Abstract
Water solubilized nanoparticles such CdSe-ZnS core-shell nanocrystals (quantum dots, QDs) have great potential in bioimaging and sensing applications due to their excellent photophysical properties. However, the efficient modification of QDs with complex biomolecules represents a significant challenge. Here we describe a straightforward arylhydrazone approach for the chemoselective covalent modification of QDs that is compatible with neutral pH and micromolar concentrations of the peptide target. The kinetics of covalent modification can be monitored spectroscopically at 354 nm in the presence of the QD and average peptide:QD ratios from 2:1 to 11:1 were achieved with excellent control over the desired valency. These results suggest that aniline catalyzed hydrazone ligation has the potencial to provide a general method for the controlled assembly of a variety of nanoparticle-biomolecule hybrids.
Keywords: Quantum dot, ligation, hydrazone, aniline, PEG, protease
Introduction
The efficient functionalization of nanoparticles (NPs) is an ongoing challenge in nanotechnology. Although semiconductor nanocrystals (quantum dots, QDs) and metallic nanoparticles have great potential in a variety of technical applications, much of their utility requires specific interactions with surfaces, organic materials or biological macromolecules. In particular, luminescent QDs have great potential in biological imaging and sensing applications due to their excellent photophysical properties that include high quantum yields, large Stokes shifts, broad absorption spectra, low levels of photobleaching, long luminescent lifetimes and photoluminescent (PL) emissions that are narrow, symmetric and size-tunable.1–6 In aqueous media, the hydrophobic semiconducting CdSe-ZnS core-shell quantum dots can be solubilized through coating with amphiphilic block copolymers7–10 or acidic thiol ligands such as mercaptoacetic acid11,12 and dihydrolipoic acid (DHLA).13 Recently, the use of hydrophilic polyethyleneglycol (PEG) appended-thiols14,15 has produced highly water soluble QDs that can be terminally functionalized with amines or carboxylic acids to facilitate non-selective labeling with functional groups present on biomolecules.15 In addition, QDs can be capped with polyCys16–18 or polyHis19 (His6 tag) peptide sequences. In particular, His6 sequences have been shown to bind the ZnS shell in DHLA and DHLA-PEG capped QDs with high affinity, making them an attractive approach to introduce functional groups into QDs.
Several strategies have been developed to introduce biological molecules onto the surface of QDs. For example, commercial streptavidin decorated QDs have been used to bind biotin linked labels,20,21 and positively charged proteins have been shown to bind negatively charged acidic QDs through electrostic interactions.13 However, these approaches are limited in utility, as they require a large protein to mediate the binding interaction. In contrast, covalent labeling enables a direct link to the solubilizing caps of the QD to be achieved. The most commonly used method is amide bond formation between carboxylic acids and amines,3,7,8,11,15,22–24 an approach that requires a significant excess of label and typically suffers from poor chemoselectivity, and a high degree of crosslinking.4,13,24 Recently, the use of a PEG-EDC coupling reagent that reduces crosslinking and aggregation in aqueous systems was reported.24 Alternatively, cysteine-maleimide reactions have found utility in peptide-QD conjugation.25,26
Despite significant advances in QD functionalization in aqueous solution, the efficient covalent modification of QDs and other NPs remains a significant technical challenge. While QDs are typically handled at μM concentrations, most conjugation reactions use a large excess of label (mM), which limits the efficiency of the process.27 In addition, since analytical methods are not available to rapidly characterize NP:peptide conjugates, these reactions are typically performed without direct feedback on the progress of the reaction. As a result, there is a need for the development of coupling strategies that can achieve enhanced reaction kinetics while maintaining chemoselectivity. Peptides are especially attractive molecules for QD modification since they bind selectively to numerous important targets and can be generated synthetically in a straightforward manner.28 These properties make them good candidates for cellular delivery, sensing applications and drug therapy.
Approach
The generation of peptide-QD complexes would be greatly facilitated by the development of a robust and convergent approach for the direct chemical conjugation of fluorescent peptide substrates directly to a reactive nanoparticle (Scheme 1). Ideally, such a method would allow unprotected peptides to be conjugated chemoselectively and without requiring an excess of peptide. Recently, we described an approach to accelerate hydrazone ligations by the nucleophilic catalyst aniline.29–31 For example, we and others have found that the high chemoselectivity of the hydrazone reaction in neutral aqueous buffer, enables the conjugation of peptides to carbohydrates on cell surfaces.32,33 Such enhanced reaction rates, especially at neutral pH, suggested that this hydrazone based approach might be utilized for the controlled and efficient assembly of peptides to nanoparticles such as QDs.
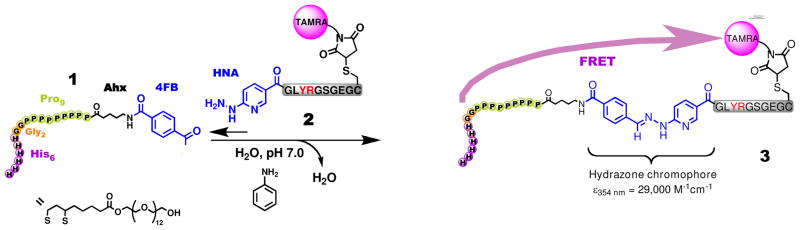
Scheme 1.
