Abstract
Background
Mandelic acid (MA), an important component in pharmaceutical syntheses, is currently produced exclusively via petrochemical processes. Growing concerns over the environment and fossil energy costs have inspired a quest to develop alternative routes to MA using renewable resources. Herein we report the first direct route to optically pure MA from glucose via genetic modification of the L-phenylalanine pathway in E. coli.
Results
The introduction of hydroxymandelate synthase (HmaS) from Amycolatopsis orientalis into E. coli led to a yield of 0.092 g/L S-MA. By combined deletion of competing pathways, further optimization of S-MA production was achieved, and the yield reached 0.74 g/L within 24 h. To produce R-MA, hydroxymandelate oxidase (Hmo) from Streptomyces coelicolor and D-mandelate dehydrogenase (DMD) from Rhodotorula graminis were co-expressed in an S-MA-producing strain, and the resulting strain was capable of producing 0.68 g/L R-MA. Finally, phenylpyruvate feeding experiments suggest that HmaS is a potential bottleneck to further improvement in yields.
Conclusions
We have constructed E. coli strains that successfully accomplished the production of S- and R-MA directly from glucose. Our work provides the first example of the completely fermentative production of S- and R-MA from renewable feedstock.
Keywords: Escherichia coli, fermentation, fine chemicals, metabolic engineering, renewable resources, R-mandelic acid, S-mandelic acid
Background
Mandelic acid (MA), an important fine chemical, has been widely used in the synthesis of cephalosporin antibiotics [1], the preparation of various other pharmaceuticals [2-4], as well as the resolution of racemic alcohols and amines [5,6]. Its stereoisomers generally are prepared using a chemical approach [7-9]. However, it has also been reported that its stereoisomers can be prepared via enzymatic routes; for example, stereospecific nitrilases are used to produce S- or R-MA [10-13] and enantioselective microbes are employed to degrade S-MA or R-MA to yield R- or S-enantiomers [14-16], respectively. In addition, a system combining the two microbes Pseudomonas polycolor and Micrococcus freudenreichii has also been explored for the production of R-MA from the racemate [17]. However, these commodity chemicals are manufactured entirely from petroleum-based feedstock such as benzaldehyde and mandelonitrile. Growing concerns over the environment and fossil resources costs have inspired a quest to develop more sustainable processes that afford these products from renewable feedstock at lower cost. Thus, biological processes based on renewable resources such as glucose that would provide direct one-step production of chiral MA in a microbial fermentation process would be of commercial interest.
Thus far, no direct biosynthetic pathway to free chiral MA has been identified in nature, although some organisms possess an MA catabolic pathway [18]. Engineering a microbe for the production of a heterologous compound requires the establishment of a new biochemical pathway. To construct synthetic pathways for producing S-MA and R-MA in E. coli, two problems need to be resolved. Firstly, suitable starting intermediates must be available. Secondly, the introduction of enzymes catalyzing reactions to produce the desired products must be feasible.
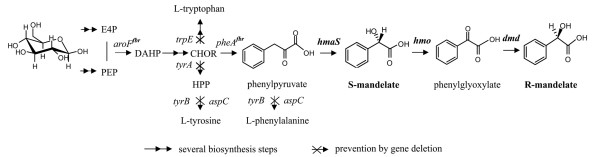
Here we describe the construction of synthetic pathways for the production of S-MA and R-MA in E. coli by combining biological entities from various organisms. For the production of S-MA, 4-hydroxymandelate synthase (HmaS) from Amycolatopsis orientalis was used to convert phenylpyruvate to S-MA [19]. To expand the S-MA synthesis to produce R-MA, two other enzyme reactions were explored: 4-hydroxymandelate oxidase (Hmo) from Streptomyces coelicolor was used to convert S-MA to phenylglyoxylate [19] and a third enzyme, D-mandelate dehydrogenase (DMD) derived from R. graminis, was used for the stereo-reduction of phenylglyoxylate to R-MA [20]. As phenylpyruvate is the direct precursor of L-phenylalanine, starting from phenylpyruvate it should be possible to construct an artificial pathway using HmaS, Hmo, and DMD to biosynthesize chiral MA directly from glucose in E. coli (Figure 1).
Figure 1.
Schematic representation of S-MA or R-MA biosynthesized from glucose in engineered E. coli. hmaS: Hydroxymandelate synthase gene from A. orientalis; hmo: Hydroxymandelate oxydase gene from S. coelicolor; dmd: D-mandelate dehydrogenase gene from R. graminis; E4P: erythrose-4-phosphate; PEP: phosphoenolpyruvate; CHOR: chorismate; HPP: hydroxyphenylpyruvate; DAHP: 3-deoxy-D-arabino-heptulosonate-7-phosphate.
In this study, we have modified E. coli strains that successfully achieved the production of S- and R-MA directly from glucose. To our knowledge, this is the first report describing the engineering of the L-phenylalanine pathway in E. coli to produce enantiomerically pure S-MA and R-MA directly from a renewable resource such as glucose.
