Summary
In the homoacetogenic bacterium Sporomusa ovata, phenol and p-cresol are converted into α-ribotides, which are incorporated into biologically active cobamides (Cbas) whose lower ligand bases do not form axial coordination bonds with the cobalt ion of the corrin ring. Here we report the identity of two S. ovata genes that encode an enzyme that transfers the phosphoribosyl group of nicotinate mononucleotide (NaMN) to phenol or p-cresol, yielding α-O-glycosidic ribotides. The alluded genes were named arsA and arsB (for alpha-ribotide synthesis), arsA and arsB were isolated from a genomic DNA library of S. ovata. A positive selection strategy using an Escherichia coli strain devoid of NaMN:5,6-dimethylbenzimidazole (DMB) phosphoribosyltransferase (CobT) activity was used to isolate a fragment of S. ovata DNA that contained arsA and arsB, whose nucleotide sequences overlapped by 8 bp. SoArsAB was isolated to homogeneity, shown to be functional as a heterodimer, and to have highest activity at pH 9. SoArsAB also activated DMB to its α-N-glycosidic ribotide. Previously characterized CobT-like enzymes activate DMB but do not activate phenolics. NMR spectroscopy was used to confirm the incorporation of phenol into the cobamide, and mass spectrometry was used to identity of the SoArsAB products.
Introduction
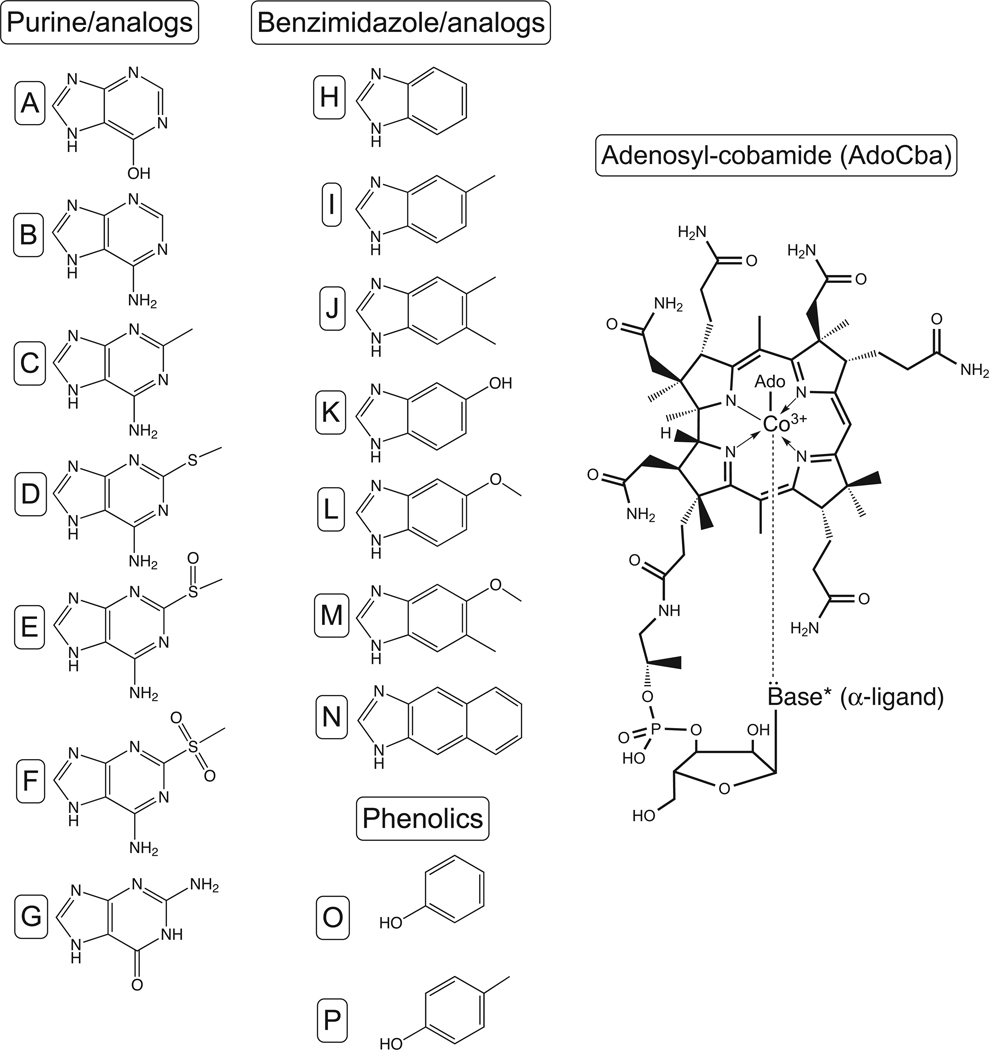
Cobamides (Cbas) are cobalt containing modified tetrapyrroles mainly involved in enzyme-catalyzed carbon skeleton rearrangements, methyl-group transfers, and reductive dehalogenation (Halpern, 1985; Kräutler et al., 2003; Marsh, 1999). The biosynthesis of cobamides is complex and is only performed by some bacteria and archaea (Warren et al., 2002). Cobamides have an upper (Coβ) ligand and a lower (Coα) ligand base (Fig. 1). Some cobamides have a 5’-deoxyadenosyl (Ado) group as the Coβ ligand that participates in radical chemistry reactions [e.g., methylmalonyl-CoA mutase (EC 5.4.99.2), ethanolamine ammonia-lyase (EC 4.3.1.7), diol dehydratase (EC 4.2.1.28), etc]. When cobamides serve as transient methyl carriers, the upper ligand is the methyl group being transferred [e.g., Cbl-dependent methionine synthase (EC 2.1.1.13), tetrahydromethanopterin S-methyltransferase (EC 2.1.1.86), etc]. The chemical nature of the lower (Coα) ligand, however, varies depending on the organism (Renz, 1999). For example, Salmonella enterica sv Typhimurium LT2 (hereafter S. enterica) synthesizes three AdoCbas that differ by their lower ligand base. One AdoCba contains 5,6-dimethylbenzimidazole (DMB) (a.k.a. adenosylcobalamin, AdoCbl, AdoB12, CoB12), another one contains adenine (a.k.a. Ado-pseudoCbl, Ado-pseudoB12, pseudo-CoB12), and a third one contains 2-methyladenine (Ado-factor A) (Johnson and Escalante-Semerena, 1992; Keck and Renz, 2000). In other organisms, cobamides may contain substituted benzimidazoles, purine analogs, or phenolic compounds (i.e., phenol, p-cresol) (Renz, 1999) (Fig. 1).
Figure 1. Structure of AdoCba and diversity of lower ligand bases found in Cbas.
Purines/analogs: A, Hypoxanthine; B, adenine; C, 2-methyladenine; D, 2-methylmercaptoadenine; E, 2-methylsulfinyladenine; F, 2-methylsulfonyladenine; G, guanine. Benzimidazole/analogs: H, benzimidazole; I, 5-methylbenzimidazole; J, 5,6-dimethylbenzimidazole; K, 5-hydroxybenzimidazole; L, 5-methoxybenzimidazole; M, 5-methoxy-6-methylbenzimidazole; N, naphthimidazole. Phenolics: O, phenol; P, p-cresol. * Phenol and p-cresol do not form coordination bonds with the Co ion of the ring.
Lower ligand bases need to be activated before they can enter biosynthetic pathways that incorporate them into AdoCbas. The general scheme for the conversion of the base to its activated α-ribotide is shown below in equation #1.
| (1) |
The activation of the lower ligand base of cobamides has been studied in S. enterica, Pseudomonas denitrificans, Clostridium sticklandii and Propionibacterium shermanii (Cheong et al., 1999; Friedmann and Harris, 1965; Friedmann and Fyfe, 1969; Trzebiatowski et al., 1994; Trzebiatowski and Escalante-Semerena, 1997).
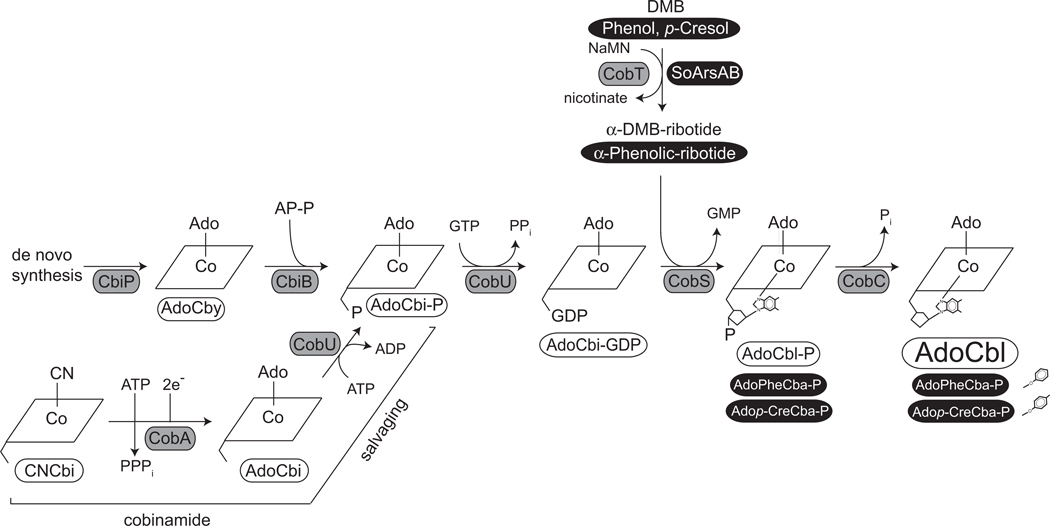
Figure 2 shows a scheme for the late steps of AdoCbl biosynthesis in S. enterica. In this bacterium the NaMN:5,6-dimethylbenzimidazole (DMB) phosphoribosyltransferase (CobT, EC 2.4.2.21) enzyme activates DMB yielding α-DMB-ribotide (a.k.a. α-ribazole-5’-P), the co-substrate for the AdoCbl-5’-P synthase (CobS, EC 2.7.8.26) that catalyzes the penultimate step of the pathway (Maggio-Hall and Escalante-Semerena, 1999; Zayas and Escalante-Semerena, 2007). Notably, SeCobT cannot activate phenolic bases such as phenol or p-cresol (Cheong et al., 2001). Figure 2 also depicts the pathway used by S. enterica to salvage exogenous cobinamide; this information is relevant to some of the experiments performed in this work.
Figure 2. Late steps in AdoCbl biosynthesis in S. enterica.
For simplicity purposes, the corrin ring is represented as a rhomboid, and lower ligands hydroxyl groups or water molecules are not shown. AdoCby, adenosylcobyric acid; AdoCbi-P, adenosylcobinamide-phosphate; AdoCbi-GDP, adenosylcobinamide-GDP; AdoCbl-P, adenosylcobalamin-phosphate; AdoCbl, adenosylcobalamin; AP-P, aminopropanol phosphate; DMB, 5, 6-dimethylbenzimidazole; NaMN, nicotinate mononucleotide. CbiP (AdoCby synthase) and CbiB (AdoCbiP synthetase) catalyze the last two steps of the de novo corrin ring biosynthetic pathway. CobU (AdoCbi kinase/AdoCbi-P guanylyltransferase), CobS (AdoCbl-P synthase), CobT, NaMN:DMB phosphoribosyltransferase, and CobC (AdoCbl-P phosphatase) comprise the nucleotide loop assembly pathway, also known as the late steps. CobA (ATP :Co(I)rrinoid adenosylytransferase) is the housekeeping corrinoid adenosylating enzyme required for de novo corrin ring biosynthesis, and for the salvaging of incomplete corrinoids like cobinamide. The phenolic substrates for ArsAB and the resulting Cbas synthesized using the α-phenolic-ribotide products are highlighted in black and white. Structures of O-glycosidic phenolic ligands are shown next to corresponding cobamide name. Phenolic ligands do not form a coordination bond with the Co ion of the ring.
