlations based on gene expression data using MCF7 and PC3 cells, the two cell lines used in the connectivity map pipeline. The resulting similarity matrix is shown in Fig. 3A, and the dendrogram produced from hierarchical clustering is shown in Fig. 3B.
Fig. 3.
Clustering characterized lethal compounds based on gene expression profiles. (A) Heat map of the similarity matrix showing the Spearman correlation between gene expression profiles of lethal compounds. (B) Dendrogram derived from hierarchical clustering of the similarity matrix in A. *Compound used at sublethal concentration. **Compound used at supralethal concentration.
There are a number of noteworthy features in the dendrogram created from gene expression profiles. Not surprisingly, sublethal concentrations of compounds do not cluster into the appropriate mechanistic class (Fig. 3B, asterisk and note that lethal concentrations in MCF7 cells of all of the compounds are listed in SI Appendix, Table S4). Looking only at compounds tested at relevant lethal concentrations, some mechanistic classes were quite well-differentiated, such as HDAC inhibitors and proteasome inhibitors, whereas others were not. Kinase inhibitors and topoisomerase inhibitors were mixed together. Interestingly, very high concentrations (relative to the EC50) of microtubule destabilizers clustered together, whereas lower but still cytotoxic concentrations did not. Overall, characterized compounds are less accurately placed into their appropriate mechanistic class when clustering is based on gene expression profiling compared with modulatory profiling.
We also clustered compounds based on their chemical structure. Apart from a small number of analogs, chemical structure did not correctly group compounds according to their known bioactivities (SI Appendix, SI Text and SI Appendix, Fig. S7).
Clustering Uncharacterized Compounds Based on Modulatory Profiles.
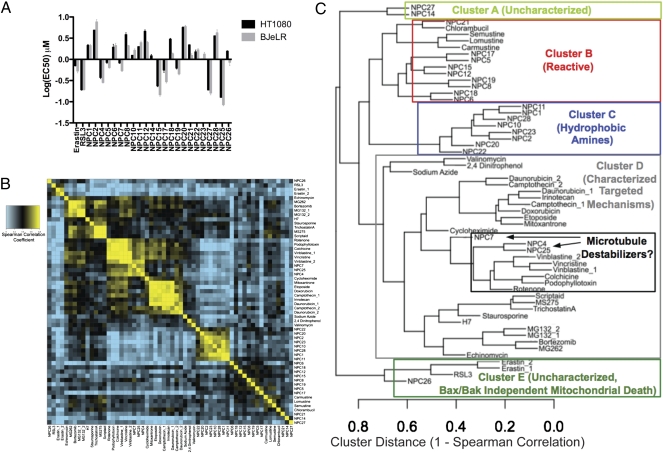
We next sought to use modulatory profiling to classify compounds lacking annotated mechanisms. We examined two compounds that we previously identified in a screen for selectively killing cells expressing a constitutively active mutant of the rat sarcoma viral oncogene homolog (RAS) (40–42) as well as 23 other compounds [novel profiling compounds (NPC)] that we found to be lethal to BJ-TERT/LT/ST/RASV12 cells (42) but for which no other characterization had been performed. The potency of these compounds in HT-1080 and BJ-TERT/LT/ST/RASV12 cells is shown in Fig. 4A, and their structures are shown in SI Appendix, Table S5. We performed modulatory profiling on these compounds (SI Appendix, Fig. S8 and SI Appendix, Table S3), calculated Spearman correlations, and hierarchically clustered the characterized and uncharacterized compounds together (Fig. 4 B and C).
Fig. 4.
Clustering uncharacterized lethal compounds based on modulatory profiles. (A) Potency of uncharacterized lethal compounds in BJ-TERT/LT/ST/RASV12 and HT-1080 cells after 48 h. Values represent the average of three replicates ± SEM. (B) Heat map of the similarity matrix showing the Spearman correlation between modulatory profiles of characterized and uncharacterized lethal compounds. (C) Dendrogram derived from hierarchical clustering of the similarity matrix in B.
Three Compounds Destabilize Microtubules.