Results
Fermentation of glucose via expression of HmaS in E. coli to produce S-MA directly
In order to construct a pathway to produce S-MA via a modified L-phenylalanine pathway in E. coli, an enzyme with S-mandelate synthesis activity is required to convert phenylpyruvate to S-MA. Thus, HmaS from A. orientalis was cloned and over-expressed in E. coli; the specific activity of HmaS was determined to be 10.18 mU/mg total protein. Production of MA in the reaction broth was identified by HPLC equipped with a chiral column, and the results illustrated that HmaS was able to convert phenylpyruvate to S-MA (Additional file 1), but not for the control (Additional file 1). Surprisingly, trace amounts of R-MA were detected as well, the results revealing that HmaS is not a rigorous stereo-inverting enzyme for catalyzing phenylpyruvate to S-MA. The enantiomeric excess (ee), > 98%, was calculated according to a previous report [15]. Therefore, this enzyme could be used further for the preparation of S-MA in E. coli.
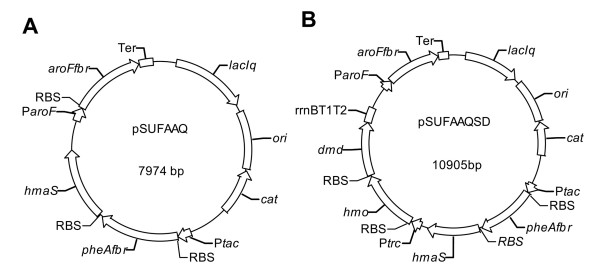
Over-expressions of deregulated 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase and chorismate mutase/prephenate dehydratase are well known strategies for increasing L-phenylalanine production in E. coli [21]. In this study, both the feedback deregulated DAHP synthase encoded by aroFfbr [22] and chorismate mutase/prephenate dehydratase encoded by pheAfbr [23] were constructed and over-expressed to increase the carbon flow into the shikimate pathway and ensure sufficient flow from chorismate to phenylpyruvate. To allow the fermentation of glucose to produce S-MA, an artificial operon with aroFfbr, pheAfbr and hmaS over-expression was created, which resulted in the recombinant plasmid pSUFAAQ (Figure 2A).
Figure 2.
Recombinant plasmids constructed in this study. (A) The recombinant plasmid pSUFAAQ used for S-MA synthesis. (B) The recombinant plasmid pSUFAAQSD used for R-MA synthesis.
To produce S-MA in E. coli, wild-type strains of E. coli W3110 harboring pSUFAAQ were cultured in fermentation medium in shake flasks. This resulted in the production of 0.092 g/L S-MA within 24 h, and a large amount of L-phenylalanine was detected as well. This proved that the artificial operon was functionally expressed in E. coli. Therefore, this strain could be used as a basis strain for further improvement to the production of S-MA directly from glucose. The biosynthetic pathway for S-MA is shown schematically in Figure 1.
Combined deletion of competing pathways to optimize the production of S-MA
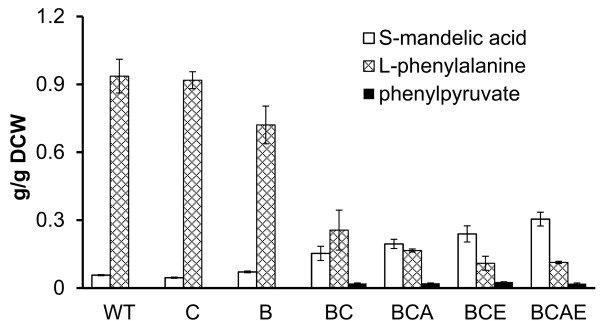
To further to optimize S-MA production, one logical possibility is the reduction of the byproducts formed. Thus, genes involved in competing side reactions would need to be deleted to avoid the consumption or degradation of the desired intermediates. Since the concentration of phenylpyruvate was the major limitation in the synthesis of S-MA, the aromatic amino acid aminotransferase encoded by tyrB and aspartate aminotransferase encoded by aspC, which are involved in the conversion of phenylpyruvate to L-phenylalanine, were deleted to minimize the loss of phenylpyruvate. The results showed that the deletion of tyrB increased S-MA production (0.07 g/gDCW) in comparison to the wild-type (0.06 g/gDCW), whereas the deletion of aspC had no positive effect on S-MA production (0.05 g/gDCW) and produced 0.92 g/gDCW L-phenylalanine. Furthermore, the production of S-MA significantly increased up to 0.15 g/gDCW (a 2.5-fold increase compared to wild-type), whereas the byproduct of L-phenylalanine (0.26 g/gDCW) was dramatically reduced in double mutants (Figure 3).
Figure 3.
Biosynthesis of S-MA in different mutants with pSUFAAQ. Different deletion mutants containing recombinant pSUFAAQ were cultivated in shake flasks for 24 h. The product S-mandelic acid, the intermediate phenylpyruvate, and the byproduct L-phenylalanine were analyzed. Data shown are means ± standard deviations, calculated from triplicate individual experiments.
To further increase the carbon flux to phenylpyruvate, the branch pathways of L-tyrosine and L-tryptophan were disrupted based on tyrB and aspC double mutants (BC) to reduce divergence from chorismate. Then we deleted the chorismate mutase/prephenate dehydrogenase encoded by tyrA and the anthranilate synthase encoded by trpE. As expected, for tyrA or/and trpE deletion based on double mutants, the production of S-MA increased to 0.30 g/gDCW (BCAE) with a 2-fold increase compared to BC and a 5-fold increase compared to wild-type.