Cobamides containing phenolic bases comprise about 16% of the total cobamides isolated from gut microbiome of humans (Allen and Stabler, 2008). Prior to this work, the identity of genes encoding enzymes responsible for the activation of phenolics during AdoCba biosynthesis was unknown, and the enzymes had not been studied.
Phenolyl- and p-cresolyl-Cba were first isolated from the Gram-negative anaerobe Sporomusa ovata (Stupperich et al., 1988; Stupperich and Eisinger, 1989b), a member of the Veillonellaceae family of the Firmicutes. Phenolyl- and p-cresolyl-Cba have two distinctive features. First, an α-O-glycosidic bond links the base to ribose, rather than the α-N-glycosidic bond observed in cobamides containing purines, purine analogs, and derivatives of benzimidazole. And second, neither phenol nor p-cresol can establish a coordination bond with the Co ion of the ring. The absence of a coordination bond between the Co ion and the lower ligand base limits the use of phenolyl- and p-cresolyl-Cba to enzymes that use cobamide coenzymes in their base-off conformation (Stupperich and Eisinger, 1989a). Enzymes using the cobamide coenzymes in their base-on conformation require coordination of the Co ion by the lower ligand base.
S. enterica is a good system to investigate the use of phenolyl- and p-cresolyl-Cba in vivo, because base-off and base-on cobamide-dependent enzymes are present in this bacterium, and conditions that require their activities for growth are known (Jeter et al., 1984; Roof and Roth, 1988). For example, the cobalamin-dependent methionine synthase (MetH) enzyme requires a base-off form of the cofactor for function (Drennan et al., 1994), whereas ethanolamine ammonia-lyase (EutBC) uses AdoCbl in its base-on form (Abend et al., 1999; Shibata et al., 1999; Shibata et al., 2010).
The limiting factor for the in vivo analysis has been the availability of cobamides unable to form the base-on conformation, such as phenolyl- and p-cresolyl-Cba. To circumvent this limitation we sought to identify the genes encoding the enzymes that activate phenol and p-cresol in S. ovata. Notably, S. ovata can also synthesize benzimidazolyl-Cba when benzimidazole is present in its environment (Stupperich and Eisinger, 1989b), indicating that this bacterium has enzymes that can synthesize α-N-glycosidic and α-O-glycosidic ribotides.
In this paper we report that the newly discovered heterodimeric enzyme ArsAB [for alpha-ribotide synthesis (Ars) proteins A and B] of S. ovata activates phenol, p-cresol, and DMB to α-phenolyl, α-p-cresolyl-ribotide, and α-DMB-ribotide, respectively. We used a heterologous complementation system to isolate the genes arsA and arsB [encoding ArsA (36.65 kDa) and ArsB (36.18 kDa)], from a S. ovata gene library. Bioinformatics analyses of the putative amino acid sequences showed that SoArsA and SoArsB were homologous to SeCobT. However, unlike SeCobT, SoArsA and SoArsB function as a heterodimer, and can activate phenol, p-cresol, in addition to DMB. We also report that the AdoCba biosynthetic machinery of S. enterica incorporates phenolic bases into the final cobamide when arsA+ and arsB+ were provided in trans. We note that neither phenolyl-nor p-cresolyl-Cba supported S. enterica growth on ethanolamine or 1,2-propanediol as a carbon and energy sources, consistent with the need of ethanolamine ammonia-lyase and diol dehydratase for a base-on AdoCba.
Results
Isolation of S. ovata genes encoding functions needed for the activation of phenol and p-cresol
Our search for S. ovata genes encoding phenol and p-cresol activating functions assumed that, in S. ovata, the activation of these compounds was catalyzed by a homolog of SeCobT. At the time of this work, the sequence of the S. ovata was not known, precluding the use of bioinformatics approaches for the identification of genes encoding SeCobT homologs. To circumvent this problem, we took a function-based approach. For this purpose, we constructed a S. ovata gene library (Fig. S1) comprised of approximately 12,500 fosmids, each of which carried one 40-kbp (average size) fragment of S. ovata genomic DNA. Since we assumed that the size of the S. ovata genome was 5 Mbp, the total amount of S. ovata DNA in the library represented 100× coverage. The search for fosmids encoding S. ovata CobT-like functions was performed using strain JE11215, a derivative of E. coli EPI300 (Epicentre, Table 1) carrying a marker-less deletion of metE (encodes the Cbl-independent methionine synthase) and a replacement of cobT with the kan+ gene encoding kanamycin resistance. In the absence of exogenous methionine or Cbl, a CobT-like function encoded on the fosmid is required for growth of strain JE11215 under aerobic conditions on minimal medium supplemented with cobinamide (Cbi), a precursor of cobamides. We describe the method used to isolate the wild type alleles of arsA and arsB in the Experimental procedures section and supplemental material. The initial search yielded seven fosmids with distinct restriction patterns that restored AdoCba synthesis in JE11215. Among these fosmids, pSolibC17 was selected for further characterization. A strategy that combined sub-cloning and complementation of function yielded a fragment of S. ovata DNA that was sequenced in its entirety (2,267 bp; GenBank Accession # JF895493; Fig. S2). The S. ovata genes encoding SeCobT-like function were named arsA and arsB for alpha ribotide synthesis; arsA and arsB were found together, with a sequence overlap of 8 bp that included the stop codon of arsA (Fig. S2). Both proteins were 350 aa long, and shared end-to-end homology with SeCobT (356 aa) (Fig. 3). A notable difference between SoArsB and SoArsA and SeCobT was an 11-residue gap located approximately in the middle of the primary sequence of SoArsB. This gap was confirmed by sequencing arsA and arsB directly from S. ovata genomic DNA.
Table 1.
Strains and plasmids used in this study
| S. enterica strains | Genotype | Source |
|---|---|---|
| TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| TR6583 derivatives | ||
| JE2501 | cobT109::MudJa cobB1176::Tn10d(tet+)b | laboratory collection |
| JE8248 | ΔcobS1313 | (Otte and Escalante-Semerena, 2009) |
| JE13867 | cobT109::MudJ cobB1176::Tn10d(tet+) / pBAD33 | |
| JE13871 | cobT109::MudJ cobB1176::Tn10d(tet+) / pARSA4 | |
| JE13872 | cobT109::MudJ cobB1176::Tn10d(tet+) / pARSB2 | |
| JE13874 | cobT109::MudJ cobB1176::Tn10d(tet+) / pARSAB4 | |
| E. coli strains EPI300 | F− mcrA Δ(mrr-hsdRMS-mcrBC) (StrR) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ– rpsL nupG trfA tonA dhfr | Epicentre |
| EPI300 derivatives | ||
| JE11098 | metE400::kan+ | |
| JE11106 | ΔmetE401 | |
| JE11215 | EPI300 ΔmetE401 cobT150::kan+ | |
| JE14470 | EPI300 ΔmetE401 cobT150::kan+ / pSolibC17 | |
| BL21(DE3) | F−, ompT, hsdSB (rB−, mB−), dcm, gal, λ(DE3) | laboratory collection |
| JE13607 | BL21 cobT150::kan+ | |
| Keio collection strains BW25113 | rrnB3 ΔlacZ4787 hsdR513 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | (Baba et al., 2006) |
| metE400::kan+ | (Baba et al., 2006) | |
| cobT150::kan+ | (Baba et al., 2006) | |
| Plasmids pBAD33 derivatives | complementation vector, ParaBAD cat+ | (Guzman et al., 1995) |
| pARSA4 | S. ovata arsA+ | |
| pARSB2 | S. ovata arsB+ | |
| pARSAB4 | S. ovata arsA+ arsB+ | |
| pTEV5 derivatives | Over-expression vector that fuses the N-terminus of the protein of interest to a H6 tag, which can be removed by rTEV protease, bla+ | (Rocco et al., 2008) |
| pARSAB7 | S. ovata arsA+ arsB+ | |
| Other plasmids | ||
| pEAK2 | bla+ recA+ | (Kouzminova and Kuzminov, 2004) |
| pCC1FOS | Fosmid construction plasmid cat+ | Epicentre |
MudJ is an abbreviation of MudI1734 (Castilho et al., 1984)
Tn10d(tet+) is an abbreviation of Tn10Δ16Δ17 (Way et al., 1984)
Figure 3. Alignment of SoArsA, SoArsB with SeCobT.
Conserved regions are highlighted. SoArsA and SoArsB are 37% identical, 57 % similar to each other; SoArsA is 37% identical, 56% to SeCobT, and SoArsB is 35% identical, 55% similar to SeCobT. The conserved predicted active site glutamyl residue is highlighted in red.
S. ovata arsA+ and arsB+ are necessary and sufficient to restore synthesis of AdoCba in a S. enterica strain lacking CobT function
We performed complementation studies in S. enterica to establish whether SeCobT-like activity was associated with arsA, arsB or their combination. For this purpose, arsA+ and arsB+ were placed individually or together under the control of the L-(+)-arabinose-inducible promoter in plasmid pBAD33 (Guzman et al., 1995), resulting in plasmids pARSA4 (arsA+), pARSB2 (arsB+), and pARSAB4 (arsA+ arsB+). These plasmids were moved individually into strain JE2501 (S. enterica metE cobT cobB), resulting in strains JE13867 (metE cobT cobB / pBAD33), JE13871 (metE cobT cobB / pARSA4), JE13872 (metE cobT cobB / pARSB2), and JE13874 (metE cobT cobB / pARSAB4) (Table 1). The cobB gene, which encodes an NAD+-dependent protein deacetylase (Starai et al., 2002), was inactivated to eliminate all CobT-like activity in the cell (Trzebiatowski et al., 1994; Tsang and Escalante-Semerena, 1998).