General strategy followed for the ligation of peptides to PEGylated QDs. Peptide 2 is ligated to QD:peptide 1 to yield the QD:peptide 3 product. The highlighted Y and R residues in peptide 3 are subsequently cleaved by addition of the proteases chymotrypsin or trypsin.
The reaction of a 4-formylbenzoyl group (4FB) and a 2-hydrazinonicotinoyl group (HYNIC) is adventagous since it yields a highly conjugated hydrazone product that has a clear optical signature with ε354 = 29,000 M−1cm−1 (Scheme 1).34,35 In addition, the high reaction rate at pH 7.0 (170 M−1s−1)36 enables the efficient coupling and labeling of biomolecules at micromolar concentrations and neutral pH. To introduce the required functional groups onto the DHLA-PEG600QDs, we designed a peptide with a benzaldehyde reactant and a His6 QD surface binding domain, separated by a polyproline spacer to extend the reactive aldehyde away from the NP/PEG surface. Specifically, the peptide, 4FB-Ahx-Pro9Gly2His6, (4FB = 4-formylbenzaldehyde, Ahx = Amino-hexanoyl) utilizes an Ahx-Pro9Gly2-linker in which the Pro9 is expected to form a Proline type II helix extending ~3 nm from the QD/PEG surface.37,38 This approach allows considerable control over the number of aldehyde groups displayed on each QD through the molar ratio added,19 and is also compatible with commercially available QD preparations.39 In addition, the 4FB/aldehyde is easily introduced into the capping peptide using established chemistries (Scheme 1).36 Similar to the dithiol capping groups, the His6 tag interacts directly with the ZnS surface shell with high affinity (Kd ~1 nM).19 To validate the approach, a protease substrate peptide 2 (HYNIC-GLYRGSGEGC-TAMRA) was designed to incorporate both a HYNIC hydrazine ligation handle and a Cys/maleimide linked TAMRA fluorophore. These QD-peptide conjugates were designed to act as fluorescently quenched protease substrates40, since the TAMRA flurophore is known FRET acceptor for QDs.41–43
Experimental procedures
Materials and Methods
All solvents and chemicals were purchased from commercial sources and used without further purification: DMF (HPLC grade) from OmniSolv, CH2Cl2 from Fisher, TFA (trifluoroacetic acid, Biograde) from Halocarbon, CH3CN from J. T. Baker. Water was purified using a Millipore MilliQ water purification system. DIEA (N,N-diisopropylethylamine) and 4-formyl-benzoic acid (4FB) from Sigma-Aldrich, HCTU (o-(1-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) and HOBt (1-hydroxybenzotriazole) from Peptides International, HATU (o-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) and Tetramethylrhodamine-5-(and-6)-maleimide from Anaspec, 6-Boc-hydrazinonicotinic acid (6-Boc-HYNIC) from Solulink Biosciences and amino acids from CS Bio. Peptides were purified by reverse phase HPLC (RP-HPLC). Analytical HPLC was carried out in a Varian ProStar Model 210 equipped with a Dynamax Absorbance Rainin Detector. Analytical injections were monitored at 220 nm. Separations were performed using an Einstein C-18 cartridge (65×15 mm) at a flow rate of 2 mL/min with a gradient from 0 to 67% of B in 30 min (eluent A: 0.1% TFA/H2O, eluent B: 0.09% TFA in 90% CH3CN/H2O). Preparative HPLC was performed in a Waters Delta Prep 4000 equipped with a Gilson UV detector model 116 and a Phenomenex column (10 μm, 90 Å, 250×21.20 mm) at a flow rate of 15 mL/min employing the following gradient: 0->85 min, 0->70% of B. Preparative injections were monitored at 220 nm. Peptides were desalted prior their use with OPC Oligonucleotide Purification Cartridge (Applied Biosystems).19
Peptides were characterized using electrospray ionization MS on a LC/MS API I Plus quadrupole mass spectrometer (Sciex). Peptides masses were calculated from the experimental mass to charge (m/z) ratios from all of the observed protonation states of a peptide by using the MacSpec software (Sciex).
UV measurements were undertaken in a Genesys 6 UV-VIS spectrometer (Thermo Electron Corporation). Fluorescence was performed in a Tecan Safire Monochromator Plate Reader (Tecan, Research Triangle Park, NC).
Peptide Synthesis
Peptides were synthesized manually using in situ neutralization cycles for Boc-solid-phase-peptide synthesis (Boc-SPPS) following procedures described in the literature.44 Peptides were synthesized on 0.1 mmol MBHA resin (4-Methylbenzhydrylamine, Peptides International, 0.65 mmol/g) using 1.0 mmol of amino acid, 1.0 mmol of HCTU/HOBt (in a 0.4 M solution in DMF 1:1, HCTU: [2-(1H-6-chlorobenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate], HOBt: 1-hydroxybenzotriazole) and 2 mmol of DIEA (N,N-diisopropylethylamine). Coupling times were 1 h. Following chain assembly, peptides were cleaved from the resin with HF and 10% of anisole for 1 h at 0ºC.