We still detected amounts of phenylpyruvate (approximately 0.02 g/gDCW) that accumulated from BC to BCAE (Figure 3), which implied that the activity of HmaS was insufficient to convert all precursors of phenylpyruvate to S-MA. We postulate that combined deletion of the competing pathways led to increased phenylpyruvate accumulation and resulted in driving HmaS to synthesize more S-MA. Therefore, HmaS might be a potential bottleneck for further optimization of S-MA production and its derivatives.
Characterization of the fermentation of recombinant strain BCAE/pSUFAAQ
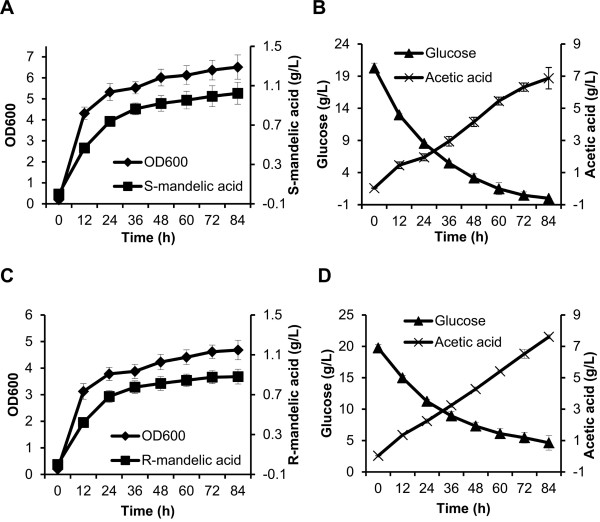
To further characterize the potential capacity of the best strain of BCAE harboring pSUFAAQ for S-MA production, shake flask fermentation was performed. During the process of fermentation, the cell density, S-MA production, glucose assimilation, and byproduct acetic acid accumulation were monitored, as shown in Figure 4A and 4B. As a result, 0.74 g/L S-MA was observed within 24 h, with a yield of 6.5% (g/g) from glucose, which reached 31% of the theoretical maximum (theoretical yield is 21% from glucose). The maximum production of S-MA reached 1.02 g/L within 84 h (Figure 4A) and, at the same time, glucose was completely consumed (Figure 4B). The optical isomer of MA in the broth was identified by HPLC using a chiral column (Additional file 2), confirming the formation of the S-isomer. Then the products in the broth were further analyzed by GC-MS (Additional file 3), confirming the product produced by recombinant strain BCAE/pSUFAAQ is S-MA. Therefore, the strain BCAE/pSUFAAQ could be used as the potential production strain in further studies.
Figure 4.
Time profiles of cell growth, S- or R- mandelic acid production, and residual glucose and acetic acid accumulation during the culture of the engineered E. coli. (A) OD600, S-mandelic acid produced, and (B) glucose consumed, byproduct acetic acid accumulated were monitored for BCAE with pSUFAAQ. (C) OD600, R-mandelic acid produced, and (D) glucose consumed, byproduct acetic acid accumulated were monitored for BCAE with pSUFAAQSD. Data shown are means ± standard deviations calculated from triplicate individual experiments.
In addition, 1.95 g/L of the byproduct acetic acid was detected in 24 h, and the titer significantly increased to 6.87 g/L within 84 h (Figure 4B). Acetic acid accumulation could cause many obstacles to the industrial production of interesting products. To minimize acetate formation, many strategies have been developed for this purpose during E. coli aerobic fermentations [24].
Use of Hmo together with DMD to convert S-MA via phenylglyoxylate to R-MA in vitro
Thus far, no enzyme with R-mandelate synthase activity has been identified that could catalyze the precursor of the amino acid to R-MA. Therefore, to promote fermentation production of R-MA in vivo, two other enzymes are required to convert S-MA via phenylglyoxylate to R-MA. In this study, Hmo from S. coelicolor and DMD from R. graminis were cloned and expressed in E. coli. Then, the specific activities of Hmo and DMD were determined to be 0.257 mU/mg and 87.44 mU/mg, respectively. These enzymes were used for further studies.
To test whether Hmo together with DMD could transform S-MA to R-MA in vitro, these two genes were cloned into a vector of pTrc99a under IPTG inducible promoter Ptrc resulting in pTrc99aSD. Then an assay for the simultaneous catalysis of S-MA to R-MA via the enzymes Hmo and DMD was performed in vitro. As shown in Additional file 1, R-MA was detected, but not for the control (Additional file 1). Thus, the conversion of S-MA via phenylglyoxylate to R-MA proved the functional expression of these two enzymes in E. coli, and paved the way for further synthesis of R-MA using their activities in vivo.
Co-expression of hmaS, hmo, and dmd in engineered E. coli to produce R-MA directly from glucose fermentation
We have demonstrated that expression of HmaS in E. coli can ferment glucose to produce S-MA and proved that the simultaneous expression of Hmo and DMD can convert S-MA via phenylglyoxylate to R-MA in vitro. Therefore, we presumed that co-expression of HmaS, Hmo, and DMD in E. coli could produce R-MA from glucose directly.