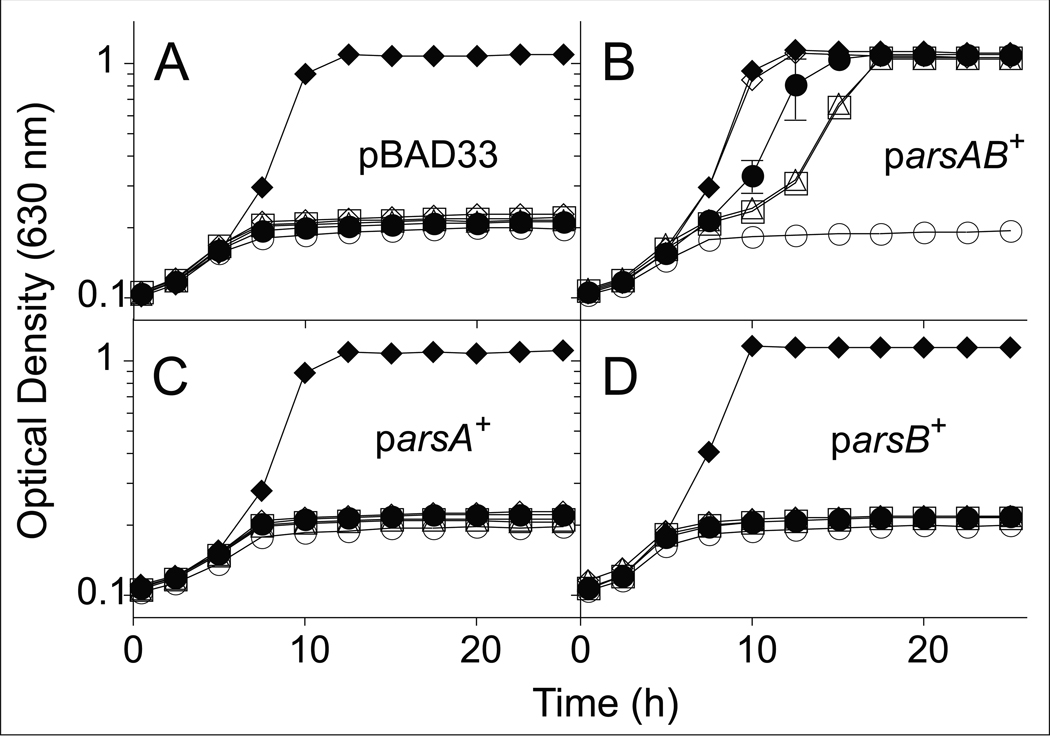
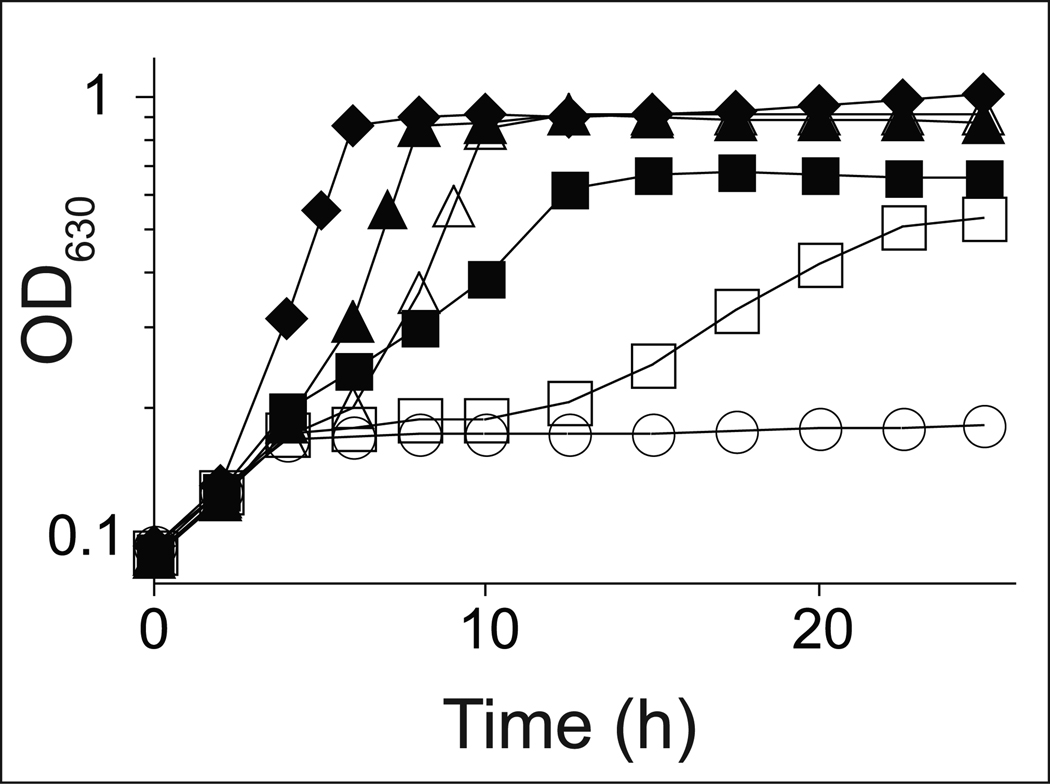
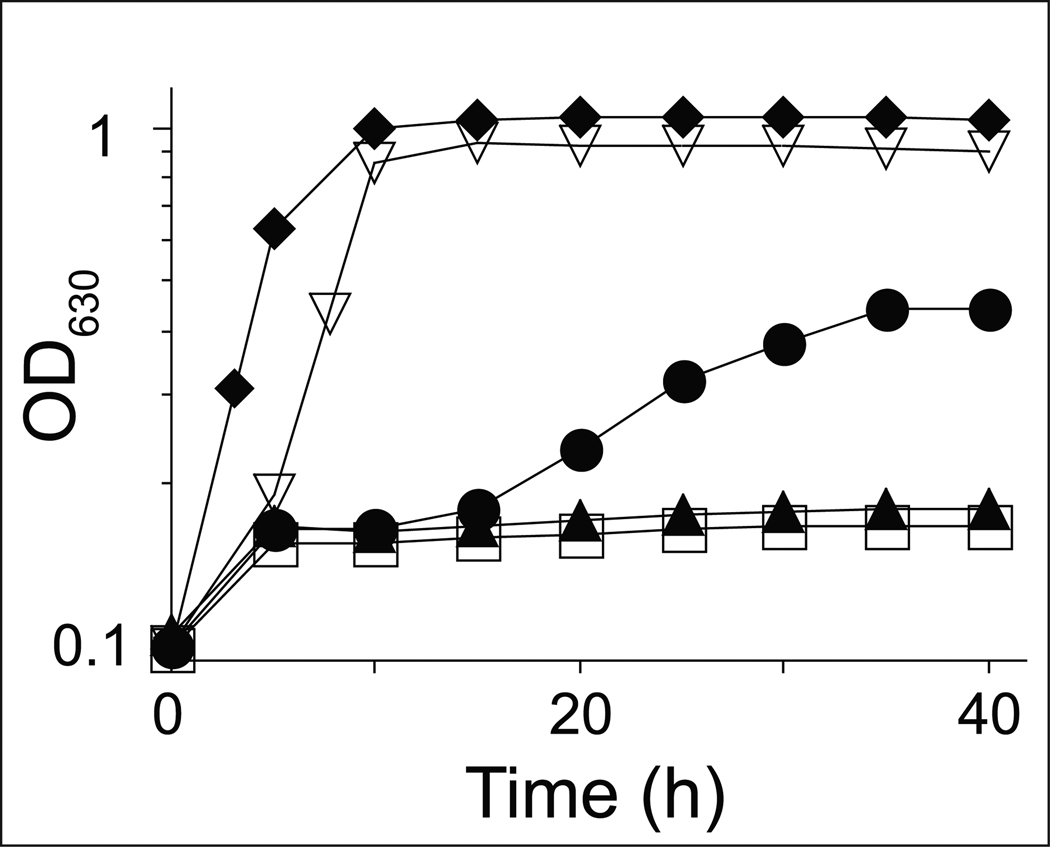
As shown in figure 4, co-expression of arsA+ and arsB+ compensated for the lack of CobT function in JE13874, allowing growth of the strain (Fig. 4B). In contrast, expression of arsA+ or arsB+ alone did not restore AdoCba biosynthesis in JE13871 and JE13872 (Fig. 4C, 4D). We note that the addition of phenol and p-cresol extended the lag time prior to the onset of exponential growth of strain JE13874 co-expressing arsA+ and arsB+ (Fig. 4B). Such an increase in the lag time was not observed when DMB substituted for phenol or p-cresol.
Figure 4. SoArsAB compensate for the lack of CobT activity during AdoCba biosynthesis in S. enterica.
Strains of S. enterica were grown in minimal medium supplemented with glycerol (22 mM); (CN)2Cbi (15 nM) was added to all cultures except to the control medium (open circles). Phenol (75 µM, open squares); p-cresol, (75 µM, open triangles), or DMB (150 µM, open diamonds); the growth response in medium lacking phenol, p-cresol or DMB is also shown (solid circles). CNCbl (15 nM, solid diamonds) was used as positive control. A. Strain JE13867 (metE cobT cobB / pBAD33); B. JE13874 (parsAB+ = pARSAB4); C. JE13871 (parsA+ = pARSA4); D. JE13872 (parsB+ = pARSB2).
S. enterica synthesizes phenolyl- and p-cresolyl-Cba when SoArsA and SoArsB substitute for CobT and phenol or p-cresol are present in the culture medium
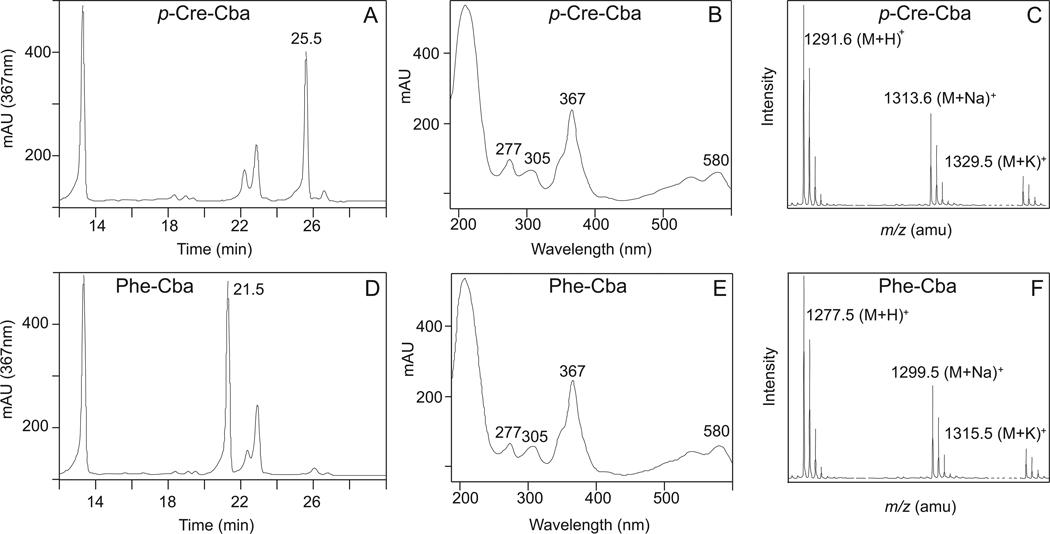
We confirmed that growth of strain JE13874 (metE cobT cobB / pARSAB4 arsA+ arsB+) under the conditions described above was due to the synthesis of phenolyl-Cba or p-cresolyl-Cba. Figure 5 (panels A, B, and C) shows the RP-HPLC separation (isolation), UV-visible spectrum and mass spectrum (identity) of p-cresolyl-Cba synthesized by strain JE13874. Equivalent data for phenolyl-Cba are presented in figure 5D, E, and F. The UV-visible spectra shown in figures 5B, E confirmed that the isolated compounds were corrinoids, but the data did not distinguish between complete and incomplete corrinoids (compare to (CN)2Cbi and CNCbl, Fig. S3). However, the mass spectra data for p-cresolyl-Cba [m/z = 1291, (M+H)+] and phenolyl-Cba [m/z = 1277, (M+H)+] (Fig. 5C, F, respectively) were consistent with the predicted formula mass of phenolyl- and p-cresolyl-Cba without an upper ligand. We did not detect any phenolic-Cba in the cell extract when phenol or p-cresol was not added to the culture medium.