Synthesis of 4FB-Ahx-Pro9Gly2His6 peptide 1
4-Formylbenzoyl (4FB) was coupled to Ahx-Pro9Gly2His6 (12 mg, 6.2×10−3 mmol) in a H2O/CH3CN solution (5 mL, 1:1) using succinimidyl 4-formylbenzoate (2.3 mg, 9.3×10−3 mmol, 1.5 equiv) and DIEA (7 equiv) yielding 11.5 mg of 1 (90%).
Synthesis of HYNIC-GLYRGSGEGC-TAMRA peptide 2
Hydrazinonicotinic acid (HYNIC) (0.15 mmol) was activated with HATU (o-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) (0.15 mmol) and DIEA (0.23 mmol) in DMF (1.5 mL) for 2 ½ min and then added over the resin (0.1 mmol). Labeling with TAMRA (N,N,N′,N′-Tetramethylrhodamine) was carried out by reacting the cysteinyl peptide (5.5 mg, 4.8×10−3, 1.5 equiv) with (5,6)-Maleimide TAMRA (2 mg, 4.1×10−3, 1 equiv) in 10 mM Tris.HCl pH 7.5, 100 mM NaCl buffer. 1.8 mg and 3.2 mg of each isomer were obtained after HPLC purification (76% both isomers).
Quantum Dot Synthesis
Quantum Dots made of CdSe and coated with a ZnS shell with an inorganic core of about 2.8 nm were synthesized according procedures previously reported.2 Following the synthesis, TOP/TOPO (trioctyl phosphine/trioctyl phosphine oxide) ligands were exchanged with DHLA-PEG600 in methanol solution under N2 atmosphere.14
Peptide:QD ligation
QDs (~1 μM) were first incubated with His6 bearing peptides 1 (15–30 μM) for 30 min at room temperature in 50–100 mM HEPES buffer pH 7.0 (350 μL total volume). Then, hydrazine-TAMRA peptide 2 was added at the desired concentration and, finally, neat aniline (~100 mM). Ligation reactions were carried out in 0.5 mL quartz cuvettes when monitored at 354 nm or in eppendorf tubes.
Reactions were conducted over 5 h and then filtered through PD-10 columns (GE Healthcare, 2.5 mL void volume) using 10 mM sodium phosphate buffer pH 7.5 as eluting buffer. The first fraction (1 mL) was collected and the absorbance at 354 nm and 555 nm was measured, affording the number of peptides 2 ligated per QD (ε350 = 520,000 M−1 cm−1). The 555 nm absorbance is unique to TAMRA fluorophore (ε555 = 65,000 M−1 cm−1) allowing its concentration to be determined as well as the equimolar bisarylhydrazone group in all ligated peptides. Using these values, the QD concentration was determined by measuring the absorbance at 354 nm and subtracting components attributed to the bisarylhydrazone and TAMRA chromophores (bisarylhydrazone ε354 = 29,000 M−1 cm−1 and TAMRA ε354 is 11% of the value at 555 nm). Predicted loadings for 3 were estimated using Keq = 2.3×106 M−1.36
Fluorescence and FRET analysis
For fluorescence analysis samples were transferred to a 96-well microtitter plate, excited at 300 nm and fluorescence spectra were collected. FRET efficiency En (n = number of TAMRA acceptors per QD) was determined according the following equation:
| (1) |
where FD and FDA represent the fluorescence intensities of the donor in the absence and presence of n dye acceptors (number of dye labeled peptide 2 ligated per QD), respectively.45 Using the Förster theory and data from FRET efficiency, distance r from center-to-center (QD-to-TAMRA) was calculated:
| (2) |
where R0 is the Förster distance corresponding to a 50% energy transfer efficiency.45 The experimental data are analyzed using a corrected FRET efficiency where effects of heterogeneity in the conjugate valence are taken into account via the Poison distribution function p(k,n).40,46 The corrected FRET efficiency was rewritten as:
| (3) |
where n is the nominal average ratio of peptides 2 per QD mixed in solution and k is the exact number of peptides 2 conjugated to the QD:1 complexes.
The corrected values were used to provide calibration curves that were used in the enzymatic assays.
Enzymatic assays
α-Chymotrypsin from bovine pancreas (specific activity of 60 units/mg) and trypsin from bovine pancreas (specific activity of 10000–15000 BAEE units/mg of protein) were purchased from Sigma-Aldrich. For inhibition assays, ovomucoid trypsin inhibitor (Sigma-Aldrich) with a specific activity of 1 mg inhibits 0.8–1.2 mg of trypsin was used.