To produce R-MA ultimately from glucose, a new artificial biosynthetic pathway was assembled in E. coli. The scheme of this new pathway is shown in Figure 1. Based on the findings described above, we constructed an artificial biosynthetic pathway containing HmaS, Hmo, and DMD, with the corresponding genes obtained from A. orientalis, S. coelicolor, and R. graminis, respectively. Then, an engineered E. coli strain BCAE with the recombinant plasmid pSUFAAQSD (Figure 2B) was created.
Batch cultivations of BCAE/pSUFAAQSD were performed in shake flasks. During the fermentation process, cell density, R-MA production, glucose assimilation, and accumulation of the byproduct acetic acid were monitored (Figure 4C and 4D). The results illustrated that 0.68 g/L of R-MA with a yield of 7.8% (g/g) from glucose was achieved within 24 h and reached 37% of theoretical maximum, and the titer of R-MA reached 0.88 g/L in 84 h. The fermentation broth was analyzed by HPLC equipped with a chiral column (Additional file 2), and the products were further identified by GC-MS (Additional file 4). Combining these data showed that the compound in the broth was confirmed as R-MA.
Therefore, in this study we demonstrated that combined expression of the three enzymes in engineered E. coli strains resulted in the first completely fermentative synthesis of R-MA directly from glucose.
HmaS might be a potential bottleneck for further improvement of chiral MA production
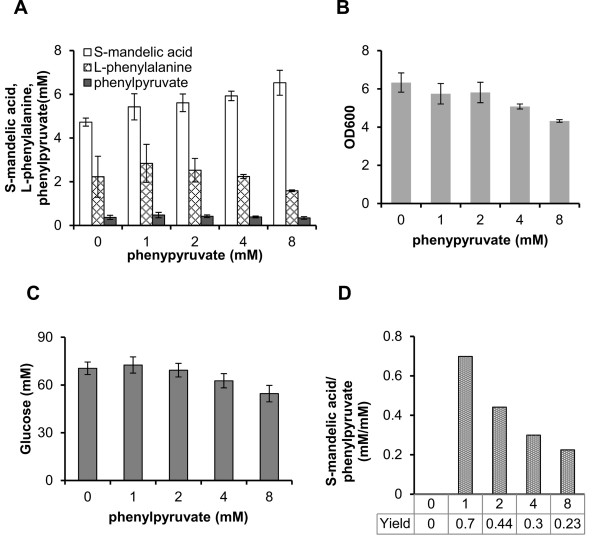
To further investigate HmaS as a potential rate-limiting enzyme preventing further improvement in the yields of S-MA and R-MA, we cultured BCAE with pSUFAAQ with various quantities of phenylpyruvate supplied. We found that the titers of S-MA gradually increased as the concentration of phenylpyruvate increased (Figure 5A). However, the increase in rate was not remarkable and a maximum of up to 38% was achieved with 8 mM phenylpyruvate supplied, whereas the cell density declined from OD600 of 6.3 to 4.3, suggesting that the higher concentrations of phenylpyruvate might be toxic to the host. It is also clear that the yield of S-MA from phenylpyruvate was obviously reduced from 0.70 (mM/mM) to 0.23 (mM/mM) with a feeding gradient of 1 mM to 8 mM (shown in Figure 5D). These results implied that the activity of HmaS was insufficient to catalyze all supplied phenylpyruvate to S-MA.
Figure 5.
Biosynthesis of S-mandelic acid with phenylpyruvate supplied. Products of engineered strains BCAE with pSUFAAQ fed with various concentrations of phenylpyruvate were analyzed in shake flasks during 24 h. S-mandelic acid, L-phenylalanine, and phenylpyruvate (A), cell growth (B), glucose consumption (C), and yields of S-mandelic acid from supplied phenylpyruvate (D) are listed. Data shown are means ± standard deviations calculated from triplicate individual experiments.
Furthermore, the kinetics parameters of HmaS were measured: the Km of HmaS was 0.45 ± 0.04 mM with phenylpyruvate as substrate, which was approximately 70-fold greater than the Km with its native substrate 4-hydroxyphenylpyruvate (6.5 ± 0.8 μM) [25]. This indicates that more phenylpyruvate is needed, compared to its native substrate, to achieve the same effect for HmaS synthesis of the corresponding products. Therefore, the Km of HmaS for phenylpyruvate needs to be modified to improve the catalytic efficiency at lower phenylpyruvate concentrations. This would be useful not only in terms of HmaS competing with other enzymes for catalyzing phenylpyruvate to S-MA, but would also be beneficial in alleviating phenylpyruvate toxicity towards the host at lower concentrations.
Therefore, in combining these data we infer that HmaS will be an important target for further strain improvement to enhance S-MA and R-MA production.
Discussion
Chiral mandelic acid, an important medical intermediate, is mainly prepared by chemical or enzymatic approaches. In this study, a new fermentative route was created using an engineered E. coli to produce optical isomers based on renewable resources. The L-phenylalanine pathway was modified and an artificial pathway using the enzymes HmaS, Hmo, and DMD from A. orientalis, S. coelicolor, and R. graminis, respectively, was constructed.
Many enzymatic methods have been reported for the preparation of stereoisomers [10,13]. However, none of them were able previously to produce mandelate enantiomers from glucose directly in a single microbial fermentation step. Thus far, substrates of enzymatic routes such as benzaldehyde and mandelonitrile have not been identified among the microbial primary metabolic pathways. If in the future some enzymes are discovered for catalyzing the primary metabolites to benzaldehyde or mandelonitrile, such multistep enzyme-catalyzed processes might be developed in the microbe for the production of mandelate isomers from glucose. However, the conversion rate would not be superior to that in this report due to the requirement for more heterogeneous enzymes and the extra reaction steps required.