Figure 5. Isolation and characterization of phenolyl-Cba (Panels A–C) and p-cresolyl-Cba (Panels D–F) synthesized by S. enterica.
Phenolyl- and p-cresolyl-Cba were synthesized by strain JE13874 (metE cobT cobB / pARSAB4 arsA+ arsB+) when phenol and p-cresol were supplemented in the growth medium. Panels A, D: Isolation of the cobamides by RP-HPLC. Panels B, E: UV-visible spectra of the isolated cobamides. Panels C, F: ESI mass spectra of the purified cobamides. Details of the analyses can be found in the Experimental procedures section.
We used a bioassay to further establish that the isolated corrinoids were complete. To do this, we used a bioassay that employed S. enterica strain JE8248 (metE cobS), a cobamide auxotroph that cannot catalyze the penultimate step of the pathway (Fig. 2) (Zayas and Escalante-Semerena, 2007). As shown in figure 6, phenolyl-Cba and p-cresolyl-Cba supported growth of strain JE8248 on glucose under conditions that required the synthesis of methionine from homocysteine to occur via the Cba-dependent methionine synthase (MetH) enzyme. However, 10-fold higher concentrations of phenolyl-Cba or p-cresolyl-Cba were needed to match the response to cobalamin. In contrast to the above results, phenolyl-Cba and p-cresolyl-Cba did not support growth of JE8248 on ethanolamine or 1,2-propanediol (data not shown), consistent with the inability of phenol or p-cresol to form a coordination bond with the Co ion of the ring.
Figure 6. Phenolyl-Cba and p-cresolyl-Cba substitute for cobalamin in S. enterica.
Strain JE8248 (metE cobS) was grown on medium supplemented with glucose (11 mM) + CNCbl (15 nM, diamonds), phenolyl-Cba (15 nM, open squares), phenolyl-Cba (150 nM, solid squares), p-cresolyl-Cba (15 nM, open triangles), or p-cresolyl-Cba (150 nM, solid triangles); no corrinoid control (circles). Growth on strain JE8248 depended on the function of MetH, the cobamide-dependent methionine synthase that converts homocysteine to methionine in this bacterium.
In S. enterica, the phenolyl moiety of phenolyl-Cba is derived from exogenous, not endogenous sources
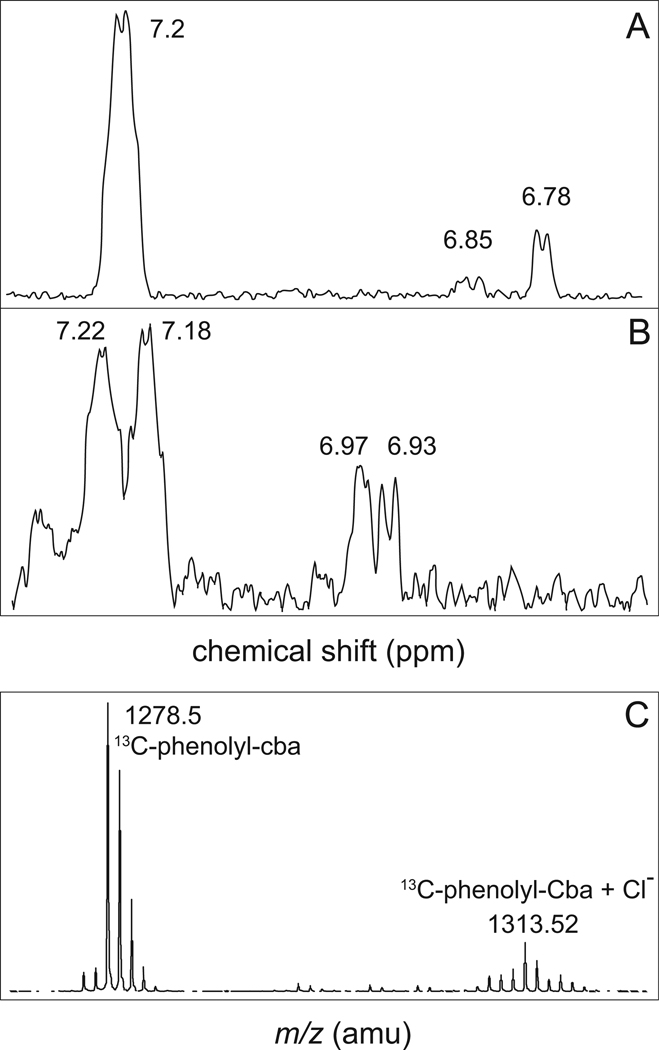
To confirm the incorporation of phenol into the cobamide, we grew strain JE13874 (metE cobT cobB / pARSB4 arsA+ arsB+) in minimal medium supplemented with [13C-1]-phenol (99% enriched). The cobamide synthesized under these conditions was isolated as described above, and was analyzed by 13C heteronuclear multiple bond correlation (HMBC) NMR spectroscopy (Bax and Summers, 1986), and mass spectrometry. The spectrum of the [13C-1]-phenolyl-Cba was compared to the spectrum of [13C-1]-phenol (Fig. 7A,B). HMBC experiments detect long-range H-C couplings while suppressing H-H correlations, therefore the chemical shifts of protons in an HMBC spectrum are the result of only long range effects of the 13C nucleus. As seen in figure 7B, [13C-1]-phenolyl-Cba showed detectable long-range H-C couplings, which suggested that [13C-1]-phenol was incorporated into the cobamide. If the 13C label were not present in the cobamide, no chemical shifts would have been detected with the HMBC experiment. The difference in chemical shifts between free [13C-1]-phenol and [13C-1]-phenolyl-Cba was due to the differences in the magnetic environment of the protons relative to the free hydroxyl group in phenol and the ether-linked ribose in phenolyl-Cba. The complexity of the signals observed in the [13C-1]-phenolyl-Cba spectrum, relative to the free [13C-1]-phenol spectrum, was likely due to diverse magnetic environments affecting the phenolyl moiety of the cobamide. As expected, the mass [13C-1]-phenolyl-Cba was one atomic mass unit greater than the mass of [12C-1]-phenolyl-Cba (Fig. 5F vs Fig. 7C).
Figure 7. HMBC NMR spectroscopy and mass spectrometry of 13C-labeled phenolyl-Cba.
A. 13C HMBC NMR spectra of [13C-1]-phenol (A), and 13C-labeled phenolyl-Cba (B). C. MALDI-TOF spectrum of the 13C-labeled phenolyl-Cba. Number of acquisitions for [13C-1]-phenol = 64,000. Number of acquisitions for 13C-labeled phenolyl-cobamide = 16,384.
S. ovata ArsA and ArsB are soluble and active as a heterodimer
Our attempts to purify SoArsA and SoArsB individually resulted in insoluble proteins (data not shown). Given the in vivo observation that cobamide biosynthesis in strain JE2501 (cobT cobB) was restored only when arsA+ and arsB+ were co-expressed, we sought to co-purify SoArsA and SoArsB (SoArsAB) by over-expressing arsA+ and arsB+ together. To facilitate protein purification, a recombinant tobacco etch virus (rTEV) protease cleavable hexahistidine (H6) tag was fused to the N-terminus of SoArsA using cloning vectors described elsewhere (Rocco et al., 2008). SoArsB encoded by the resulting construct (pARSAB7, Table 1) was not tagged.
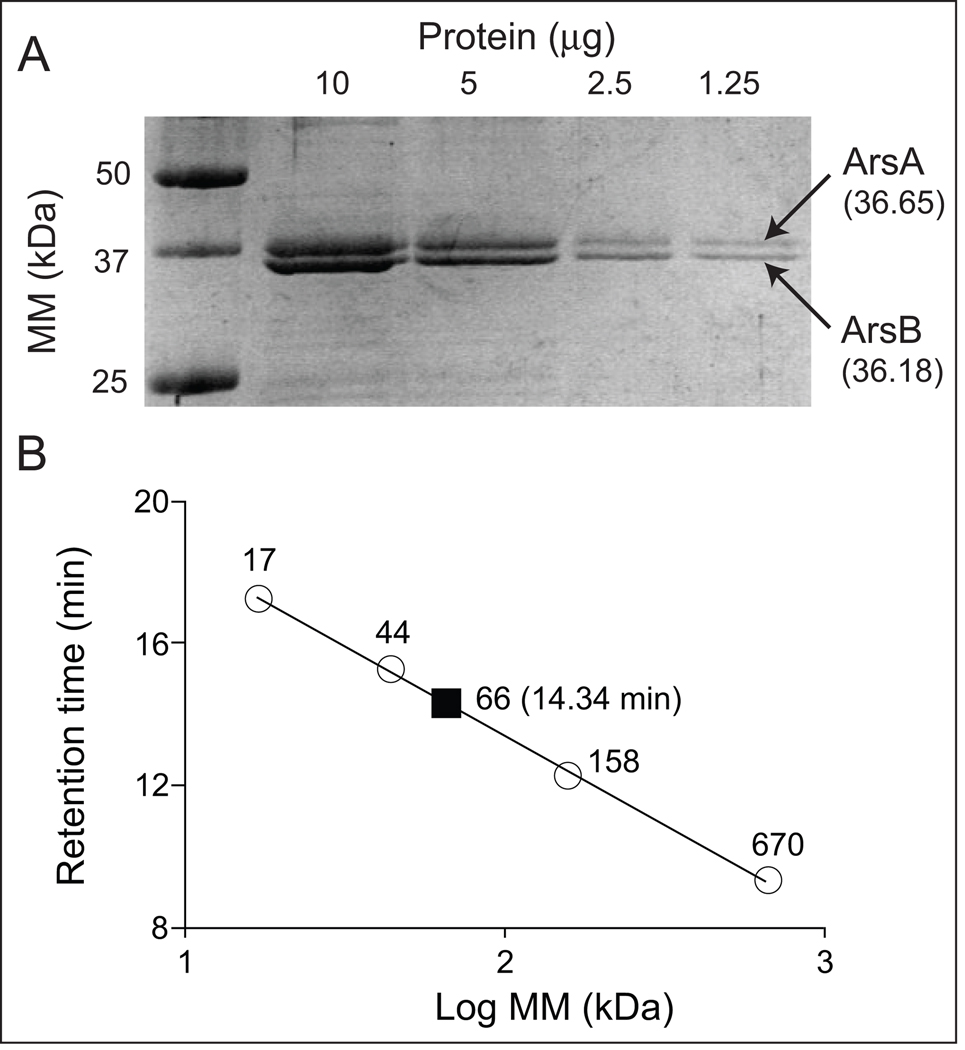
Two protein bands were resolved by SDS-PAGE after nickel affinity purification and rTEV protease treatment of soluble proteins (Fig. 8A). Since SoArsB was not tagged but co-purified with SoArsA, we hypothesized that the proteins interacted. Results of size exclusion chromatography experiments supported this idea (Fig. 8B). SoArsAB heterodimers had a retention time of 14.34 min, which corresponded to a mass of ~66 kDa. Results of peptide tandem mass spectrometry MALDI TOF (MS/MS) fingerprinting analysis of a tryptic digest of the fraction containing ArsAB confirmed that the two soluble proteins were SoArsA and SoArsB (Table S1). In addition, the fraction containing purified SoArsAB had NaMN:DMB phosphoribosyltransferase activity with a specific activity of 19 ± 3 nmol/min/mg of protein (Table S2).
Figure 8. Purity and oligomeric state of SoArsAB.