QD-peptide 3 conjugates with different ratios of ligated peptide were used as standard curve and substrate for FRET-based proteolytic assays as described.40 For all samples the dispersions were subjected to gel filtration to remove excess aniline prior to peforming the proteolytic assays. QD:peptide substrates (1:11, 0.1 μM QDs) were exposed to increasing concentrations of α-chymotrypsin (0.15–20 μM) and trypsin (0.04–42 μM) in 0.5XPBS buffer pH 7.5 at 30°C for 10 min incubations. The reactions were quenched with the addition of the alkylating agent α-iodoacetamide (final concentration 0.36 mg/mL), and changes in the FRET signature were monitored by tracking the QD emission at 537 nm (peak emission). Under these conditions, initial proteolytic rates could be measured. By comparison of the values from the QD PL following proteolysis with the calibration curves allow us to infer the number of QD:peptide complex that are intact and to transform the FRET recovery signal to velocity units. Using standard Michaelis-Menten kinetic analysis for excess enzyme conditions, maximal apparent velocity Vmax.app. and the apparent Michaelis constant KMapp. were determined.47
| (4) |
where [P] and [E] designate the product (digested substrate) and enzyme concentrations, respectively. The inhibitor dissociation constant Ki was calculated assuming a competitive inhibition mechanism (ovamucoid trypsin inhibitor (I) is known to act competitively):
| (5) |
where Vimax.app and KiMapp. correspond to the Michaelis-Menten parameters in the presence of an inhibitor. Assuming that KiMapp. ~ KMapp., then the Ki can be derived from:
| (6) |
Agarose gel electrophoresis
QDs (~1 μM) were assembled with 4FB-Ahx-Pro9Gly2His6 (~15 equiv) and several equivalents of peptide HYNIC-GLYRGSGEGC-TAMRA were ligated (0, 2, 4, 6, 8, 12 or 16). Reactions were carried out in 50–100 mM HEPES pH 7.0 and ~100 mM aniline for 5 h. Then, they were filtered through PD-10 columns (GE Healthcare) using 10 mM sodium phosphate buffer pH 7.5 and the absorbance at 354 and 555 nm registered affording the number of peptides 2 ligated per QD. Finally, ~20 picomol of QDs (~0.5 μM) conjugated with TAMRA ligated peptides were mixed with loading buffer (30% glycerol in H2O, no staining dyes), loaded into 2% agarose gel and run in 1XTBE (90 mM Tris borate, 2 mM EDTA, pH 8.3) buffer at ~10 V/cm for 10 to 20 minutes. Fluorescent images were collected on a Kodak 440 Digital Image Station (Rochester, NY) using 365 nm excitation and long-pass cut-off filters at 523 or 590 nm.
Results and Discussion
Assembly of benzaldehyde functionalized QDs
Water solubilized DHLA-PEG600 QDs (537 nm emission) were modified with synthetic peptide 1 (4FB-Ahx-Pro9Gly2His6) using the established method of direct His tag binding to the ZnS QD surface.19 They will be referred to as 4FB-1-QDs. It has been shown that His6 binding domain enables not only a strong and rapid binding to the NPs, but also the controlled functionalization of up to ~ 50 His6 peptides per ZnS shell.40,48–50 In order to test the utility of aniline catalyzed hydrazone ligation for the covalent modification of QDs, we self-assembled ~1 μM QD with between 15 and 30 copies of 1 (15–30 μM) depending on the individual experiment. Although the QD nanoparticles can accommodate larger numbers of peptides, this lower loading ensured a quantitative loading of the His6 peptides.48 The resulting concentrations of displayed peptide were ~15–30 μM and are consistent with concentrations frequently encountered when handling biological molecules and NPs.
Direct monitoring of peptide-QD ligation by UV/Vis
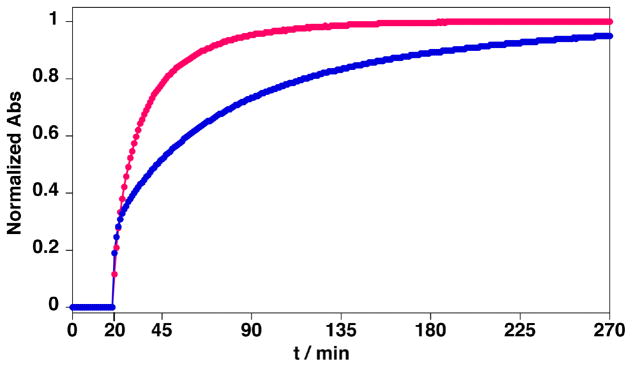
The ligation reaction was initiated by adding HYNIC peptide 2 (14 μM, pH 7.0, 23°C) to the 4FB-1-QDs (0.9 μM QDs, 21 μM 1). As shown in Figure 1, the hydrazone reaction is very slow at pH 7.0 in the absence of aniline catalyst (Figure 1,
 , t = 0 to 20 min) as monitored by hydrazone chromophore formation (the ligation rate constant at pH 7 is less than 1. 0 M−1s−1).36 Addition of ~100 mM aniline at t = 20 min dramatically increased hydrazone product formation, approaching equilibrium after ~4 h. When compared to the reaction in the absence of quantum dots (Figure 1,
, t = 0 to 20 min) as monitored by hydrazone chromophore formation (the ligation rate constant at pH 7 is less than 1. 0 M−1s−1).36 Addition of ~100 mM aniline at t = 20 min dramatically increased hydrazone product formation, approaching equilibrium after ~4 h. When compared to the reaction in the absence of quantum dots (Figure 1,
 ) the reaction appears to proceed at a similar rate for the first 30% of the reaction (~ 5 min) and subsequently proceeds at a slightly slower rate. This profile is consistent with an increase in steric hindrance as the QDs become highly functionalized. Nevertheless, the QD reaction does approach a similar level of total conjugation (~ 90 % of the added peptide 2 is conjugated) in the 4 hr time window shown. This demonstrates that a high yielding, covalent conjugation of peptides to hydrophilic QDs can be achieved. It should be noted that, since the hydrazone ligation is reversible, a maximal conversion of 95% was expected based on Keq = 2.3×106 M−1 for the hydrazone linkage at pH 7.36 This suggests that the local enviroment of the hydrazone extending from the surface of the QD is not dramatically altered compared to bulk solvent.