Due to a lack of R-mandelate synthase, which could catalyze phenylpyruvate to R-MA directly, a relatively complex artificial pathway was constructed in this work for the production of R-MA. If R-mandelate synthase were available, the method for preparation of R-MA described here would be optimized using R-mandelate synthase only. We tried to find this kind of enzyme in some microorganisms using bioinformatics means. Unfortunately, thus far we have not been able to find it due to limited resources; however, it should be possible to construct enzymes with R-mandelate synthase activity through directed evolution.
Although we have successfully achieved the preparation of S-MA and R-MA in engineered E. coli, the byproduct of L-phenylalanine was still formed by the double mutant. It could be that the branched-chain amino acid aminotransferase encoded by ilvE is able to catalyze phenylpyruvate to L-phenylalanine [26], so the deletion of ilvE should eliminate L-phenylalanine formation to optimize the production of S-MA and R-MA in the future.
We observed the accumulation of large quantities of acetic acid during the fermentation process (Figure 4B and 4D). Acetic acid excretion by E. coli during aerobic growth on glucose is a major obstacle to recombinant protein production and chemical products, and gene disruptions have been successfully employed to minimize acetic acid formation and increase the production level of desired chemicals [27-30]. In this work, to improve MA production, we have deleted acetate-forming genes including poxB, pta, ackA, and acs to reduce acetic acid formation. Results showed that single gene deletions did not reduce acetate accumulation, suggesting that alternative pathways were also involved in acetate production (Additional file 5). As expected, simultaneous deletion of poxB, ackA and acs greatly minimized acetate accumulation (Additional file 6). However, the acetate reduction led to no increase in the carbon flux to MA production, which might be caused by significantly reduced cell growth rates.
In order to improve the production of S-MA, two genes (ppsA and tktA) [31] were co-expressed in the engineered S-MA-producing strains to enhance the supply of precursors of PEP and E4P. In the resulting strains the productivity of S-MA improved whereas the titer did not improve (data not shown). This result indicated that some other bottleneck exists downstream of the S-MA synthesis pathway.
HmaS is not only a key enzyme for S-MA synthesis but is also the first step in R-MA synthesis. We have previously investigated the effects of different mutants on S-MA production. The results implied that the activity of HmaS was insufficient to convert all phenylpyruvate to S-MA, leading to the accumulation of phenylpyruvate in the culture. To further explore the activity of HmaS, we also conducted phenylpyruvate feeding experiments. Although higher concentrations of phenylpyruvate slightly enhanced the yield of S-MA, the conversion rate of HmaS was significantly reduced with increasing phenylpyruvate. Furthermore, we measured the kinetic parameters of HmaS and found that its affinity towards the non-natural substrate phenylpyruvate was much lower than that towards its natural substrate [25]. This suggests that, in order to synthetize S-MA, HmaS needs a higher concentration of the substrate phenylpyruvate as the driving force. Therefore, combining these data we infer that HmaS will potentially be the bottleneck for further improvement of S-MA and R-MA synthesis. Thus, the catalytic activity of HmaS for converting phenylpyruvate to S-MA should be improved first through protein engineering to further improve the titers and yields of S-MA and R-MA production.
Conclusions
We have established a functional artificial pathway in E. coli and developed the first completely fermentative route for producing optically pure S-MA and R-MA from glucose, though further strain improvement and fermentative process optimization, including metabolic flux analysis, will be needed to improve the production of S-MA and R-MA. The successful construction of S-MA- and R-MA-producing E. coli in this study provides a solid basis for the development of other engineered bacteria, which could yield other commercially attractive bio-products or their derivatives from renewable resources.
Methods
Bacterial strains and culture media
Strains and plasmids used in this study are listed in Table 1. Antibiotics were used when needed at the following concentrations: Ampicillin (100 mg/L), Chloramphenicol (25 mg/L), Kanamycin (50 mg/L), Tetracycline (15 mg/L), Spectinomycin (50 mg/L), Apramycin (50 mg/L).
Table 1.