A. SDS-PAGE separation of purified tag-less SoArsAB. Predicted molecular masses for SoArsA and SoArsB were 36.65, 36.18 kDa, respectively. B. Gel filtration chromatography of tag-less SoArsAB. The retention time of protein standards of known molecular masses were plotted against their retention time. SoArsAB (solid square) eluted at 14.34 min, which corresponded to a mass of ~66 kDa.
SoArsAB synthesize α-phenolyl-, α-p-cresolyl-, and α-DMB-ribotide
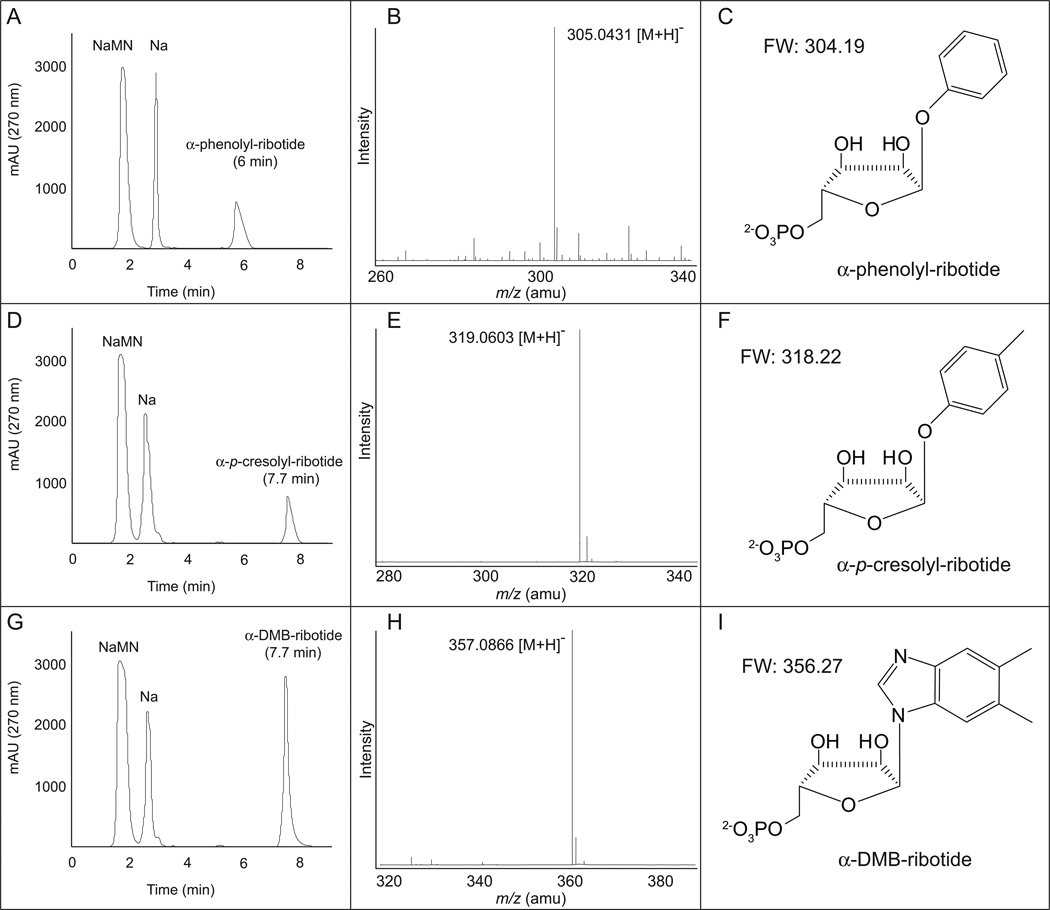
The homology of SoArsA and SoArsB to SeCobT suggested that SoArsAB had NaMN:phenol/p-cresol phosphoribosyltransferase activity. Data reported in Table S2 show that SoArsAB had phosphoribosyltransferase activity, and that it activated phenol and DMB with similar efficiency. Out of the conditions tested, SoArsAB was most active in a glycine pH 9 buffer. The product of SoArsAB when phenol, p-cresol, or DMB was used as substrate was purified by RP-HPLC and identified by mass spectrometry (Fig. 9). To confirm the products of SoArsAB were ribotides, each of the products was dephosphorylated using alkaline phosphatase (see Experimental procedures), re-purified by RP-HPLC and their mass was assessed by mass spectrometry (Fig. S4). In all instances, the observed masses were in agreement with the calculated formula masses. To test whether the SoArsAB products were biologically active, we supplemented the growth medium of S. enterica strain JE2501 (cobT cobB) with (CN)2Cbi and RP-HPLC-purified α-phenolyl- and α-p-cresolyl-ribotide. Five-fold higher concentrations of α-phenolyl- and α-p-cresolyl-ribotides were required to support growth, relative to the concentration of α-DMB-ribotide required (Fig. 10).
Figure 9. SoArsAB dependent synthesis of α-phenolyl-ribotide (panels A–C), α-p-cresolyl-ribotide, (panels D–F), and α-5,6-dimethylbenzimidazolyl (DMB)-ribotide (panels G–I).
Conditions for the in vivo synthesis of α-ribotides are described under Experimental procedures. Panels A, D, G, RP-HPLC resolution of reaction substrates from products; panels B, E, H, ESI mass spectra of α-ribotides synthesized by SoArsAB; panels C,F,I, structure and formula weight of α-ribotides synthesized by SoArsAB.
Figure 10.
Growth of strain JE2501 (metE cobT cobB) on NCE minimal medium supplemented with glucose (11 mM) with no corrinoids (squares); (CN)2Cbi (15 nM, solid triangles); (CN)2Cbi (15 nM) + α-DMB-ribotide (15 µM, diamonds); (CN)2Cbi (15 nM) + α-p-cresolyl-ribotide (75 µM, inverted triangles); or (CN)2Cbi (15 nM) + α-phenolyl-ribotide (75 µM, circles). Under the conditions used, growth of JE2501 demanded the conversion of Cbi to Cba by the nucleotide loop assembly enzymes (see Fig. 2), and the use of the resulting Cba by the methionine synthase (MetH) enzyme.
The tandem chromosomal arrangement of S. ovata arsA and arsB and the 11-residue deletion of SoArsB are uncommon
We searched the finished and unfinished genome databases for other tandem genes encoding CobT homologs similar to the one we found in Sporomusa ovata. We searched for CobT (Pfam domain DBI_PRT pfam02277) sequences in Pfam (http://pfam.sanger.ac.uk/) for tandem gene sequences within the genomes having multiple CobT sequences. We identified consecutive genes encoding for CobT homolog (EC 2.4.2.21) sequences in the Integrated Microbial Genomes (IMG, http://www.jgi.doe.gov/) database using the EC number as a filter. We also searched the orthologs neighborhood regions in IMG once a tandem cobT homolog sequence was identified. At present, only some species of Veillonella and Dialster invisus share the tandem organization of the S. ovata arsA and arsB genes. Interestingly, the 11-residue deletion in SoArsB is found only in Dialister invisus ArsB, but not in Veillonella parvula ArsB (Fig. S5).
Discussion
The arsA and arsB genes of S. ovata encode a novel enzyme that activates phenolic bases during AdoCba biosynthesis
To date, the only organism known to synthesize phenolic cobamides is Sporomusa ovata. However, the identity of the enzymes responsible for the activation of phenol and p-cresol remained unknown. Using a combination of in vivo and in vitro approaches we isolated two genes of S. ovata whose products convert phenol and p-cresol to their corresponding α-ribotides, which are precursors in the biosynthesis of phenolic cobamides (Fig. 2). To reflect the function associated with their gene products, we propose the name arsA and arsB, which stands for alpha-ribotide synthesis.
The SoArsAB enzyme has sufficient NaMN:Base phosphoribosyltransferase activity to functionally replace the CobT enzyme of S. enterica in vivo to satisfy the methionine requirement of the cell. There are, however, notable differences between SeCobT and SoArsAB. First, SeCobT is active as a homodimer, while SoArsAB is active as a heterodimer. Second, SoArsAB can activate phenol, p-cresol, and DMB, that is, SoArsAB catalyze the formation of α-O- and α-N-glycosidic bonds. In contrast, SeCobT cannot activate phenol or p-cresol. On the other hand, SeCobT and SoArsAB have common features. First, they share a common ancestor, as shown by the end-to-end homology and the degrees of identity and similarity between SoArsA and SoArsB with SeCobT (Fig. 3). And second, based on what is known about SeCobT, it is likely that the mechanism of catalysis of SoArsAB is similar to that of SeCobT, as suggested by the conservation of the putative catalytic glutamyl residue at the C-terminus of both proteins (Fig. 3, S2). Mutational, structural and kinetic analyses of SoArsAB are needed to better understand the functional differences between this enzyme and SeCobT.
At present, the assignment of the starting methionine of SoArsA is tentative. As shown in figure 3, there is a methionine residue at position #5 in the sequence. Although the SoArsA protein we isolated has the sequence shown in figure 3, it is possible that in vivo the starting methionine of SoArsA is Met5. We are currently eliciting antibodies against SoArsAB so we can isolate the complex from S. ovata and can resolve this issue.
We are interested in learning about the structural features that allow SoArsAB to have broader base specificity than SeCobT. For example, we need to determine whether or not the heterodimeric state of SoArsAB reflects on how substrate specificity is achieved. At present, we do not know whether SoArsAB can activate purine and benzimidazole analogs (Fig. 1).
The 11-residue deletion in SoArsB (also present in D. invisus ArsB) is unique among CobT homologs (Fig. S5). It is unclear how this deletion impacts enzyme activity, substrate specificity, or any other aspect of SoArsAB function. These questions await further biochemical and kinetic analyses of SoArsAB function.
Limitations of the use of S. enterica and E. coli for the in vivo analysis of phenolic cobamide function
The use of S. enterica and E. coli as heterologous systems for the isolation and analysis of SoArsAB function was successful. However, there are limitations to the use of these systems. For example, it is clear that the SoArsAB enzyme supported Cba-dependent growth of a S. enterica cobT strain in the absence of any exogenous base (i.e., phenolics or DMB; Fig. 4B, solid circles). This result shows that S. enterica makes substrates that SoArsAB can use; the data also show that DMB is the preferred substrate for SoArsAB in this system. Although these complications do not change the conclusion that SoArsAB activates phenolic bases and DMB, they make the in vivo analysis of SoArsAB function more challenging. Another limitation of these heterologous systems is revealed by the concentration dependent growth response observed with different cobamides (Fig. 6). There are at least two possible interpretations of these results. First, transport of phenolic cobamides by the BtuBCDEF system responsible for the assimilation of corrinoids in S. enterica and E. coli (Cherezov et al., 2006; Hvorup et al., 2007; Van Bibber et al., 1999) is less efficient for phenolic cobamides than for cobalamin. And second, it is possible that the cobamide-dependent MetH enzyme has less affinity for phenolic cobamides than for cobalamin. Lastly, it appears like either S. enterica transports α-ribotides with different efficiencies, or the observed differences in growth as a function of the exogenous α-ribotide provided reflects different levels of functionality of the cobalamin 5’-P synthase (CobS) enzyme that consumes α-ribotides (Fig. 10).