NPC4, NPC7, and NPC25 are not structurally related to each other or to known microtubule-destabilizing agents (Fig. 5A); nonetheless, the modulatory profiles of these three compounds were similar to each other and to the profiles of known microtubule-destabilizing agents (Fig. 5B). We showed by immunofluorescence that all three compounds showed a near total loss in acetylated tubulin within 60 min in a fashion similar to colchicine, a known microtubule destabilizer (Fig. 5C), confirming the prediction from their modulatory profiles that they would depolymerize microtubules. Characterized and uncharacterized compounds from other clusters did not have an effect on microtubule stability (Fig. 4C and SI Appendix, Figs. S6 and S9).
Fig. 5.
Previously uncharacterized compounds destabilize microtubules. (A) Structures of the well-characterized microtubule destabilizers vinblastine, podophyllotoxin, and colchicine, the previously reported destabilizer rotenone, and the three compounds predicted to destabilize microtubules based on their modulatory profiles. (B) Heat map of the Spearman correlations between the modulatory profiles of the uncharacterized compounds NPC4, NPC7, and NPC25 and the characterized compounds colchicine, vinblastine, carmustine, trichostatin A, MG132, and doxorubicin. (C) Time course of TC-7 cells stained for acetylated tubulin after treatment with 1 μM colchicine, 28 μM NPC4, 28 μM NPC7, or 14 μM NPC25. Staining for total tubulin and acetylated tubulin after a 60-min treatment with vehicle (DMSO) is shown at the top right. Representative images were chosen from each time point.
Compounds That Act Nonspecifically Through Reactivity or Biophysical Properties.
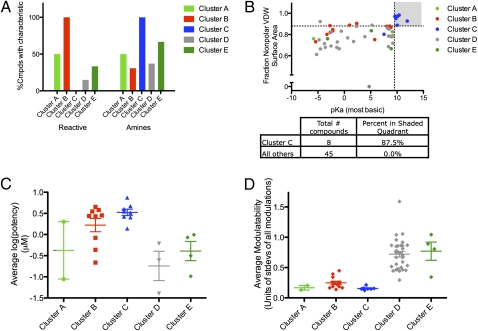
The majority of the uncharacterized compounds fell into two clusters, which are labeled B and C in Fig. 4C. Examination of these compounds’ structures (SI Appendix, Table S5) showed that cluster B contained a large number of compounds with reactive functionalities, including known alkylating agents. To quantify the relative reactivity of this cluster, we applied an established set of predictive reactivity filters (43); 10 of 13 compounds were scored as reactive by this filter, but the other 3 compounds are activated chloroarenes, also known electrophiles (44, 45). All 13 members of this cluster are therefore likely to be reactive in a biological milieu. In contrast, only 6 of the other 40 compounds were flagged as reactive by the filter (Fig. 6A). We examined analogs of one member of cluster B to further demonstrate the importance of reactivity on lethality (SI Appendix, SI Text and SI Appendix, Fig. S10 A and B).
Fig. 6.
Two clusters of previously uncharacterized lethal compounds act nonspecifically. (A) Distribution of reactive compounds and amines throughout the clusters from Fig. 4C. Reactivity was calculated by adding activated chloroarenes to filters described previously (43). (B) Fraction nonpolar van der Waals surface area vs. pKa of the most basic residue. Fraction nonpolar surface area was calculated with the built-in parameter in the software MOE. The most basic pKa was calculated with the web-based software SPARC (SI Appendix, SI Methods). Using a one-way ANOVA and Tukey's multiple comparison test, compounds in cluster C are significantly different in their fraction nonpolar surface area from those compounds in cluster D (P < 0.01) and significantly more basic than those compounds in cluster B or D (P < 0.01). (C) Average potency of uncharacterized compounds. The average of the potency in HT-1080 and BJ-TERT/LT/ST/RASV12 cells is shown. Using a one-way ANOVA and Tukey's multiple comparison test, the compounds in clusters B and C are significantly less potent than the compounds in cluster D, and the compounds in cluster C are significantly less potent than the compounds in cluster E (P < 0.05). (D) Average modulatability of compounds. Modulatability was calculated by taking the mean of the absolute value of all of the normalized changes in a compound's modulatory profile. Lines represent cluster averages ± SEM. Using a one-way ANOVA and Tukey's multiple comparison test, compounds in clusters A, B, and C each are significantly less modulatable than those compounds in clusters D and E (P < 0.05).
Compounds in cluster C are all amines with a relatively large nonpolar surface area (Fig. 6 A and B and SI Appendix, Table S5), suggesting that they act nonspecifically in a detergent-like fashion. Consistent with this observation, we found that the compounds in this cluster act rapidly, causing rounding up of cells within 40 min of treatment and subsequent loss of cell adhesion (SI Appendix, Fig. S10C).