Strains and plasmids used in this study
| Strains | Genotype | Reference |
|
A. orientalis HCCB10007 |
Vancomycin-producing bacteria | [37] |
|
S. coelicolor A3(2) M145 |
Wild type SCP1- SCP2- | [38] |
|
R. graminis ATCC20804 |
Rhodotorula graminis di Menna, anamorph, mutant derived from KGX39 | ATCC |
| DH5α | lacZΔM15 endA1 recA1 relA1 gyrA96 deoR nupG λ- | TaKaRa |
| JM105 | endA1 glnV44 sbcB15 rpsL thi-1 Δ(lac-proAB) [F' traD36 proAB+ lacIq lacZΔM15] hsdR4 (rK-mK+) | Pharmacia |
| BL21(DE3) | F- omp gal dcm lon hsdSB (rB-mB-) λ(DE3) | Novagen |
| JW1256-1 | F- λ- ΔlacZ4787 (::rrnB-3), ΔtrpE::FRT-kan-FRT, rph-1 hsdR514 | CGSC |
| N3087 | tyrA16::Tn10 rpsL31(strR) | CGSC |
| WT | Wild type E. coli W3110 F- λ- rph-1 INV (rrnD rrnE) | CGSC |
| B | W3110 ΔtyrB::FRT | This study |
| C | W3110 ΔaspC::FRT | This study |
| BC | W3110 ΔtyrB::FRT, ΔaspC::FRT | This study |
| BCA | W3110 ΔtyrB::FRT, ΔaspC::FRT, tyrA16::Tn10 | This study |
| BCE | W3110 ΔtyrB::FRT, ΔaspC::FRT, ΔtrpE::FRT | This study |
| BCAE | W3110 ΔtyrB::FRT, ΔaspC::FRT, tyrA16::Tn10, ΔtrpE::FRT | This study |
| Plasmids | Genotype | Reference |
| pET24a | kan PT7 expression vector | Novagen |
| pEThmaSAo | pET24a derivative, hmaSAo from A. orientalis | This study |
| pET24admd | pET24a derivative, dmd from R. graminis | This study |
| pTrc99a | bla Ptrc lacIq pBR322 ori expression vector | Pharmacia |
| pTrc99ahmo | pTrc99a derivative, hmo | This study |
| pTrc99aSD | pTrc99a derivative, hmo, dmd | This study |
| pKK223-3 | bla Ptac pBR322 ori expression vector | Pharmacia |
| pKKpheAfbr | pKK223-3 derivative, pheAfbr from E. coli | This study |
| pSU2718 | Hae II fragment of pUC18 containing lacZα and MCS, p15A replicon with chloramphenicol acetyl transferase gene (cat) | [39] |
| pSUaroFfbr | pSU2718 derivative, aroFfbr from E. coli | This study |
| pSUFA | pSU2718 derivative, aroFfbr, pheAfbr | This study |
| pSUFAQ | pSU2718 derivative, aroFfbr, pheAfbr, lacIq | This study |
| pSUFAAQ | pSU2718 derivative, aroFfbr, pheAfbr, lacIq, hmaSAo | This study |
| pSUFAAQSD | pSU2718 derivative, aroFfbr, pheAfbr, lacIq, hmaSAo, hmo, dmd | This study |
| pIJ790 | araC ParaBAD gam bet exo repA101ts oriR101 cat | [33] |
| pIJ773 | bla FRT-aac(3)IV-oriT-FRT | [33] |
| pIJ778 | bla FRT-aadA-oriT-FRT | [33] |
| BT340 | ts bla cat | [33] |
LB medium was used for culturing the general strains for cloning. The engineered E. coli strains were cultivated in shake flasks in fermentation medium to test S- or R-MA production in vivo.
The fermentation medium contained the following components: D-glucose (20 g/L), KH2PO4 (1.0 g/L), (NH4)2SO4 (16 g/L), MgSO4 7H2O (1.0 g/L), L-tyrosine (0.2 g/L), L-tryptophan (0.1 g/L), L-aspartic acid (3 g/L), FeSO4 7H2O (0.01 g/L), MnSO4 H2O (0.008 g/L), CaCO3 (20 g/L). The stock solution of D-glucose, CaCO3, and MgSO4 was autoclaved separately whereas L-tryptophan, FeSO4, and MnSO4 were sterilized using a 0.22-μm filter.
Plasmid construction
The gene encoding feedback resistant mutant aroFfbr P148L [22] including the native promoter was obtained by overlap PCR, and the genomic DNA of E. coli W3110 was used as the PCR template. Two overlapping DNA fragments were amplified using primer pairs P148L-F/aroFSacI-R and P148L-R/aroFSacI-F, respectively. Then, using the two overlapping DNA fragments as the templates, another PCR reaction was performed with primer pair aroFSacI-F/aroFSacI-R and the whole length mutant aroFfbr gene was obtained. The resulting product was digested with Sac I and cloned into pSU2718 cut with the same enzyme, creating pSUaroFfbr.
The gene encoding feedback resistant mutant pheAfbr G309C [23] was constructed with the primers pheA-M-F/pheA-M-R and pheA-MN-FM/pheA-M-RM using overlap PCR (as described above). The purified PCR fragments of mutant pheAfbr were double digested with EcoR I and Hind III and then cloned into pKK223-3, creating pKKpheAfbr. Then the gene pheAfbr with the promoter Ptac and RBS (ribosome binding site) was amplified from pKKpheAfbr with primers PTacpheA*-F and PTacpheA*-R. The resulting PCR product was ligated into pSUaroFfbr vector, creating pSUFA.
The gene lacIq was amplified from pTrc99a with primers lacIq-F and lacIq-R. Then, it was cut with Nhe I and ligated into the same site of pSUFA, creating pSUFAQ.
The gene hmaS was amplified from the genomic DNA of A. orientalis HCCB10007 (GenBank: HQ679900) using the primers hmaSAo-F and hmaSAo-R. The PCR fragments were digested with Nde I and Hind III, and cloned into pET24a vector, generating pEThmaSAo. hmaS with the RBS of pET24a was amplified by PCR using primers pETrbs+hmaSAo-F and pETrbs+hmaSAo-R. The corresponding PCR fragments were purified and cut with Sal I and BamH I, then cloned into pSUFAQ, generating the recombinant plasmid pSUFAAQ for the production of S-MA (the scheme of plasmid pSUFAAQ can be seen in Figure 2A).
