Abstract
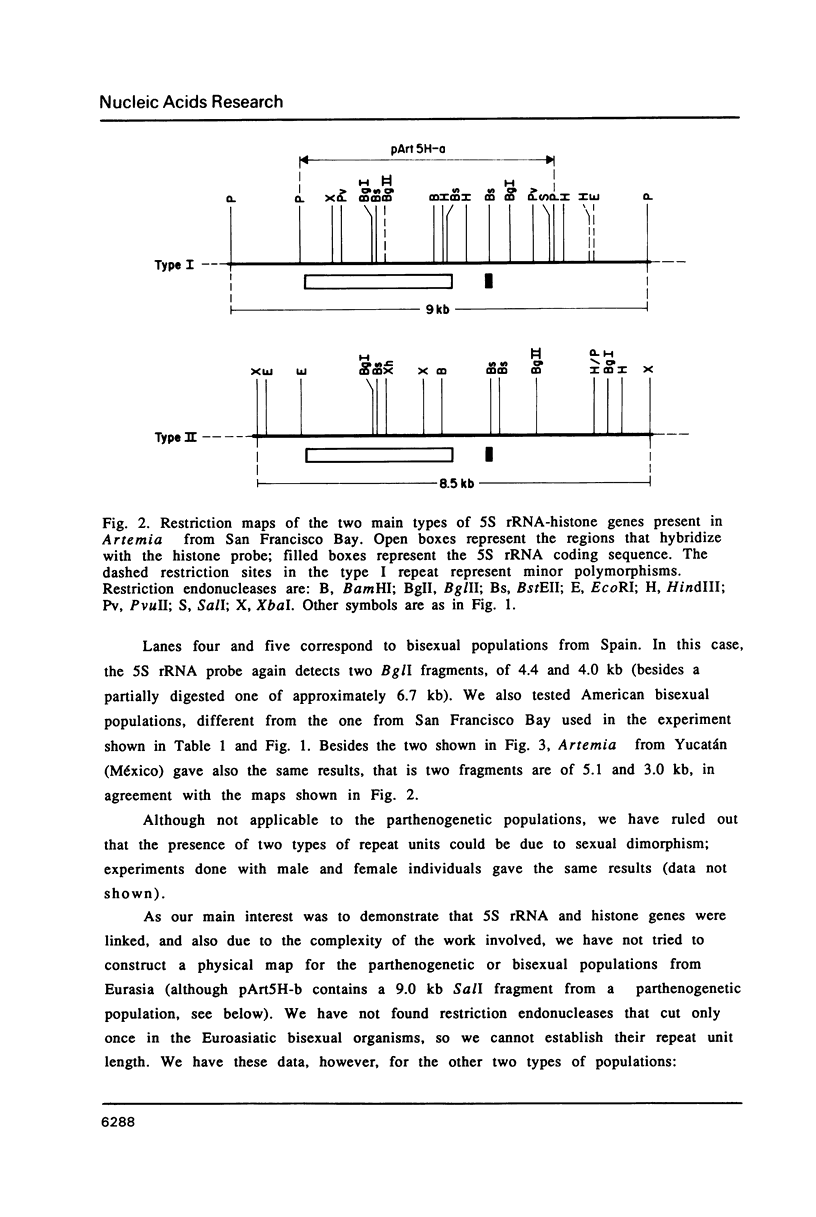
5S rRNA genes are linked to the histone genes in the 13 populations of the crustacean Artemia that we have studied. In all cases, two types of repeat units are found. Southern blot analysis of all populations shows that they can be grouped into three classes: a) American bisexuals; b) Eurasian bisexuals, and c) parthenogenetic organisms (all from Eurasia). Restriction analysis of a bisexual population from San Francisco Bay shows that the two repeat units are of 9.0 and 8.5 kb (with minor heterogeneities of restriction sites). In parthenogenetic organisms, the two repeat units are of approximately 12 kb. Sequencing data from the region of the 5S rRNA from the San Francisco Bay population, shows that in both types of units, the single 5S rRNA gene (315 bp in length), is located 430 bp downstream the 3' regulatory sequences of the H2A gene, the last gene in the histone cluster. We have isolated three clones that contain 5S rRNA sequences. Two of them (one from an American bisexual and the other from a parthenogenetic population) contain histone and 5S rRNA genes, both with the same transcriptional polarity. The third clone, lacking histone genes, is likely to be an orphon derived from the parthenogenetic population.