Small molecules are more likely to act specifically at low concentrations, whereas at higher concentrations, they can act more promiscuously (46). We examined the potency of the compounds in the different lethal compound clusters (Fig. 6C), omitting the characterized compounds to avoid selection bias. Consistent with our expectation, the compounds in clusters B and C that appear to act nonspecifically were the least potent.
Although a compound targeting a specific cellular pathway is susceptible to modulation, compounds causing nonspecific cellular damage are unlikely to be susceptible to such modulation. Thus, nonspecifically acting compounds should have modulatory profiles consisting of minimal changes. To test this theory, we computed the average of the absolute value of each profile and called this value the modulatability score. We compared the average modulatability of the different lethal compound clusters (Fig. 6D). As expected, cluster D (containing compounds known to target specific cellular processes) had high modulatability, whereas compounds in clusters B and C, which appear to act through reactivity or biophysical mechanisms, had consistently low modulatability (this finding can also be seen in the heat map shown in SI Appendix, Fig. S8).
There were two clusters of uncharacterized compounds remaining: clusters A and E. Cluster E had potent compounds with high modulatability. Cluster A had one potent and one less potent compound, both with low modulatability. We investigated further the compounds in cluster E, because based on these metrics, they are more likely to act through modulation of specific cellular targets.
Cluster of Compounds Induces BAX-/BAK-Independent Mitochondrial Cell Death.
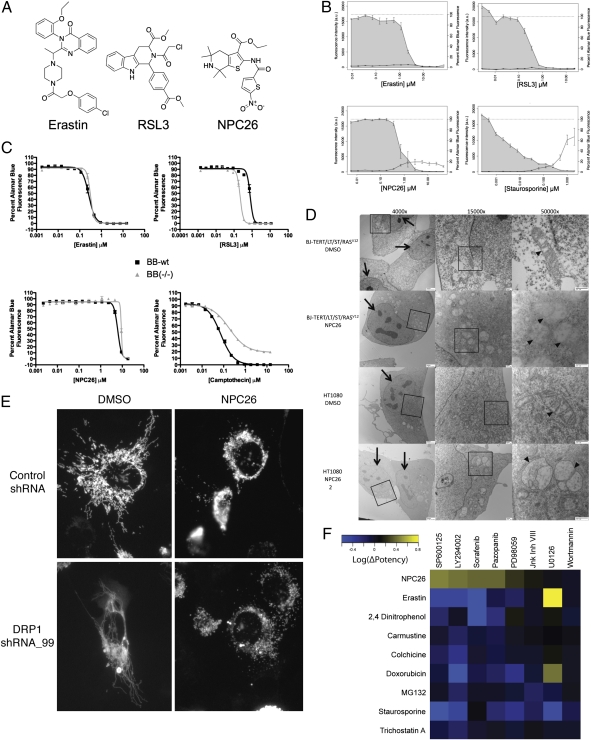
Cluster E in Fig. 4C contains three compounds. We previously found that two of these compounds, erastin and Ras-selective-lethal compound 3 (RSL3), induce a nonapoptotic, iron-dependent form of death that involves the generation of reactive oxygen species. Further confirming this finding in the present context, we observed that erastin and RSL3 caused no detectable caspase activation, which was measured by cleavage of a fluorogenic caspase substrate (Fig. 7B).
Fig. 7.
Previously uncharacterized compounds induce BAX-/BAK-independent mitochondrial cell death. (A) Structures of erastin, RSL3, and NPC26, which are the three compounds in cluster E. (B) Caspase activity in HT-1080 cells treated for 12–15 h with erastin, RSL3, NPC26, and staurosporine. Caspase activity was measured by the cleavage of a fluorogenic caspase substrate and is shown on the left y axis (points represent the mean of three replicates ± SEM). Cell viability is shown by the shaded region and on the right y axis (points represent the mean of three replicates ± SEM). (C) Lethality of erastin, RSL3, NPC26, and camptothecin in WT or Bax−/−Bak−/− MEFs after 48 h of treatment. Data represent the mean of three replicates ± SEM. (D) EM images taken of BJ-TERT/LT/ST/RASV12 cells after a 3-h treatment with either DMSO or 9 μM NPC26 and EM images taken of HT-1080 cells after a 6-h treatment with either DMSO or 9 μM NPC26. Arrows show the nuclei, and arrowheads show mitochondria. Boxes show the portion of the image shown at higher magnification in the next image to the right. (E) Fluorescence images of HT-1080 cells expressing a mitochondrially targeted dsRed construct infected with either shRNA targeting DRP1 or a control nontargeting shRNA treated with either DMSO or 9 μM NPC26 for 6 h. (F) Heat map depicting the ability of various kinase inhibitors to alter death induced by NPC26 and other lethal compounds. Protection is depicted in yellow, and sensitization is depicted in blue. Experiments were performed in HT-1080 cells.