) the reaction appears to proceed at a similar rate for the first 30% of the reaction (~ 5 min) and subsequently proceeds at a slightly slower rate. This profile is consistent with an increase in steric hindrance as the QDs become highly functionalized. Nevertheless, the QD reaction does approach a similar level of total conjugation (~ 90 % of the added peptide 2 is conjugated) in the 4 hr time window shown. This demonstrates that a high yielding, covalent conjugation of peptides to hydrophilic QDs can be achieved. It should be noted that, since the hydrazone ligation is reversible, a maximal conversion of 95% was expected based on Keq = 2.3×106 M−1 for the hydrazone linkage at pH 7.36 This suggests that the local enviroment of the hydrazone extending from the surface of the QD is not dramatically altered compared to bulk solvent.
Figure 1.
UV monitoring of the chemical ligations. (
 ): between 1 (21 μM) self-assembled to QDs (0.9 μM) and 2 (14 μM) in presence of ~100 mM of aniline. (
): between 1 (21 μM) self-assembled to QDs (0.9 μM) and 2 (14 μM) in presence of ~100 mM of aniline. (
 ): between 1 (22 μM) and 2 (14.9 μM) in presence of 100 mM of aniline and absence of QDs. Aniline was added to reaction mixtures at 20 min. Note that the contributions from the QDs and TAMRA to the absorption has been subtracted before normalization.
): between 1 (22 μM) and 2 (14.9 μM) in presence of 100 mM of aniline and absence of QDs. Aniline was added to reaction mixtures at 20 min. Note that the contributions from the QDs and TAMRA to the absorption has been subtracted before normalization.
Since the hydrazone ligation reaction proceeds to near completion, one would expect that the average peptide labeling ratio coud be controlled by varying the number of equivalents of HYNIC peptide substrate per QD. To test this premise, a batch of QDs (1.1 μM, 1 equiv) was assembled with 1 (16.5 μM, 15 equiv). To this 4FB-1-QD complex, varying equivalents of peptide 2 were added (0, 2, 4, 8 or 12 equiv). Gratifyingly, efficient conversion was achieved, even at a concentration of 2.2 μM of peptide 2 (Table 1). After buffer exchange by gel filtration (PD-10 column) to remove excess aniline, the final ratio of QD:ligated peptide could be calculated by measuring the absorbance at 354 nm (arylhydrazone, QD and TAMRA) and at 555 nm (TAMRA), (Table 1). It is important to note that at the concentrations used, the QD absorbance is low enough (ε350nm = 520,000 M−1cm−1) to allow for accurate measurements of the hydrazone contribution to the overall absorbance, enabling a quantitative determination of hydrazone formation. For example the measured hydrazone contribution lowest labeling (two hydrazones at ε354nm = 29,000 M−1cm−1 each) represents ≥ 10% of the total absorbance, which enabled an accurate determination of hydrazone formation.
Table 1.
Ratio of peptide 3 per QD achieved by incubating 4FB-1-QD (15:1) with increasing equivalents of peptide 2 (Keq = 2.3×106 M−1) in the presence of aniline (~100 mM).
| [QD] (μM) | [1] (μM) | [2] (μM) | Ratio 3:QD (predicted) | Ratio 3:QD (observed) |
|---|---|---|---|---|
| 1.1 | 16.5 | 0 | 0 | 0 |
| 1.1 | 16.5 | 2.2 | 1.9 | 1.9 (100%) |
| 1.1 | 16.5 | 4.4 | 3.9 | 3.9 (100%) |
| 1.1 | 16.5 | 8.8 | 7.6 | 6.0 (79%) |
| 1.1 | 16.5 | 13.2 | 11.0 | 11.0 (100%) |
The physical association of the peptide:QD complex was further confirmed by agarose gel electrophoresis. The QDs used here were surface functionalized with neutral PEG ligands, which means that their migration gel should be slow. We found that the QDs conjugated with an increasing number of peptides migrated faster in the gel, Figure 2. This increase in the mobility shift for the conjugates can be attributed to a net negative charge of the attached peptides. (Although the peptides have equal numbers of positive and negatively charged side chains, there should be a slight net negative charge at pH 8.3). Also, it is clearly observed (Figure 2, bottom panel) that the fluorescence emitted by the QDs and the TAMRA dye colocalize, and the emission of the QDs is weaker for bands corresponding to higher peptide 2-TMRA ratios (Figure 2, top and bottom panels). This QD PL reduction can be attributed to FRET interactions, which is only possible if both donor (QD) and acceptor (TAMRA) are in close proximity (i.e., QD-peptide 2-TAMRA conjugate formation). The quenching efficiency increases with the number of TAMRA acceptors per QD up to a 6:1 ratio, and is consistent with the spectroscopic data in Figure 3 (see below). Finally, the integrity of the ligated peptide 3 on the QD was confirmed by gel filtration of the QD:peptide complex, cleavage of the ligated peptides from the QD surface with 0.5 M imidazole and identification by LC/MS (SI).