Full text
PDF



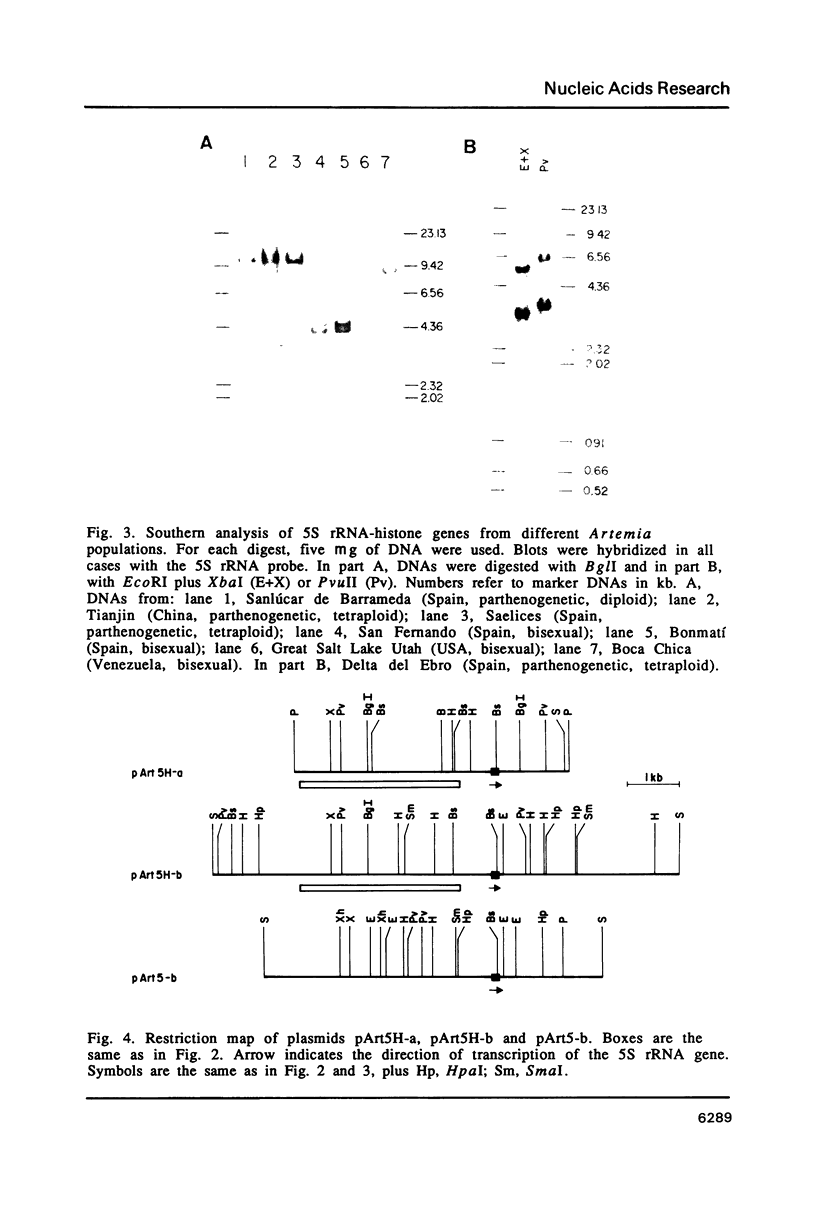


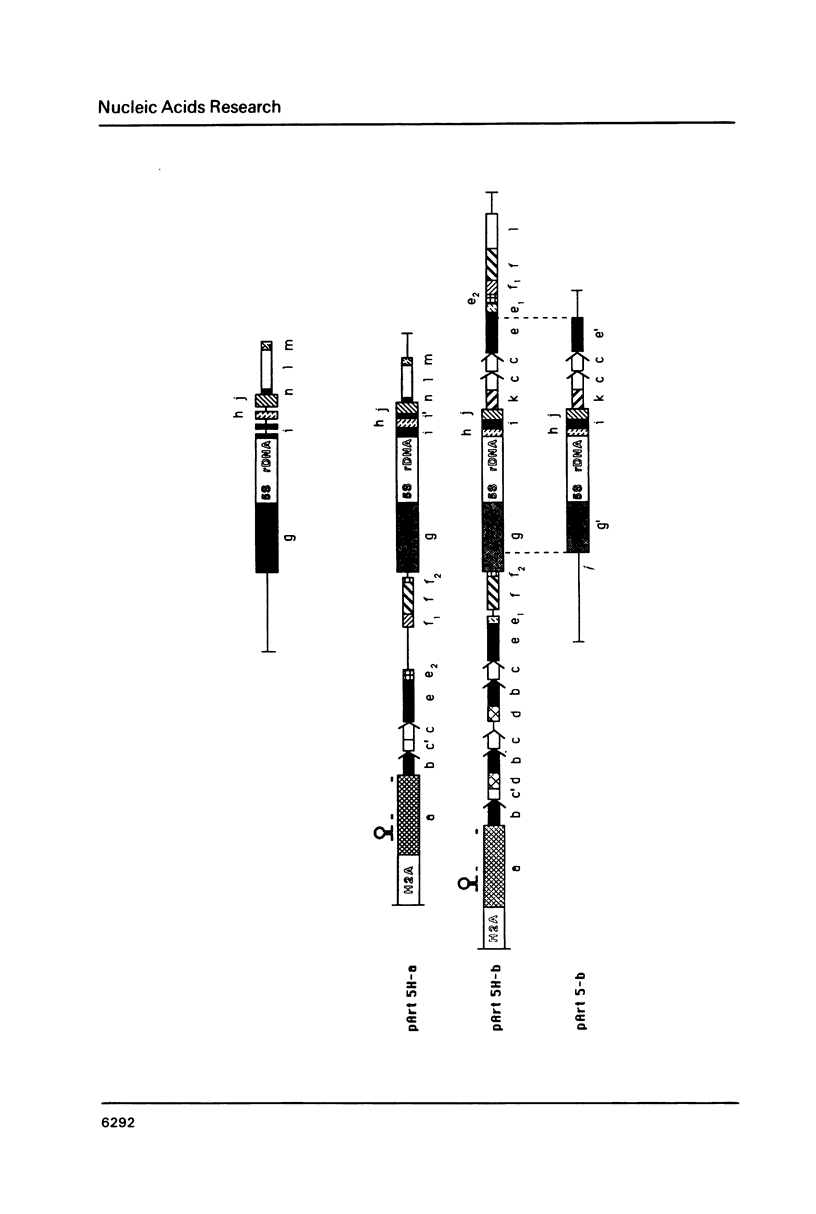





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Sprecher C., Graves R., Marzluff W. F., Skoultchi A. I. Regulated expression of a chimeric histone gene introduced into mouse fibroblasts. Mol Cell Biol. 1985 Sep;5(9):2316–2324. doi: 10.1128/mcb.5.9.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. T., Vaughn J. C., Perry B. A., Bagshaw J. C. Interspersion of histone and 5S RNA genes in Artemia. Gene. 1987;51(1):61–67. doi: 10.1016/0378-1119(87)90474-4. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Schedl P., Tschudi C., Pirrotta V., Steward R., Gehring W. J. The 5S genes of Drosophila melanogaster. Cell. 1977 Dec;12(4):1057–1067. doi: 10.1016/0092-8674(77)90169-6. [DOI] [PubMed] [Google Scholar]
- Bagshaw J. C., Skinner H. B., Burn T. C., Perry B. A. Nucleotide sequence of the 5S RNA gene and flanking regions interspersed with histone genes in Artemia. Nucleic Acids Res. 1987 Apr 24;15(8):3628–3628. doi: 10.1093/nar/15.8.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Cruces J., Sebastián J., Renart J. Restriction mapping of the rRNA genes from Artemia larvae. Biochem Biophys Res Commun. 1981 Jan 30;98(2):404–409. doi: 10.1016/0006-291x(81)90854-8. [DOI] [PubMed] [Google Scholar]
- Cruces J., Wonenburger M. L., Díaz-Guerra M., Sebastián J., Renart J. Satellite DNA in the crustacean Artemia. Gene. 1986;44(2-3):341–345. doi: 10.1016/0378-1119(86)90200-3. [DOI] [PubMed] [Google Scholar]
- Das G., Henning D., Wright D., Reddy R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 1988 Feb;7(2):503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L., De Baere R., Vandenberghe A., De Wachter R. The sequence of 5S ribosomal RNA of the crustacean Artemia salina. Nucleic Acids Res. 1981 Oct 10;9(19):5141–5144. doi: 10.1093/nar/9.19.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Drouin G., Hofman J. D., Doolittle W. F. Unusual ribosomal RNA gene organization in copepods of the genus Calanus. J Mol Biol. 1987 Aug 20;196(4):943–946. doi: 10.1016/0022-2836(87)90417-7. [DOI] [PubMed] [Google Scholar]