Death independent of caspase activity can be dependent on mitochondrial outer membrane permeabilization mediated by BCL2-associated X protein (BAX) or BCL2-antagonist/killer (BAK) (47). We therefore tested the lethality of RSL3 and erastin in WT and Bax−/−Bak−/− double KO mouse embryonic fibroblasts (MEFs). The absence of BAX and BAK did not suppress death induced by erastin, and it sensitized cells to death induced by RSL3 (Fig. 7C). Thus, modulatory profiling correctly predicted that erastin and RSL3 act through a form of cell death that is nonapoptotic and, in fact, is independent of the core apoptotic machinery—caspases, BAX, and BAK.
We then asked whether the other compound in cluster E, NPC26, also acts through a nonapoptotic cell death process. We repeated a subset of the changes in the modulatory profile in two additional cell lines and found good consistency between the cell lines (SI Appendix, Fig. S11A), suggesting that the modulatory profile is a good marker of the compound's action independent of cell line. Within the profile, however, there were some immediate mechanistic differences apparent between NPC26 and erastin/RSL3. Erastin and RSL3 are virtually inactive in the presence of all of the reactive oxygen species scavengers tested [α-tocopherol, Trolox, butylated hydroxyanisole (BHA), and butylated hydroxytoluene (BHT)], the iron chelator deferoxamine, and the MAPK/ERK Kinase (MEK) inhibitor U0126. NPC26 was only slightly inhibited by α-tocopherol, Trolox, and deferoxamine, and it was insensitive to the presence of BHA, BHT, or U0126 (SI Appendix, Table S3). Thus, reactive oxygen species, iron, and MEK signaling play a less central (if any) role in NPC26-induced death. NPC26 caused a small increase in cleavage of a fluorogenic caspase substrate (Fig. 7B), but death induced by NPC26 was not blocked by caspase inhibitors (SI Appendix, Fig. S11B) or affected by the absence of BAX and BAK (Fig. 7C). These observations led us to the hypothesis that NPC26 induced a different but perhaps distantly related death process from the process induced by erastin and RSL3.
We had previously found by EM that erastin does not affect nuclear morphology but does cause a loss of mitochondrial structural integrity (41). To test the hypothesis that NPC26 induces a distantly related form of death, we performed EM to determine if NPC26 induced similar features. The images, shown in Fig. 7D, reveal no change in nuclear morphology and a drastic mitochondrial phenotype. By 3 h in BJ-TERT/LT/ST/RASV12 and 6 h in HT-1080 cells, mitochondria have become circular and lost their cristae completely. We investigated these mitochondrial morphological changes further by expressing a mitochondrially targeted red fluorescent protein (DsRed2-mito). NPC26 induced a conversion from elongated to punctate mitochondria, a phenotype that has been previously observed during apoptosis (48) and by death induced by inhibitors of oxidative phosphorylation and uncoupling agents (49). However, typical apoptotic-inducing stimuli failed to induce similar mitochondrial fragmentation in HT-1080 cells (SI Appendix, Fig. S11C). Unlike with inhibitors of oxidative phosphorylation, NPC26-induced mitochondrial fragmentation is independent of the activity of the dynamin-related protein (DRP1) (Fig. 7E and SI Appendix, Fig. S11D). NPC26 is also not a direct uncoupling agent (SI Appendix, Fig. S11E). Thus, similar to erastin and RSL3, NPC26 induces a distinct form of BAX-/BAK-independent mitochondrial cell death.
NPC26-Induced Death Depends on a Unique Kinase Signaling Pathway.