Potential use of S. ovata enzymes for the study of cobamide specificity
Although the S. ovata predominantly synthesizes phenolic cobamides (Stupperich and Eisinger, 1989b; Stupperich et al., 1989), we now know that SoArsAB can also activate DMB (Table S2), thus S. ovata has the capability of making cobalamin. It is of interest to identify growth conditions that demand synthesis of different cobamides in S. ovata. It is possible that S. ovata regulates the synthesis of phenolic cobamides versus other cobamides by controlling the activity or expression of arsAB in response to environmental cues. However, it is also possible that S. ovata synthesizes phenolic cobamides due to the lack of an endogenous source of DMB or other lower ligands. Previous studies revealed that a 40-kDa protein that contained a bound phenolic cobamide was induced by the presence of methanol in S. ovata. These findings suggested that the above-mentioned protein was involved in methanol and 3,4-dimethoxybenzoate metabolism in this bacterium (Stupperich et al., 1992; Stupperich and Konle, 1993). Of note is the reported inhibition of S. ovata growth by exogenous DMB (Stupperich et al., 1990). Whether the observed inhibition by DMB was due to the specific growth conditions that demanded the synthesis of phenolic cobamides remains an open question.
On the basis of the ability of SoArsAB to activate DMB, it is possible that there are conditions where exogenous DMB is not inhibitory to S. ovata growth, and under such conditions this bacterium synthesizes cobalamin. It is also possible, however, that the ability of SoArsAB to activate DMB is a remnant of the evolution of the enzyme that is of no use to S. ovata, either because this bacterium does not synthesize DMB, and because it does not encounter this base in its environment.
Results from in vivo analyses showed that phenolic cobamides supported growth of a S. enterica strain requiring the function of MetH (methionine synthase, Fig. 6). Since MetH is known to bind cobalamin in the base-off form (Drennan et al., 1994), it is not surprising that phenolic cobamides, whose lower ligand cannot coordinate to the cobalt atom, supported MetH function. We note that there were differences in the growth behavior when the medium was supplemented with Cbl or different phenolic cobamides (Fig. 6). These results can be explained by decreased binding affinities of MetH for phenolic cobamides relative to its affinity for Cbl.
Phenolic cobamides did not support the growth of S. enterica on 1,2-propanediol or ethanolamine (data not shown). These results were consistent with reports of diol dehydratase and ethanolamine ammonia-lyase requiring axial coordination of the lower ligand base to the cobalt atom for function (Abend et al., 1999; Shibata et al., 2010; Yamanishi et al., 1998), a coordination bond that phenolic cobamides cannot form. A previous study, which showed that p-cresolyl-Cba was a competitive inhibitor for the 1,2-propanediol dehydratase is consistent with the involvement of a base-on coenzyme in the function of this enzyme (Poppe et al., 1997).
We predict that the genome of S. ovata encodes a homolog of the cobinamide amidohydrolase (CbiZ) enzyme (Woodson and Escalante-Semerena, 2004), which is involved in lower ligand remodeling (Gray and Escalante-Semerena, 2009). The existence of a CbiZ-like activity in S. ovata would be a good complement to the broad specificity of SoArsAB for its base substrate, and would provide S. ovata with the ability to synthesize diverse cobamides when the need for a specific cobamide arises. Unfortunately, the pace at which this work can be performed is limited by the lack of a S. ovata genome sequence. There is, however, information in the literature about Cba-dependent methyltransferases from S. ovata (Stupperich et al., 1992; Stupperich and Konle, 1993). Nevertheless, we are now in a good position to explore the specificity of these enzymes for different cobamides.
Experimental procedures
Strain constructions
All strains used in this study were derivatives of Salmonella enterica sv Typhimurium strain LT2 or Escherichia coli K12. Strain genotypes are described in Table 1. Chromosomal mutations were introduced into E. coli by phage P1-mediated transduction (Cronan et al., 1969). Plasmids were introduced into S. enterica by electroporation (O'Toole et al., 1993), and into E. coli by heat-shock transformation (Hanahan et al., 1991). The pEAK2 temperature-sensitive plasmid carrying the recA gene was transformed into E. coli EPI300 strain prior to P1-mediated transductions (Kouzminova and Kuzminov, 2004). The Flp recombinase encoded in plasmid pCP20 was used to resolve the kan gene encoding kanamycin resistance in strains from the Keio collection (Datsenko and Wanner, 2000).
Construction of a S. ovata gene library and identification of S. ovata arsA, arsB genes
Sporomusa ovata type strain (ATCC® 35899, DSM 2662) was obtained from ATCC and grown as described (Möller et al., 1984). S. ovata genomic DNA was isolated as described (Pospiech and Neumann, 1995), and was sheared with a 200-µL pipet tip. A gene library was constructed using the Copy Control Fosmid Library Production Kit (Epicentre) (Fig. S1). Briefly, sheared DNA was resolved on 10×15cm 0.75% low melting point agarose gel. Forty-kb fragments were excised from the gel, melted and digested with agarase enzyme provided in the fosmid production kit before ethanol precipitation. The ends of the DNA fragments were blunted and phosphorylated with a mixture of T4 DNA polymerase, T4 polynucleotide kinase, ATP and dNTPs (provided in the kit) before ligation to Eco72I blunt-ended plasmid pCC1FOS, and packaging into empty lambda phage heads in vitro. Each fosmid contained approximately 40 kb of S. ovata genomic DNA. The lambda phage suspension was used to infect strain JE11215 (E. coli EPI300 ΔmetE cobT400::kan+) plating on LB + chloramphenicol. Assuming that the S. ovata genome is 5 Mb, we obtained about 100X coverage of the genome with approximately 12,500 clones. Twelve pools of ~1,000 clones were combined and inoculated into liquid culture, and plasmids were induced to multi-copy by the addition of L-(+)arabinose (1.3 mM). Approximately 104 cells were plated onto minimal medium. Strain JE11215 requires methionine in the absence of a complete cobamide. Derivatives of strain JE11215 harboring a fosmid were selected for growth on NCE minimal glycerol (22 mM) plates supplemented with (CN)2Cbi (150 nM), thiamine (3 µM), leucine (150 µM), L-(+)-arabinose (1.3 mM); the same culture medium supplemented with phenol (75 µM) or p-cresol (75 µM) was also used; plates were incubated at 30°C for two days. Twenty-eight plasmids that supported growth of JE11215 on the selective media described above were screened by restriction analysis using NcoI (Fermentas). Plasmids were incubated with NcoI for 2 h, and restriction fragments were resolved in a 1% (w/v) agarose gel at 100 V for 1 h (Voytas, 2000). Seven plasmids with different restriction patterns were analyzed further; the pCC1FOS vector backbone has one NcoI restriction site. One of these plasmids (hereafter referred to as pSolibC17) was fragmented by sonication and the ends blunted as described above. Two 5-kb fragments were isolated by agarose gel electrophoresis (see above), followed by DNA extraction using Promega’s Gel and PCR Cleanup Kit™. DNA fragments were ligated into Eco72I blunt-ended plasmid pCC1FOS and introduced into strain JE11215 by heat-shock transformation. Inheritance of the plasmid carrying S. ovata CobT-like functions supported growth on minimal medium containing glycerol, (CN)2Cbi and DMB, phenol or p-cresol. The fragment of S. ovata DNA in pSolibC17 was sequenced using primers flanking the Eco72I insertion site (5'-GGATGTGCTGCAAGGCGATTAAGTTG-3' and 5'-CTCGTATGTTGTGTGGAATTGTGAGC-3’), and subsequent sequences were obtained by primer walking. Plasmid pSolibC17 encoded two CobT homologs, namely SoArsA and SoArsB. A scheme for the above procedure can be found in figure S1 (supplemental information).
Culture media, conditions and analyses
Strains were grown at 37°C in no-carbon essential (NCE) minimal medium (Vogel and Bonner, 1956) supplemented with glucose (11 mM), glycerol (22 mM), ethanolamine (90 mM), or 1,2-propanediol (90 mM) as carbon and energy source where indicated. Minimal medium also contained MgSO4 (1 mM), trace minerals (Balch et al., 1979). When added, ampicillin, kanamycin or chloramphenicol was present at 100 µg/mL, 50 µg/mL, or 12.5 µg/mL, respectively. For all growth experiments, strains were grown in triplicate in 96-well microtiter dishes; samples (2 µL) of overnight cultures grown in lysogeny broth (LB) (Bertani, 1951, 2004) was used to inoculate 198 µL of fresh medium plus supplements. Growth was analyzed using a computer-controlled BioTek ELx808-1 Ultra microplate reader (BioTek Instruments). Cell density measurements at 630 nm were acquired every 1800 s for 36 h; plates were shaken for 1745 s between readings. Data were analyzed using the GraphPad Prism v4 software package (GraphPad Software).
Plasmid constructions
Plasmid pARSA4
arsA+ was amplified from plasmid pSolibC17 with primers 5’-CGAGCTCGTAATGGAGGTTATTATGAGTTTACTGC-3’ and 5’-CTCTAGACGCTGACACTGTTCCATCGC-3’. The resulting fragment was cloned into the cloning vector pGEM (Promega) yielding plasmid pARSA2. The latter was cut with SacI and XbaI and ligated into the same sites in vector pBAD33 (Guzman et al., 1995) yielding plasmid pARSA4. Plasmid pARSA4 was used in functional complementation studies.
Plasmid pARSB2
arsB+ was amplified from plasmid pSolibC17 with primers 5'-CGAGCTCGCTCTATGTGGCCATTAAGC-3’ and 5’-CTCTAGAGCTTGCTAATCTCTAACATCCTTG-3’. The resulting fragment was cloned into pGEM (Promega) yielding plasmid pARSB1. The latter was cut with SacI and XbaI, and the fragment was ligated into the same sites in pBAD33, yielding plasmid pARSB2. Plasmid pARSB2 was used in functional complementation studies.
Plasmid pARSAB4
arsA+ and arsB+ were amplified together from plasmid pSolibC17 with primers 5’-CGAGCTCGTAATGGAGGTTATTATGAGTTTACTGC-3 and 5’-CTCTAGAGCTTGCTAATCTCTAACATCCTTG-3’. The resulting fragment was cloned into the pGEM (Promega), yielding plasmid pARSAB2. The latter was cut with SacI and XbaI and ligated into the same sites in pBAD33, yielding plasmid pARSAB4. Plasmid pARSAB4 was used in functional complementation studies.
Plasmid pARSAB7
arsA+ and arsB+ were amplified together from plasmid pSolibC17 with primers 5'-AGCTCGCCCGGGGATGGAGGTTATTATGAGTTTACTGCAAGC-3’ and 5'-GCAGCTAGCGCTTGCTAATCTCTAACATCCTTGC-3’. The fragment was cut with SmaI and NheI enzymes and ligated into the StuI and NheI sites of plasmid pH6T, yielding plasmid pARSAB7. Plasmid pH6T is a derivative of cloning vector pET31b, which directs the synthesis of proteins of interest fused to an N-terminal hexahistidine (H6) tag and an rTEV protease cleavage site. pH6T lacks two tyrosine residues in the sequence before the rTEV recognition site in pKLD37 (Rocco et al., 2008). Plasmid pARSAB7 directed the synthesis of H6-ArsA and tag-less SoArsB. Plasmid pARSAB7 was used to overproduce SoArsAB proteins.