- Gallego M. E., Díaz-Guerra M., Cruces J., Sebastián J., Renart J. Characterization of two types of rRNA gene repeat units from the crustacean Artemia. Gene. 1986;48(1):175–182. doi: 10.1016/0378-1119(86)90363-x. [DOI] [PubMed] [Google Scholar]
- Hart R. P., Folk W. R. Structure and organization of a mammalian 5 S gene cluster. J Biol Chem. 1982 Oct 10;257(19):11706–11711. [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Krol A., Carbon P., Ebel J. P., Appel B. Xenopus tropicalis U6 snRNA genes transcribed by Pol III contain the upstream promoter elements used by Pol II dependent U snRNA genes. Nucleic Acids Res. 1987 Mar 25;15(6):2463–2478. doi: 10.1093/nar/15.6.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morton D. G., Sprague K. U. In vitro transcription of a silkworm 5S RNA gene requires an upstream signal. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5519–5522. doi: 10.1073/pnas.81.17.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S., Di Liegro C., Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987 Oct 9;51(1):81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R. Histone genes: not so simple after all. Cell. 1984 Oct;38(3):624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Olsen G. J., Woese C. R. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986 May 9;45(3):325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Garcia A. D. Transcription of the Drosophila melanogaster 5S RNA gene requires an upstream promoter and four intragenic sequence elements. Mol Cell Biol. 1988 Mar;8(3):1266–1274. doi: 10.1128/mcb.8.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B. Surprises in polymerase III transcription. Cell. 1988 Jan 29;52(2):153–154. doi: 10.1016/0092-8674(88)90500-4. [DOI] [PubMed] [Google Scholar]
- Tyler B. M. Transcription of Neurospora crassa 5 S rRNA genes requires a TATA box and three internal elements. J Mol Biol. 1987 Aug 20;196(4):801–811. doi: 10.1016/0022-2836(87)90406-2. [DOI] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]