Figure 2.
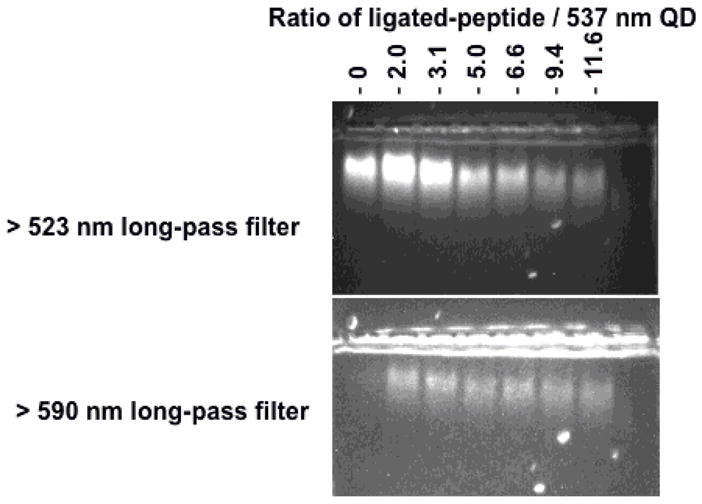
Agarose gel electrophoresis of 537 nm emitting QDs self-assembled with 15-His6-carboxybenzaldehyde peptides after ligation to an increasing ratio of TAMRA substrate peptide as indicated. Using a 523 nm long pass filter allows the simultaneous visualization of the emissions from QDs and TAMRA while use of a 590 nm filter allows only TAMRA signal to be visualized. Note clear evidence of the FRET induced quenching of the QD signal observed when the number of ligated peptides increases (top panel).
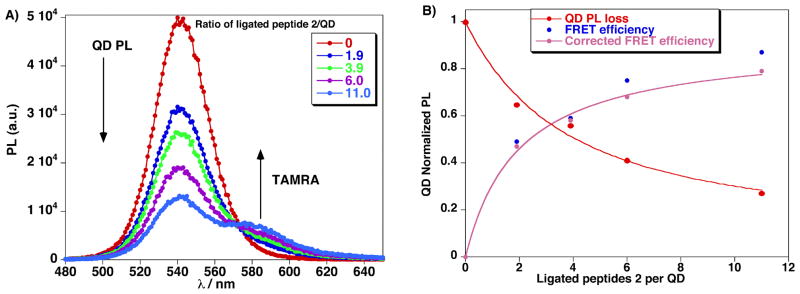
Figure 3.
A) Composite PL spectra of QD-TAMRA-hydrazine peptides 3 conjugates formed from reacting increasing numbers of peptide 2 with QDs preassembled with peptide 1 at (15:1). B) QD photoluminescence loss (red), FRET efficiency (blue) and FRET efficiency corrected (pink) vs. ratio of number of ligated peptides 2 per QD (537 nm) assembled with 15 equiv of peptide 1.
FRET quenching of QDs
The fluorescence emission of each peptide:QD sample clearly displayed evidence of efficient FRET. As shown in Figure 3A, QD donor PL at 537 nm decreases while the sensitized emission of the TAMRA at 587 nm increases in a manner that tracks the ratio of TAMRA attached per QD. An approximately 60% FRET efficiency is measued at a 4:1 TAMRA-peptide:QD ratio; increasing the ratio to 11:1 increases the FRET quenching efficiency to ~80%. These data demonstrates that controlling the peptide:QD ratio is critical for the development of FRET-based sensing of specific biological processes, such proteolytic activity in the present case. Using a quantum yield of 0.2 for the hydrophilic QDs a Förster distance R0 of ~5 nm is estimated for this donor-acceptor pair40,51 and analysis of the resulting FRET data (Figure 3B) indicates that the acceptor dye is located at ~5.8 nm from the QD center, which matches the calculated value accounting for the QD radius and peptide extension.19,39,52 We should emphasize that the QDs experience a substantial loss in their emission in the presence of the aniline catalyst.49 Removal of aniline by PD-10 gel filtration lead to a full recovery of QD signal and provided samples that could be used for the FRET analysis of proteolytic enzyme activity.