We next asked whether the signaling pathways controlling NPC26-induced death are unique from those pathways controlling death induced by characterized compounds or erastin. We first tested a collection of kinase inhibitors for their ability to inhibit NPC26-induced death (compounds and activity are shown in SI Appendix, Table S6). We then tested active inhibitors and related inactive inhibitors for their ability to inhibit erastin and a representative subset of the characterized lethal compounds. The results (Fig. 7F) show that NPC26-induced death depends on kinase signaling pathways distinct from the other compounds.
Although this pattern serves as a measure of uniqueness, it did not allow us to identify a specific kinase involved, because no two redundant inhibitors were effective in preventing death. For example, one JNK inhibitor (SP600125) prevented death, whereas another inhibitor did not (JNK Inh VIII). Similar results were obtained with MEK inhibitors (PD98059 protects and U0126 does not protect) and PI3K inhibitors (LY294002 protects and wortmannin does not protect). Such inhibitors have significant promiscuity within the kinase family, raising two possibilities for these results: (i) the compounds that suppress NPC26-induced death inhibit the same off-target kinase or (ii) the compounds that suppress NPC26-induced death inhibit multiple kinases, and the pathway is robust to the inhibition of any single kinase.
Structure Activity Relationships of NPC26.
Because of the high potency and modulatability of its cluster, modulatory profiling predicted that NPC26 would exert its effect through a specific process. To further confirm this hypothesis, we tested structural analogs of NPC26 (17 purchased and 7 synthesized analogs) and scored their activity based on their ability to induce cell death that could be inhibited by the kinase inhibitor SP600125 (SI Appendix, Table S7). In addition, we resynthesized NPC26, providing the most rigorous confirmation that the assigned structure is correct.
Analog testing provided evidence supporting a specific mechanism for NPC26-induced death. Subtle structural changes that would not be predicted to alter the compound's reactivity or lipophilicity eliminated activity. For example, although analog 26A14 is active, analog 26A3, in which a seven-membered ring in 26A14 is replaced by a six-membered ring, is inactive (SI Appendix, Fig. S11F).
In addition, we tested an analog containing a linker with a protected amine attached at the end. This analog (SI Appendix, Table S7, SRS1-78) retained activity, suggesting the potential for the use of NPC26 as a probe for future affinity purification of its target. In summary, NPC26 acts through a nonapoptotic, mitochondrial-driven mechanism, and it has a modulatory profile and structure activity relationship to suggest a specific molecule target and mechanism. Thus, modulatory profiling is useful not only for classifying compounds that act through known cell death mechanisms and targets but also for flagging for additional study compounds that are likely to act through previously unidentified cell death mechanisms.
Discussion
Misregulation of cell death plays a central role in tumorigenesis, neurodegeneration, damage from infarction, and a host of other human diseases. Greater understanding of the scope of cell death pathways that can be activated in cells and gaining pharmacological control over such pathways hold the promise of improving human health. In this study, we present modulatory profiling as a methodology for categorizing diverse inducers of cell death and identifying inducers of previously uncharacterized cell death processes, with the goal of ultimately creating a global view of cell death mechanisms.
Over the past decade, technologies have enabled the collection of large-scale molecular profiles and led to their use in characterizing cellular states and perturbations to those states. These technologies include gene expression and copy number variation using DNA microarrays, protein expression and metabolite profiling using MS, enzyme activity using fluorescence probes, and cell morphology using high-content imaging (50). Although these technologies have proven invaluable for investigating a wide range of basic biological mechanisms and disease processes, it can be difficult to separate causal changes from correlative changes using such methods. For example, deletion, knockdown, or inhibition of many of the proteins that are altered in abundance in a specific cell state has no consequence, because these changes are products of the cell state rather than regulators of it. A great deal of effort is being put to identifying the smaller number of driver mutations responsible for producing cell states of interest (51–53). Such techniques are computationally and data intensive, and they require expert adaptation and fine tuning to each new biological domain. Thus, despite these technological and computational advances, there remains a need for the development of high-dimensionality profiles of functionally relevant measurements that can be used to characterize cell states.