The gene hmo was amplified from the genomic DNA of S. coelicolor A3(2) M145 (GenBank: AL939115) with the primers hmo-F and hmo-R. The PCR fragments were purified, then digested with Nco I and Kpn I, and cloned into pTrc99a vector, creating pTrc99ahmo.
The gene dmd was amplified from the total cDNA of R. graminis ATCC20804 (GenBank: AJ001428) with the primers dmd-F and dmd-R, and cloned into the Nde I and EcoR I sites of pET24a to create pET24admd. Then, the fragments including dmd gene and the RBS of pET24a were amplified with the primers pETrbs+dmd-F and pETrbs+dmd-R. The corresponding PCR fragments were cloned into the Kpn I and Pst I sites of pTrc99ahmo to generate pTrc99aSD. Then the DNA fragments containing genes hmo and dmd with Ptrc promoter were obtained by PCR with the primers pTrcSD-F and pTrcSD-R, and inserted into the BamH I site of pSUFAAQ to create pSUFAAQSD for the production of R-MA (the scheme of plasmid pSUFAAQSD can be seen in Figure 2B).
Plasmids were isolated using the AxyPrep Plasmid Miniprep Kit (Axygen, Hangzhou, China). AxyPrep DNA Gel Extraction Kit was used to isolate DNA fragments from agarose gels (Axygen, Hangzhou, China). All PCR fragments were sequencing identified by BioSune (Shanghai, China). DNA polymerase KOD plus for PCR reactions was purchased from ToYoBo (Shanghai, China). All restriction enzymes and Taq DNA polymerase were purchased from Fermentas (Shanghai, China). Rapid DNA ligase and alkaline phosphatase were obtained from TaKaRa (Shanghai, China). Oligonucleotides were ordered from BioSune (Shanghai, China).
The plasmids constructed in this work are listed in Table 1, and the oligonucleotides used are given in Table 2.
Table 2.
Primers used for gene cloning in this study
| Primer Name | Nucleotide Sequence | Restriction site |
| P148L-F | 5'-TTAGATCTGAATAGCCCGCAATACCTGGGC- 3' | |
| P148L-R | 5'-GCTATTCAGATCTAACGCTTCCGTCGCCAGTGG - 3' | |
| aroFSacI-F | 5'-AACGAGCTCACCGGAAAGTCCTCGGGCATAAG - 3' | Sac I |
| aroFSacI-R | 5'-AACGAGCTCCGACTTCATCAATTTGATCGCGTAA - 3' | Sac I |
| pheA-M-F | 5'-GGGAATTCTATGACATCGGAAAACCCGTTAC - 3' | EcoR I |
| pheA-M-R | 5'- ATCCGGAAGCTTTTCATCAGG - 3' | Hind III |
| pheA-MN-FM | 5'- AACAAGCCTGTGCGCTGG - 3' | |
| pheA-M-RM | 5'- TCAACCAGCGCACAGGCTTGTTGC - 3' | |
| PTacpheA*-F | 5'- GATCCGAAGCTTATCGACTGCACG - 3' | Hind III |
| PTacpheA*-R | 5'- AGTCGACGCTTTTCATCAGGTTGG - 3' | Sal I |
| lacIq-F | 5'- CGCTAGCCCTGACGGGCTTGTCTG - 3' | Nhe I |
| lacIq-R | 5'- CGCTAGCTTCCGATGGCTGCCTG - 3' | Nhe I |
| hmaSAo-F | 5'- CGCATATGCAGAATTTCGAGATCGACTAC - 3' | Nde I |
| hmaSAo-R | 5'- CAAGCTTAACGTACGTCATCGCCG - 3' | Hind III |
| hmo-F | 5'-GATATACCATGGGCAGCAGCCATC-3' | Nco I |
| hmo-R | 5'-CGGTACCTGGTCATCCGTGGCTCCTG-3' | Kpn I |
| pETrbs+hmaSAo-F | 5'- CCGTCGACAAATAATTTTGTTTAACTTTAAG - 3' | Sal I |
| pETrbs+hmaSAo-R | 5'- CGGCCGGATCCTTGAAGATCTC - 3' | BamH I |
| dmd-F | 5'-CCATATGCCTCGCCCTCGCGTC-3' | Nde I |
| dmd-R | 5'-GGAATTCAGTAGGCGCGAAAAGCG-3' | EcoR I |
| pETrbs+dmd-F | 5'-CGGTACCTTTGTTTAACTTTAAGAAGG-3' | Kpn I |
| pETrbs+dmd-R | 5'-ACTGCAGTGGTGGTGGTGGTGGTGCT-3' | Pst I |
| pTrcSD-F | 5'-AGGATCCAAATCACTGCATAATTCG-3' | BamH I |
| pTrcSD-R | 5'-TGGATCCGTTATTGTCTCATGAGCG-3' | BamH I |
Underlined text denotes the restriction site sequences, and bold type indicates the start condons.