Biosynthesis and isolation of phenolyl- and p-cresolyl-Cba
Biosynthesis
An overnight culture of strain JE13874 (cobT cobB / pARSAB4 arsA+ arsB+) was grown in LB supplemented with chloramphenicol, and was used to inoculate (1 ml/L of fresh medium) into minimal NCE medium containing glycerol (55 mM), (CN)2Cbi (300 nM), phenol or p-cresol (75 µM), L-(+)-arabinose (1.3 mM), and chloramphenicol (12.5 µg/mL). Two 1-liter cultures were grown for two days and harvested by centrifugation (Beckman/Coulter Avanti J20-XPI refrigerated centrifuge, equipped with a JLA-8.1000 rotor; 15 min at 4 °C; 6,000 × g), and kept at −80 °C until used.
Extraction
Frozen cells were resuspended in 30 mL of a 2:1 mixture of methanol and KH2PO4 (50 mM, pH 6.5) + KCN (5 mM). Once resuspended, cells were placed in a MaxQMini 4000 Barnstead/Lab-Line e Class shaking incubator heated to 65 °C; cells were continuously shaken (220 rpm) for 2 h. Cell debris was removed by centrifugation (Beckman/Coulter Avanti J25-I refrigerated centrifuge, equipped with a JA-25.50 rotor; 1 h at 4°C; 43,000 × g). The supernatant was filtered (Whatman™ Puradisc™ 25 mm PES filter, 0.45 mm pore size) and dried under vacuum using a Savant SPD111V concentrator running at room temperature. The pellet was resuspended in KH2PO4 buffer A [(50 mM, pH 6.5) + KCN (5 mM)] and applied onto a C18 Sep-Pak (Waters) previously conditioned with 10 mL of 100% methanol followed by 10 mL of dH2O. Corrinoids bound to the column were washed with 20 mL of dH2O before elution with 100% methanol. Corrinoids dissolved in methanol were concentrated under vacuum. The pellet was resuspended in buffer A.
Isolation
Cobamides were resolved by reverse-phase high performance liquid chromatography (RP-HPLC) using a Beckman Coulter System Gold 126 system equipped with a Phenomenex Synergi Hydro-RP (150 by 4.6 mm) using a modified system I mobile phase described elsewhere (Blanche et al., 1990). Briefly, the column was equilibrated at 23% buffer B [KH2PO4 (50 mM, pH 8.0) + KCN (5mM) + 50% (v/v) acetonitrile) and 77% buffer C [KH2PO4 (100 mM, pH 6.5) + KCN (10 mM)] at a flow rate of 1 mL/min and developed on a linear gradient to 53% buffer B for 43.2 min. The column was further developed to 100% buffer B for 5 min. Corrinoids were detected at 367 nm. Concentrations of phenolyl- and p-cresolyl-Cba were estimated using the molar extinction coefficient of (CN)2Cbi at 367 nm (21,800 M−1 cm−1).
Mass spectrometry of phenolyl- and p-cresolyl-cobamide
Corrinoids with a retention time of 25.1 and 21.5 min were collected, desalted and dried under vacuum. The molecular mass of each of these corrinoids was determined on a 4800 Matrix-Assisted Laser Desorption/Ionization-Time of Flight-Time of Flight (MALDI TOF-TOF) mass spectrometer (Applied Biosystems) scanning 700–4,000 Da mass range using 1000 shots acquired from 20 randomized regions of the sample spot at 3800 intensity of OptiBeam™ on-axis Nd:YAG laser with 200Hz firing rate and 3 to 7 ns pulse width in positive reflectron mode.
[13C-1]-phenol labeling of cobamides and NMR spectroscopy
A cobamide was extracted and purified from strain JE13874 (metE cobT cobB / pARSAB4 arsA+ arsB+) grown in minimal glycerol medium containing (CN)2Cbi and 99%-enriched [13C-1]-phenol (75 µM) (Cambridge Isotope Laboratories). 13C-labeled phenolyl-Cba was isolated, desalted, and dried under vacuum as described under Experimental procedures. The sample was resuspended in D2O (Cambridge Isotope Laboratories) to an approximate concentration of 7.5 µM, which was calculated using the molar extinction coefficient of (CN)2Cbi (see above). Authentic [13C-1]-phenol dissolved in D2O was used as standard. Both samples were subjected to a 13C heteronuclear multiple bond correlation (HMBC) experiment on a 750.13 MHz, Avance III Bruker NMR spectrometer to detect long-range 1H - 13C connectivity with one bond couplings suppressed with a low-pass J-filter; no decoupling was performed during acquisition using gradient pulses for selection (Bax and Summers, 1986).
Purification of SoArsAB proteins
Wild type SoArsA and SoArsB proteins were overproduced in strain JE13607 (E. coli BL21(DE3) cobT::kan+) from plasmid pARSAB7. Proteins were over-produced in E. coli BL21(DE3) cobT::kan+ to ensure that native EcCobT did not interfere. Proteins synthesis was induced in early-log-phase cultures (OD600 ~0.5) by the addition of isopropyl-1-thio-β-D-galactopyranoside (IPTG) to a final concentration of 0.5 mM; after the addition of IPTG, cultures were grown for 16 h at 30 °C. Cultures were harvested by centrifugation (15 min at 4 °C; 6,000 × g) in a Beckman/Coulter Avanti J20-XPI centrifuge equipped with a JLA-8.1000 rotor. Cells were stored at −80 °C until used. Frozen pellets were resuspended in buffer D [HEPES, (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid) buffer (50 mM, pH 8) containing NaCl (300 mM), imidazole (10 mM), and lysozyme (1 mg/mL)]. Cells were broken by sonication (60 s, 28% duty, 2 s pulses, setting 9) with a 500 Sonic Dismembrator (Fisher Scientific). Cell debris was removed by centrifugation (45 min at 4°C; 43,000 × g). Clarified extract was applied onto a 1-mL Ni-charged column (Ni-NTA, Qiagen), after loading, the column was washed with 10 ml buffer D containing twice as much imidazole (20 mM). SoArsAB was eluted off the column with buffer D containing 300 mM imidazole. Fractions containing SoArsAB were pooled and H7-rTEV protease (Blommel and Fox, 2007) was added at a SoArsAB: H7-rTEV ratio of 1:100 to remove the H7 tag. H7-rTEV-treated protein was loaded onto a Ni-charged column to separate H7-rTEV and other contaminants from tag-less SoArsAB in the flow through. SoArsAB was dialyzed in HEPES buffer (50mM, pH 8) containing NaCl (200 mM), glycerol (20%, v/v), flash frozen with liquid N2, and stored at −80°C until used.
Determination of the oligomeric state of active SoArsAB
Purified SoArsAB sample (1 ml, 1 mg/mL) was applied onto an ÄKTA FPLC Explorer system (Amersham Biosciences) equipped with a Superdex 200 HR 10/30 (GE Healthcare) column. The column was developed with Na2HPO4 buffer (50 mM, pH 8) containing NaCl (200 mM). To generate a standard curve, the log of the molecular mass of selected proteins (Bio-Rad Laboratories gel filtration standards: thyroglobulin, 670 kDa; globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; vitamin B12, 1,355 Da) was plotted against their retention time. The retention time of SoArsAB was interpolated to obtain an approximate mass of the SoArsAB complex.
Peptide fingerprinting of purified SoArsAB
Purified SoArsAB proteins were reduced with 25 mM dithiotreitol (DTT in 25 mM NH4HCO3) for 30min at 56°C, alkylated in the dark with 55 mM iodoacetamide (IAA in 25mM NH4HCO3) at room temperature for 30 min, washed twice in H2O for 30 s, equilibrated in 25 mM NH4HCO3 for 1 min, dehydrated for 5 min in acetonitrile (ACN)/H2O/NH4HCO3 (50%:50%:25mM) then once more for 30 s in 100% ACN, dried again and rehydrated with 20 µL of trypsin solution [10 ng/µL trypsin Gold (PROMEGA) in 25 mM NH4HCO3 / 0.01% ProteaseMAX (w/v) (PROMEGA)]. Additional 30 µL of digestion solution [25 mM NH4HCO3 / 0.01% ProteaseMAX (w/v)] was added to facilitate complete rehydration and excess overlay needed for peptide extraction. SoArsAB digestion was conducted for 3 h at 42°C, after which peptides generated by the digestion protocol were transferred to a new tube and acidified with 2.5% trifluoroacetic acid (TFA) to 0.3% final. Degraded ProteaseMAX was removed via centrifugation [max speed, 10 min] and the peptides were solid-phase extracted (ZipTip® C18 pipette tips Millipore, Billerica, MA). Peptides were eluted off the C18 column with 1 µL of ACN/H2O/TFA (60%:40%:0.2%) into 0.5 ml Protein LoBind tube (Eppendorf), 0.5 µL was deposited onto the Opti-TOF™ 384 Well plate (Applied Biosystems) and re-crystalized with 0.50 µL of matrix [10 mg/ml α-cyano-4hydroxycinnamic acid in acetonitrile/H2O/TFA (60%:40%:0.2%)]. Peptide mass fingerprint analysis was performed on a 4800 Matrix-Assisted Laser Desorption/Ionization-Time of Flight-Time of Flight (MALDI TOF-TOF) mass spectrometer (Applied Biosystems). The peptide fingerprint was generated scanning 700–4,000 Da mass range using 1000 shots acquired from 20 randomized regions of the sample spot at 3800 intensity of OptiBeam™ on-axis Nd:YAG laser with 200Hz firing rate and 3 to 7 ns pulse width in positive reflectron mode. Raw data was deconvoluted using GPS Explorer™ software and submitted for peptide mapping search analysis against NCBInr E. coli user modified database and translated So arsA and So arsB sequences with an in-house licensed Mascot search engine (Matrix, Science, London, UK http://www.matrixscience.com/) with cysteine carbamidomethylation and methionine oxidation as variable modifications.
SoArsAB in vitro activity assay
SoArsAB phosphoribosyltransferase activity was assayed in vitro using a modification of the described SeCobT assay (Claas et al., 2010). The reaction was performed in buffer (100 mM) at the indicated pH, and the reaction mixture (35 µL) contained 120 ng SoArsAB/mL, NaMN (3 mM), non-labeled phenol (1.9 mM), and [14C-U]-phenol (0.1 mM, 60 mCi/mmol). In reactions where DMB substituted for phenol, the reaction mixture contained non-labeled DMB (1.98 mM) and [14C-2]-DMB (20 µM, 43 mCi/mmol). The effect of pH on the activity was investigated using several buffer systems: i) at pH 7, 3-(N-morpholino)propanesulfonic acid (MOPS); ii) at pH 8, HEPES; at iii) pH 9 and 10 glycine-NaOH. The reaction was initiated by the addition of phenol or DMB to reaction mixtures pre-incubated at 37 °C for 10 min. Ten-µL samples were removed after 5, 10, and 15 min, and the reaction was stopped by mixing the sample with 2 µL of KCl (3 M); the entire mixture (12 µL) was spotted on a silica TLC plate (10 × 10 cm, PE SIL G/UV, Whatman™). The TLC plate was developed with a 3:2 (v/v) chloroform:methanol mobile phase, allowed to dry for 10 min, and exposed to a MultiPurpose Phosphor Screen (Packard). Radioactivity was detected using a Cyclone Storage Phosphor System (Packard) equipped with OptiQuant v 04.00 software (Packard). The detection range of the instrument was established using known amounts of 14C-phenol or 14C-DMB for reactions using either substrate.