We developed modulatory profiling to address this deficiency, which is particularly acute with regards to small molecule-induced cell states, particularly cell death. Small molecules, even approved drugs, can have pleiotropic effects on cells (54, 55). Identifying which of the many effects is relevant in a given context is important and nontrivial. Rotenone provided an illustrative example of this problem within the data presented here. Rotenone has been used extensively as an inhibitor of mitochondrial complex I (56), but it has also been shown to destabilize microtubules (33, 34). Using a measurement such as gene expression profiling, it is not obvious which of these two effects will be most strongly represented. If one is interested in the way in which rotenone induces a downstream phenotype such as cell death, it is also not necessarily the case that the lethal effect will be the one with the strongest gene expression signature. However, modulatory profiling, by using functional measurements, highlighted the lethal mechanism of rotenone in the system tested.
Other systems have been developed using collections of functional assays to investigate small molecule activities, including comparing pairwise interactions of antibiotics in bacteria (57, 58), profiling the hypersensitivity of yeast haploid deletion mutants (59, 60), profiling cytotoxicity across 60 cancer cell lines (61, 62), and most recently, measuring changes in cancer cell survival in the presence of various shRNAs (63). Although these other systems have provided valuable insights, they also have certain drawbacks. Nonapoptotic cell death differs markedly even between mammalian cells and other metazoans, such as D. melanogaster and C. elegans (47), making the use of models in bacteria and yeast less relevant. The NCI60 approach has produced useful signatures for finding compounds with similar modes of action, but the use of 60 genetically diverse cell lines does not give any specific information about cell death mechanisms and is less amenable to genetic perturbations and the use of neurons and other specialized cell types. The advantages of modulatory profiling are that (i) it uses quantitative, functional assays that measure and perturb cell death, the process that is specifically under study; (ii) it uses a full dilution series of each compound, eliminating the possibility of making measurements at irrelevant concentrations; and (iii) the assays are performed in human cells, the species in which we ultimately want to apply our findings.
However, we made some choices that proved somewhat limiting. We used exclusively small molecules as death-inducing agents. Although small molecules are tractable experimentally, we were limited by the number of existing classes of characterized lethal compounds. It will be desirable to extend modulatory profiling to include larger numbers of other stimuli, particularly gene knockdown and overexpression. In addition, we chose two fibroblast cell lines that are well-suited for the type of miniaturized assays required for performing high-throughput assays; however, the use of these two cell lines is likely to prevent the analysis of more specialized pathways. We chose to focus, therefore, on capturing the more common mechanisms. Last, we showed here the utility of modulatory profiling as applied only to cell death. It would be desirable to extend modulatory profiling to other cell fates and processes.
We measured viability with Alamar blue, a fluorogenic dye that measures cellular reductive potential. Although this reagent has shown excellent correlation with other viability metrics, both in our hands and in the hands of others (42, 64), in certain cases, it can give false positives and negatives (65). It should be emphasized, however, that to disrupt the output of the profiling, a modulator would have to cause a viability-independent effect on Alamar blue reduction to varying extents for different lethal compounds. Such a finding is expected to be a rare event.
As a proof of principle, we showed that modulatory profiling correctly classified characterized compounds according to their known mechanisms of action. It did this without ambiguity, despite using a relatively naïve clustering algorithm. Clustering compounds based on gene expression data using the same algorithm was less accurate. Although more sophisticated methods for analyzing gene expression data have shown an improved ability to identify compounds with the same mechanism of action (37, 39), it should be emphasized that modulatory profiling accomplished accurate clustering without sophisticated computational methods.
These studies identified three microtubule destabilizers from structural classes that had not previously been shown to destabilize microtubules. It also allowed us to identify two groups of compounds that likely act nonspecifically, one through chemical reactivity and the other through detergent-like properties. Such classification is highly desirable, because these compounds are uninteresting as both probes and pharmaceuticals. Identifying such compounds without modulatory profiling would not have been possible without also removing a number of specifically acting characterized compounds and some highly interesting previously uncharacterized compounds. Finally, modulatory profiling identified compounds that kill cells through pathways distinct from characterized pathways. Expanding the number of characterized cell death pathways could contribute greatly to our basic understanding of cellular fates and provide avenues for cancer cell-specific therapies. Erastin and RSL3 were originally identified in our laboratory (40–42), and the current work serves to show the uniqueness of their mode of death relative to a large number of other compounds.
One uncharacterized compound, NPC26, had a modulatory profile similar to the profiles of erastin and RSL3. Although the mechanism of action of this compound has important differences from the mechanisms of erastin and RSL3, it also induces a BAX-/BAK-independent death that involves severe disturbances to mitochondria. The death process can be prevented by a unique set of kinase inhibitors, and structural activity relationships testing shows that this compound acts specifically and can be modified for use as an affinity probe.