Fluorescent detection of trypsin and chymotrypsin
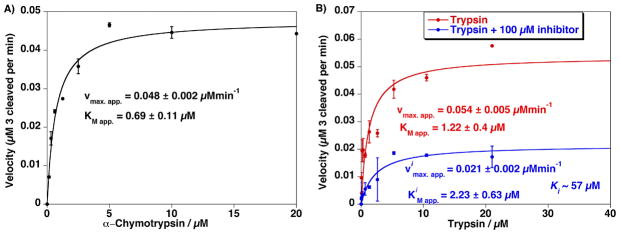
The utility of this new bioconjugation method for QDs was further demonstrated by carrying out enzymatic assays using a ligated peptide on the quantum dot. The TAMRA peptide 3 is a protease substrate that can be cleaved C-terminal to the Arg residue by trypsin or C-terminal to the Tyr residue by chymotrypsin. This cleavage releases the TAMRA dye away from the nanocrystal into bulk solution, reducing the FRET efficiency and which leads to recovery of the QD photoluminescence. The peptide 3:QD substrate (at an 11:1 ratio) was treated independently with each enzyme by monitoring changes in the fluorescence emission at 537 nm. Using Michaelis-Menten kinetic analysis under the condition of excess enzyme, the maximal apparent velocities Vmax.app. = 0.048 and 0.054 μMmin−1 and the Michaelis constants KMapp. 0.69 and 1.22 μM were determined for α-chymotrypsin and trypsin, respectively (Figure 4).40,53 The proteolytic ativity of trypsin was monitored in the presence of ovoinhibitor revealing a dissociation constant of Ki~57 μM assuming competitive inhibition. Overall, these results demonstrate that TAMRA-labeled peptides conjugated to DHLA-PEG600 QDs using a Pro9 spacer are sufficiently close to the NP surface to quench the QD luminescence via FRET, yet extend sufficiently beyond the PEG coating to allow for effective proteolytic cleavage. The ability to efficiently couple QDs with control over the QD-substrate ratio, combined with the broad pH stability provided by the DHLA-PEG QD coating, suggests that these NPs may have utility in cellular applications.
Figure 4.
A) Proteolytic activity from assaying an increasing concentration of Chymotrypsin versus a constant amount of QD–TAMRA substrate peptide 3 (0.1 μM QD). Estimated Vmax.app. and KMapp. are indicated. B) Proteolytic activity from assaying an increasing concentration of trypsin versus a constant amount of 537 nm QD–TAMRA substrate peptide 3 (0.1 μM QD) in the absence and presence of 100 μM ovomucoid trypsin inhibitor. Note the change in Vimax.app. while the KiMapp. values remain relatively unchanged which is consistent with a competitive inhibition process. Ki value of ~57 μM was estimated from the assay data in the presence of inhibitor. Error bars represent standard deviation from the mean of triplicate measurements.
Conclusions
A direct and highly efficient ligation strategy using aniline catalyzed hydrazone ligation in neutral aqueous buffer has been demonstrated for attaching peptides to hydrophilic CdSe-ZnS QDs. QD-peptide bioconjugates were prepared in a relatively straightforward manner without use of excess peptide and the resulting labeled NPs were available for use after a single gel filtration step. Importantly, the reactions are highly chemoselective and proceed quantitatively at micromolar concentrations which allows for control over the ratio of peptides ligated per QD; something that is not straightforward using alternative QD-bioconjugation approaches. Further, ratiometric control of peptide valence can be exerted through either the self-assembly of the coating peptide 1 or through the equivalents of HYNIC-peptide 2 ligated. The resulting hydrazone linkages have been shown to be stable under physiological conditions including human plasma and absorb uniquely at 354 nm,54 which enables the monitoring and direct quantitation of peptides attached to QDs. The QD-peptide conjugates assembled using this coupling strategy were further developed as a sensor (via FRET) for the specific detection of proteolysis. This constitutes a major improvement over some of the currently available multistep and difficult to control amide forming reactions. The ‘modular’ nature of the ligation strategy, i.e., interchangeability of functional peptide sequences and linker structure, is an important asset of this approach.
Cumulatively, these remarkable properties suggest aniline catalyzed hydrazone ligation will be a general method for the controlled assembly of a variety of QD-biomolecule hybrids. Since the hydrazone reaction has been shown to be fully chemoselective for all functional groups found in proteins, DNA and carbohydrates, this approach should find wide utility in QD-biomolecule conjugation.32 The modular nature of the NP assembly should facilitate optimization of linker length and valency to accommodate these larger biomolecules. In additon, since the ligation reaction is rapid at low micromolar concentrations, biomolecules of limited solubility and large size should be compatible with this strategy, although steric factors may limit the number of macromolecules per QD.
Supplementary Material
Acknowledgments
This work was supported by NIH GM059380 (P.E.D.), ONR, NRL-NSI and DTRA (I.M.). J.B.B-C is supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme and D.F. acknowledges a NRL-NRC fellowship.