Preparation and separation of α-ribotides
The above assay was scaled up to 200 µL and incubated for 6 h; none of the reagents was radiolabeled. α-Ribotides synthesized by SoArsAB were resolved on a Beckman Coulter System Gold® 126 HPLC system equipped a Phenomenex Synergi Hydro-RP (150 by 4.6 mm) column as reported elsewhere (Gray and Escalante-Semerena, 2010). Products were collected and dried under vacuum. The concentration of α-ribotides was estimated using the molar extinction coefficients (in water) of phenol (1373 M−1 cm−1, at 270 nm), p-cresol (1700 M−1 cm−1, at 277 nm), and DMB (5670 M−1 cm−1, at 278 nm). α-Ribotides were dephosphorylated using alkaline phosphatase (Fast AP, Fermentas) according to the manufacturer’s recommendations. Protein was removed by filtration using an Amicon® Ultra-0.5 centrifugal filter (MWCO = 10kDa), and α-ribosides were separated by RP-HPLC using a described protocol (Gray and Escalante-Semerena, 2010). Retention times (min) were as follows: i) α-phenolyl-ribotide, 6 min; ii) α-p-cresolyl-ribotide, 7.7 min; iii) α-DMB-ribotide, 7.7 min; iv) α-phenolyl-riboside, 8.5 min; v) α-p-cresolyl-riboside, 10.4 min α; vi) α-DMB-riboside, 9 min. After RP-HPLC purification, α-ribosides were desalted using a C18 Sep-Pak and dried under vacuum.
Mass spectrometry of α-ribosides and α-ribotides
The molecular masses of α-ribosides and α-ribotides were determined by electrospray ionization (ESI) mass spectrometry on an Agilent LC/MSD TOF spectrometer.
Supplementary Material
Acknowledgments
This work was supported by PHS grant R37-GM40313 to J.C.E.-S. We thank A. Kuzminov for the gift of plasmid pEAK2. Mass spectra were obtained by Melissa Boersma and Grzegorz Sabat at the Biotechnology Center of the University of Wisconsin-Madison. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/ NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
References
- Abend A, Bandarian V, Nitsche R, Stupperich E, Retey J, Reed GH. Ethanolamine ammonia-lyase has a "base-on" binding mode for coenzyme B12. Arch Biochem Biophys. 1999;370:138–141. doi: 10.1006/abbi.1999.1382. [DOI] [PubMed] [Google Scholar]
- Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A, Summers MF. Proton and C-13 Assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J Amer Chem Soc. 1986;108:2093–2094. [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F, Thibaut D, Couder M, Muller JC. Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography. Anal Biochem. 1990;189:24–29. doi: 10.1016/0003-2697(90)90038-b. [DOI] [PubMed] [Google Scholar]
- Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho BA, Olfson P, Casadaban MJ. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Escalante-Semerena JC, Rayment I. The three-dimensional structures of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella typhimurium complexed with 5,6-dimethybenzimidazole and its reaction products determined to 1.9Å resolution. Biochemistry. 1999;38:16125–16135. doi: 10.1021/bi991752c. [DOI] [PubMed] [Google Scholar]
- Cheong CG, Escalante-Semerena JC, Rayment I. Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica. J Biol Chem. 2001;276:37612–37620. doi: 10.1074/jbc.M105390200. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Yamashita E, Liu W, Zhalnina M, Cramer WA, Caffrey M. In meso structure of the cobalamin transporter, BtuB, at 1.95 Å resolution. J Mol Biol. 2006;364:716–734. doi: 10.1016/j.jmb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas KR, Parrish JR, Maggio-Hall LA, Escalante-Semerena JC. Functional analysis of the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) enzyme, involved in the late steps of coenzyme B12 biosynthesis in Salmonella enterica. J Bacteriol. 2010;192:145–154. doi: 10.1128/JB.01159-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Jr, Birge CH, Vagelos PR. Evidence for two genes specifically involved in unsaturated fatty acid biosynthesis in Escherichia coli. J Bacteriol. 1969;100:601–604. doi: 10.1128/jb.100.2.601-604.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- Friedmann HC, Harris DL. The Formation of alpha-glycosidic 5'-nucleot ides by asingle displacement trans-N-glycosidase. J Biol Chem. 1965;240:406–412. [PubMed] [Google Scholar]
- Friedmann HC, Fyfe JA. Pseudovitamin B12 biosynthesis. Enzymatic formation of a new adenylic acid, 7-α-D-ribofuranosyladenine 5'-phosphate. J Biol Chem. 1969;244:1667–1671. [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides. Mol Microbiol. 2009;74:1198–1210. doi: 10.1111/j.1365-2958.2009.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. A new pathway for the synthesis of alpha-ribazole-phosphate in Listeria innocua. Mol Microbiol. 2010;77:1429–1438. doi: 10.1111/j.1365-2958.2010.07294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern J. Mechanisms of coenzyme B12-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP. Asymmetry in the structure of the ABC transporter binding protein complex BtuCD-BtuF. Science. 2007;317:1387–1390. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- Jeter RM, Olivera BM, Roth JR. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol. 1984;159:206–213. doi: 10.1128/jb.159.1.206-213.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Escalante-Semerena JC. Identification of 5,6-dimethylbenzimidazole as the Coα ligand of the cobamide synthesized by Salmonella typhimurium. Nutritional characterization of mutants defective in biosynthesis of the imidazole ring. J Biol Chem. 1992;267:13302–13305. [PubMed] [Google Scholar]
- Keck B, Renz P. Salmonella typhimurium forms adenylcobamide and 2-methyladenylcobamide, but no detectable cobalamin during strictly anaerobic growth. Arch Microbiol. 2000;173:76–77. doi: 10.1007/s002030050011. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA, Kuzminov A. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol Microbiol. 2004;51:1279–1295. doi: 10.1111/j.1365-2958.2003.03924.x. [DOI] [PubMed] [Google Scholar]
- Kräutler B, Fieber W, Osterman S, Fasching M, Ongania K-H, Gruber K, Kratky C, Mikl C, Siebert A, Diekert G. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is Norpseudo-B12, a new type of natural corrinoid. Helvetica Chimica Acta. 2003;86:3698–3716. [Google Scholar]
- Maggio-Hall LA, Escalante-Semerena JC. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc Natl Acad Sci U S A. 1999;96:11798–11803. doi: 10.1073/pnas.96.21.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh EN. Coenzyme B12 (cobalamin)-dependent enzymes. Essays Biochem. 1999;34:139–154. doi: 10.1042/bse0340139. [DOI] [PubMed] [Google Scholar]
- Möller B, Ossmer R, Howard BH, Gottschalk G, Hippe H. Sporomusa, a new genus of Gram-negative anaerobic-bacteria including Sporomusa-Sphaeroides spec-nov and Sporomusa-Ovata spec-nov. Arch Microbiol. 1984;139:388–396. [Google Scholar]
- O'Toole GA, Rondon MR, Escalante-Semerena JC. Analysis of mutants of defective in the synthesis of the nucleotide loop of cobalamin. J Bacteriol. 1993;175:3317–3326. doi: 10.1128/jb.175.11.3317-3326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte MM, Escalante-Semerena JC. Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the hyperthermophilic archaeon Methanocaldococcus jannaschii. Biochemistry. 2009;48:5882–5889. doi: 10.1021/bi8023114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe L, Stupperich E, Hull WE, Buckel T, Rétey J. A base-off analogue of coenzyme-B12 with a modified nucleotide loop--1H-NMR structure analysis and kinetic studies with (R)-methylmalonyl-CoA mutase, glycerol dehydratase, and diol dehydratase. Eur J Biochem. 1997;250:303–307. doi: 10.1111/j.1432-1033.1997.0303a.x. [DOI] [PubMed] [Google Scholar]
- Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- Renz P. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of other bases found in natural corrinoids. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc.; 1999. pp. 557–575. [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Masuda J, Tobimatsu T, Toraya T, Suto K, Morimoto Y, Yasuoka N. A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Structure Fold Des. 1999;7:997–1008. doi: 10.1016/s0969-2126(99)80126-9. [DOI] [PubMed] [Google Scholar]
- Shibata N, Tamagaki H, Hieda N, Akita K, Komori H, Shomura Y, Terawaki SI, Mori K, Yasuoka N, Higuchi Y, Toraya T. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J Biol Chem. 2010;285:26484–26493. doi: 10.1074/jbc.M110.125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Eisinger HJ, Kräutler B. Diversity of corrinoids in acetogenic bacteria. p-Cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur J Biochem. 1988;172:459–464. doi: 10.1111/j.1432-1033.1988.tb13910.x. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Eisinger HJ. Biosynthesis of para-cresolyl cobamide in Sporomusa ovata. Arch Microbiol. 1989a;151:372–377. [Google Scholar]
- Stupperich E, Eisinger HJ. Function and the biosynthesis of unusual corrinoids by a novel activation mechanism of aromatic compounds in anaerobic bacteria. Adv Space Res. 1989b;9:117–125. [Google Scholar]
- Stupperich E, Eisinger HJ, Kräutler B. Identification of phenolyl cobamide from the homoacetogenic bacterium Sporomusa ovata. Eur J Biochem. 1989;186:657–661. doi: 10.1111/j.1432-1033.1989.tb15256.x. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Eisinger H-J, Schurr S. Corrinoids in anaerobic bacteria. FEMS Microbiol Lett. 1990;87:355–360. [Google Scholar]
- Stupperich E, Aulkemeyer P, Eckerskorn C. Purification and characterization of a methanol-induced cobamide-containing protein from Sporomusa ovata. Arch Microbiol. 1992;158:370–373. doi: 10.1007/BF00245367. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Konle R. Corrinoid-dependent methyl transfer reactions are involved in methanol and 3,4-dimethoxybenzoate metabolism by Sporomusa ovata. Appl Environ Microbiol. 1993;59:3110–3116. doi: 10.1128/aem.59.9.3110-3116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowski JR, O'Toole GA, Escalante-Semerena JC. The cobT gene of Salmonella typhimurium encodes the NaMN: 5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-alpha-D-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J Bacteriol. 1994;176:3568–3575. doi: 10.1128/jb.176.12.3568-3575.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowski JR, Escalante-Semerena JC. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J Biol Chem. 1997;272:17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- Van Bibber M, Bradbeer C, Clark N, Roth JR. A new class of cobalamin transport mutants (btuF) provides genetic evidence for a periplasmic binding protein in Salmonella typhimurium. J Bacteriol. 1999;181:5539–5541. doi: 10.1128/jb.181.17.5539-5541.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Voytas D. Agarose Gel Electrophoresis. Greene Publishing Associates & Wiley Interscience: New York; 2000. [Google Scholar]
- Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Escalante-Semerena JC. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc Natl Acad Sci USA. 2004;101:3591–3596. doi: 10.1073/pnas.0305939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi M, Yamada S, Muguruma H, Murakami Y, Tobimatsu T, Ishida A, Yamauchi J, Toraya T. Evidence for axial coordination of 5,6-dimethylbenzimidazole to the cobalt atom of adenosylcobalamin bound to diol dehydratase. Biochemistry. 1998;37:4799–4803. doi: 10.1021/bi972572a. [DOI] [PubMed] [Google Scholar]
- Zayas CL, Escalante-Semerena JC. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: Evidence that dephosphorylation of adenosylcobalamin-5'-phosphate by the CobC phosphatase is the last step of the pathway. J Bacteriol. 2007;189:2210–2218. doi: 10.1128/JB.01665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.