In summary, we have developed a system of modulatory profiling, and we have shown its ability to map cell death pathways and identify small molecules operating through known and unknown mechanisms. Use of this system should improve our understanding of cell death and provide small molecule tools to dissect and control diverse biological and disease processes.
Experimental Procedures
Cell Lines.
BJ-TERT/LT/ST/RASV12 and HT-1080 cells were cultured as described previously (42). The WT and Bax−/−Bak−/− MEFs were provided by Craig Thompson (Memorial Sloan-Kettering Cancer Center) (SI Appendix, SI Methods).
Cell Survival Assays.
Cells were trypsinized, counted, and combined with modulators or vehicle and seeded into 384-well plates. Cell death-inducing agents were then added. After 48 h, Alamar blue was added to a final concentration of 10%. After 16 h of incubation, the fluorescence intensity was determined using a Victor 3 plate reader. All assays were performed in at least triplicate.
Determination of Changes in Potency and Efficacy.
Background was subtracted from raw fluorescence measurements, and values were normalized to vehicle or modulator-only controls. Four-parameter logistic best-fit dose curves were constructed for each of the replicates using GraphPad Prism. The change in potency was defined as the log ratio of the concentration of compound in the presence of modulator to the concentration in the absence of modulator required to produce a level of cell survival equal to the one-half maximal reduction in viability produced in the absence of the modulator. Efficacy changes were defined as the difference between the curves in the presence vs. the absence of modulator at the highest concentration of lethal compound tested. The efficacy measurement was deemed unreliable when no measurement was taken at a concentration close to the bottom of the curve, and a missing value was assigned in its place (additional details on calculating the parameters and using them to cluster the compounds are in SI Appendix, SI Methods).
Preprocessing and Clustering Based on Gene Expression Profiles.
Microarray data were downloaded from the Broad's Connectivity Map website (http://www.broadinstitute.org/cmap/) as cell intensity files (CEL files). For the experiments included in our analysis, cells were treated with compound for 6 h before lysis and mRNA collection. More detailed descriptions of the experimental protocols are available on the website and in the publication about the project (37). We performed probe set summarization using the MAS5 algorithm (66). Values were thresholded, and consistently low or invariant probe sets were removed. Values were then normalized to a batch-matched vehicle-only control, and redundant probe sets for the same gene were averaged together (SI Appendix, SI Methods has additional details of processing and clustering).
Supplementary Material
Acknowledgments
We thank Todd Golub and Justin Lamb at the Broad Institute for generating and providing the microarray data, Craig Thompson for the Bax−/−Bak−/− double KO MEFs, and Lloyd Greene, Todd Golub, and Andrea Califano for comments on the manuscript. A.J.W. was supported in part by a National Institutes of Health Medical Scientist Training Program grant to Columbia University. B.R.S. is an Early Career Scientist of the Howard Hughes Medical Institute and is supported by additional funding from the Arnold and Mabel Beckman Foundation, New York Science, Technology, and Academic Research (NYSTAR), and National Institutes of Health Grants R01CA097061, R01GM085081, and RC2CA148308.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 16151.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106149108/-/DCSupplemental.