Footnotes
SUPPORTING INFORMATION. HPLC and Ms spectra of the peptides 1, 2 and 3 (stripped off the QDs), UV and fluorescence spectra of the 537 nm QDs and TAMRA and UV raw trace of the ligation reaction between 1 and 2. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.Alivisatos AP. Science. 1996;271:933–937. [Google Scholar]
- 2.Dabbousi BO, RodriguezViejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J Phys Chem B. 1997;101:9463–9475. [Google Scholar]
- 3.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 4.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 5.Bailey RE, Smith AM, Nie SM. Physica E. 2004;25:1–12. [Google Scholar]
- 6.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerion D, Pinaud F, Williams SC, Parak WJ, Zanchet D, Weiss S, Alivisatos AP. J Phys Chem B. 2001;105:8861–8871. [Google Scholar]
- 8.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 9.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, Ge NF, Peale F, Bruchez MP. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 10.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, Keller S, Radler J, Natile G, Parak WJ. Nano Lett. 2004;4:703–707. [Google Scholar]
- 11.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 12.Ding SY, Jones M, Tucker MP, Nedeljkovic JM, Wall J, Simon MN, Rumbles G, Himmel ME. Nano Lett. 2003;3:1581–1585. [Google Scholar]
- 13.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 14.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. J Am Chem Soc. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinaud F, King D, Moore HP, Weiss S. J Am Chem Soc. 2004;126:6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerman ME, Chan WCW, Laakkonen P, Bhatia SN, Ruoslahti E. Proc Natl Acad Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez N, Winter JO, Shieh F, Saunders AE, Korgel BA, Schmidt CE. Talanta. 2005;67:462–471. doi: 10.1016/j.talanta.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 19.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. J Phys Chem C. 2007;111:11528–11538. [Google Scholar]
- 20.Howarth M, Takao K, Hayashi Y, Ting AY. Proc Natl Acad Sci USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prinzen L, Miserus R-JJHM, Dirksen A, Hackeng TM, Deckers N, Bitsch NJ, Megens RTA, Douma K, Heemskerk JW, Kooi ME, Frederik PM, Slaaf DW, van Zandvoort MAMJ, Reutelingsperger CPM. Nano Lett. 2007;7:93–100. doi: 10.1021/nl062226r. [DOI] [PubMed] [Google Scholar]
- 22.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 23.Clapp AR, Goldman ER, Mattoussi H. Nat Protoc. 2006;1:1258–1266. doi: 10.1038/nprot.2006.184. [DOI] [PubMed] [Google Scholar]
- 24.Shen HY, Jawaid AM, Snee PT. ACS Nano. 2009;3:915–923. doi: 10.1021/nn800870r. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M, Nakatani E, Gronenberg LS, Tokimoto T, Wirth MJ, Hruby VJ, Roberts A, Lynch RM, Ghosh I. Bioconjugate Chem. 2007;18:323–332. doi: 10.1021/bc0601929. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Thakar R, Snee PT. J Am Chem Soc. 2008;130:3744–3745. doi: 10.1021/ja711252n. [DOI] [PubMed] [Google Scholar]
- 27.Dirksen A, Dawson PE. Curr Opin Chem Biol. 2008;12:760–766. doi: 10.1016/j.cbpa.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Ghosh I. Biopolymers. 2007;88:325–339. doi: 10.1002/bip.20655. [DOI] [PubMed] [Google Scholar]
- 29.Dirksen A, Hackeng TM, Dawson PE. Angew Chem, Int Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 30.Dirksen A, Dirksen S, Hackeng TM, Dawson PE. J Am Chem Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 31.Sander EG, Jencks WP. J Am Chem Soc. 1968;90:6154–6162. [Google Scholar]
- 32.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. Nat Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nauman DA, Bertozzi CR. Biochim Biophys Acta. 2001;1568:147–154. doi: 10.1016/s0304-4165(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz DA. Solulink Biosciences, Inc; 6, 800, 728. US Patent. 2004
- 35.Flavell RR, Kothari P, Bar-Dagan M, Synan M, Vallabhajosula S, Friedman JM, Muir TW, Ceccarini G. J Am Chem Soc. 2008;130:9106–9112. doi: 10.1021/ja801666z. [DOI] [PubMed] [Google Scholar]
- 36.Dirksen A, Dawson PE. Bioconjugate Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stryer L, Haugland RP. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arora PS, Ansari AZ, Best TP, Ptashne M, Dervan PB. J Am Chem Soc. 2002;124:13067–13071. doi: 10.1021/ja0208355. [DOI] [PubMed] [Google Scholar]
- 39.Dennis AM, Bao G. Nano Lett. 2008;8:1439–1445. doi: 10.1021/nl080358+. [DOI] [PubMed] [Google Scholar]
- 40.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Nat Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 41.Willard DM, Carrillo LL, Jung J, van Orden A. Nano Lett. 2001;1:469–474. [Google Scholar]
- 42.Medintz IL, Konnert JH, Clapp AR, Stanish I, Twigg ME, Mattoussi H, Mauro JM, Deschamps JR. Proc Natl Acad Sci USA. 2004;101:9612–9617. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi L, De Paoli V, Rosenzweig N, Rosenzweig Z. J Am Chem Soc. 2006;128:10378–10379. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- 44.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. Int J of Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 45.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]
- 46.Pons T, Medintz IL, Wang X, English DS, Mattoussi H. J Am Chem Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 47.Bowden AC. Fundamentals of Enzyme Kinetics. Portland Press Ltd; London: 1995. Revised Ed. [Google Scholar]
- 48.Prasuhn DE, Deschamps JR, Susumu K, Stewart MH, Boeneman K, Blanco-Canosa JB, Dawson PE, Medintz IL. Small. 2010;6:555–564. doi: 10.1002/smll.200901845. [DOI] [PubMed] [Google Scholar]
- 49.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 50.Delehanty JB, Medintz IL, Pons T, Brunel FM, Dawson PE, Mattoussi H. Bioconjugate Chem. 2006;17:920–927. doi: 10.1021/bc060044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pons T, Medintz IL, Wang X, English DS, Mattoussi H. J Am Chem Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 52.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 53.Medintz IL, Pons T, Trammell SA, Grimes AF, English DS, Blanco-Canosa JB, Dawson PE, Mattoussi H. J Am Chem Soc. 2008;130:16745–16756. doi: 10.1021/ja805456x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dirksen A, Yegneswaran S, Dawson PE. Angew Chem, Int Ed. 49:2023–2027. doi: 10.1002/anie.200906756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.