References
- 1.Wyllie AH. Cell death: A new classification separating apoptosis from necrosis. In: Bowen ID, Lockshin RA, editors. Cell Death in Biology and Pathology. London: Chapman & Hall; 1981. pp. 9–34. [Google Scholar]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dive C, et al. Analysis and discrimination of necrosis and apoptosis (programmed cell death) by multiparameter flow cytometry. Biochim Biophys Acta. 1992;1133:275–285. doi: 10.1016/0167-4889(92)90048-g. [DOI] [PubMed] [Google Scholar]
- 4.Duvall E, Wyllie AH. Death and the cell. Immunol Today. 1986;7:115–119. doi: 10.1016/0167-5699(86)90152-0. [DOI] [PubMed] [Google Scholar]
- 5.Taylor RC, Cullen SP, Martin SJ. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 6.Vercammen D, et al. Dual signaling of the Fas receptor: Initiation of both apoptotic and necrotic cell death pathways. J Exp Med. 1998;188:919–930. doi: 10.1084/jem.188.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 8.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitomi J, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 13.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 16.Luke CJ, et al. An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysosomal injury. Cell. 2007;130:1108–1119. doi: 10.1016/j.cell.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 18.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofius D, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 22.Kroemer G, et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2008;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bredesen DE. Key note lecture: Toward a mechanistic taxonomy for cell death programs. Stroke. 2007;38(Suppl):652–660. doi: 10.1161/01.STR.0000257802.82826.a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang J, Koen YM, Hanzlik RP. Bioinformatic analysis of xenobiotic reactive metabolite target proteins and their interacting partners. BMC Chem Biol. 2009;9:5. doi: 10.1186/1472-6769-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng BY, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson JK, et al. A comparison of the lytic properties of two ether-linked lipids, one with and one without antineoplastic activity. Biochem Cell Biol. 1994;72:297–303. doi: 10.1139/o94-042. [DOI] [PubMed] [Google Scholar]
- 28.Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 30.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 31.Coan KE, Shoichet BK. Stability and equilibria of promiscuous aggregates in high protein milieus. Mol Biosyst. 2007;3:208–213. doi: 10.1039/b616314a. [DOI] [PubMed] [Google Scholar]
- 32.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 33.Brinkley BR, Barham SS, Barranco SC, Fuller GM. Rotenone inhibition of spindle microtubule assembly in mammalian cells. Exp Cell Res. 1974;85:41–46. doi: 10.1016/0014-4827(74)90210-9. [DOI] [PubMed] [Google Scholar]
- 34.Marshall LE, Himes RH. Rotenone inhibition of tubulin self-assembly. Biochim Biophys Acta. 1978;543:590–594. doi: 10.1016/0304-4165(78)90315-x. [DOI] [PubMed] [Google Scholar]
- 35.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 36.Waring JF, et al. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol Appl Pharmacol. 2001;175:28–42. doi: 10.1006/taap.2001.9243. [DOI] [PubMed] [Google Scholar]
- 37.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 38.Boshoff HI, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 39.Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci USA. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 41.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oprea TI. Property distribution of drug-related chemical databases. J Comput Aided Mol Des. 2000;14:251–264. doi: 10.1023/a:1008130001697. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh PB, Whitehouse MW. 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole: A new fluorigenic reagent for amino acids and other amines. Biochem J. 1968;108:155–156. doi: 10.1042/bj1080155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anwair MA, et al. Lipophilicity of aminopyridazinone regioisomers. J Agric Food Chem. 2003;51:5262–5270. doi: 10.1021/jf0343938. [DOI] [PubMed] [Google Scholar]
- 46.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tait SW, Green DR. Caspase-independent cell death: Leaving the set without the final cut. Oncogene. 2008;27:6452–6461. doi: 10.1038/onc.2008.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 49.De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol. 2005;15:678–683. doi: 10.1016/j.cub.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 50.Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Multi-parameter phenotypic profiling: Using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8:567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 53.Lim WK, Lyashenko E, Califano A. Master regulators used as breast cancer metastasis classifier. Pac Symp Biocomput. 2009;2009:504–515. [PMC free article] [PubMed] [Google Scholar]
- 54.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 56.Lindahl PE, Oberg KE. Mechanism of the physiological action of rotenone. Nature. 1960;187:784. doi: 10.1038/187784a0. [DOI] [PubMed] [Google Scholar]
- 57.Farha MA, Brown ED. Chemical probes of Escherichia coli uncovered through chemical-chemical interaction profiling with compounds of known biological activity. Chem Biol. 2010;17:852–862. doi: 10.1016/j.chembiol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 59.Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 60.Parsons AB, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 61.Paull KD, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 62.Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 63.Jiang H, Pritchard JR, Williams RT, Lauffenburger DA, Hemann MT. A mammalian functional-genetic approach to characterizing cancer therapeutics. Nat Chem Biol. 2011;7:92–100. doi: 10.1038/nchembio.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nociari MM, Shalev A, Benias P, Russo C. A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J Immunol Methods. 1998;213:157–167. doi: 10.1016/s0022-1759(98)00028-3. [DOI] [PubMed] [Google Scholar]
- 65.Hamid R, Rotshteyn Y, Rabadi L, Parikh R, Bullock P. Comparison of alamar blue and MTT assays for high through-put screening. Toxicol In Vitro. 2004;18:703–710. doi: 10.1016/j.tiv.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 66.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]