Construction of deletion mutants of E. coli
The deletion mutants of genes aspC and tyrB were obtained by a one-step inactivation protocol [32]. The disruption cassettes were amplified from vector pIJ773 or pIJ778 by PCR and then transformed into E. coli W3110 that contained the plasmid pIJ790 [33]. The resulting strain with the apramycin resistant cassette inserted into the gene aspC site was designated as W3110ΔaspC. The tyrB deleted mutant W3110ΔtyrB was constructed using the spectinomycin resistant cassette by the method described above. Then the multi-deletion mutants were generated by P1 transduction [34] from these single-deletion mutants. The P1 phage lysate prepared from W3110ΔaspC was used to infect W3110ΔtyrB to produce double-deletion mutant W3110Δ(tyrB, aspC). Using the same protocol, we successfully obtained tri-deletion mutants of W3110Δ(tyrB, aspC, trpE) and W3110Δ(tyrB, aspC, tyrA) by transfer of trpE mutation from JW1256-1 or tyrA mutation from N3087 using P1 phage to W3110Δ(tyrB, aspC), respectively. W3110Δ(tyrA, tyrB, aspC, trpE) was generated by transfer of tyrA mutation from N3087 to W3110Δ(tyrB, aspC, trpE). The disruption cassettes of aspC::FRT-apra-FRT, tyrB:: FRT-spec-FRT, trpE:: FRT-kan-FRT were eliminated by pCP20 [33], except for tyrA16::Tn10. Then the resistance markers of W3110ΔaspC, W3110ΔtyrB, W3110Δ(tyrB, aspC), W3110Δ(tyrB, aspC, trpE), W3110Δ(tyrB, aspC, tyrA), and W3110Δ(tyrA, tyrB, aspC, trpE) were removed using FLP-mediated excision of the disruption cassette [33], resulting in strains C, B, BC, BCE, BCA, and BCAE, respectively. All deletion mutants were verified by PCR analysis. All of the E. coli mutants constructed in this study are described in Table 1, and the primers used are listed in Table 3.
Table 3.
Primers used for gene deletion and verification in this study
| Primer Name | Nucleotide Sequence | Characterization |
| tyrB-KO-F | 5'-TTTAACCACCTGCCCGTAAACCTGGAGAACCATCGCGTGATTCCGGGGATCCGTCGACC- 3' | Deletion primer |
| tyrB-KO-R | 5'-ACTGCAGGCTGGGTAGCTCCAGCCTGCTTTCCTGCATTATGTAGGCTGGAGCTGCTTC- 3' | Deletion primer |
| tyrB-V-F | 5'- CTGTTGCTAATTGCCGTTCG - 3' | Verification primer |
| tyrB-V-R | 5'- CACGTAGAACGATGGCATCA - 3' | Verification primer |
| aspC-KO-F | 5'-CGGACTTCCCTTCTGTAACCATAATGGAACCTCGTCATGATTCCGGGGATCCGTCGACC- 3' | Deletion primer |
| aspC-KO-R | 5'-AGCCCGCTTTTCAGCGGGCTTCATTGTTTTTAATGC TTATGTAGGCTGGAGCTGCTTC- 3' | Deletion primer |
| aspC-V-F | 5' - CCTGCGTTTTCATCAGTAATAGTTGG - 3' | Verification primer |
| aspC-V-R | 5' - CCTTATCCGGCCTACAAAATCG - 3' | Verification primer |
| tyrA-V-F | 5' - TATCCGTAACCGATGCCTGC - 3' | Verification primer |
| tyrA-V-R | 5' - GGGAAATCACCCGTTCAATG - 3' | Verification primer |
| trpE-V-F | 5' - CGTACTGAAAGGTTGGTGGCG - 3' | Verification primer |
| trpE-V-R | 5' - AGGAGAAAGCATCAGCACCG - 3' | Verification primer |
Italics represent the sequences homologous to the gene to be deleted, underlined text denotes the sequence homologous to plasmid pIJ773 or pIJ778 for amplification of the resistance cassette.
Preparation of cell extract for enzymatic assays
E. coli BL21(DE3) strains harboring the plasmids pEThmaSAo, pET24admd, pTrc99ahmo or pTrc99aSD were cultivated in tubes containing 4 mL LB medium with appropriate antibiotics at 37°C at 220 rpm overnight. Then 0.5 mL of culture broth were added to 50 mL of LB medium in 250-mL shake flasks that contained appropriate antibiotics at 30°C at 220 rpm. When OD600 reached 0.4~0.6, the cells were induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 4 h, cells were harvested and washed with 200 mM potassium phosphate buffer (pH 7.5). Then the cell pellet was resuspended in 5 mL 200 mM potassium phosphate buffer (pH 7.5), and the cells were disrupted by French Press (Constant cell disruption systems, United Kingdom) at 25 kpsi. The lysate was centrifuged at 12000 rpm for 20 min (Eppendorf Centrifage 5810R) and the supernatant was used to determine activity. As a control, E. coli BL21(DE3) containing plasmid pET24a or pTrc99a was treated in the same manner. The total protein concentration of the supernatant was determined by the Bradford method [35], using bovine serum albumin (BSA) as a standard.
Enzyme assays
The HmaS assay was performed as described previously [19], with the exception that the 5-mL reaction mixture contained 200 mM potassium phosphate buffer (pH 7.5), 5 mM phenylpyruvate, 44 mM ascorbate, 0.3 mM FeSO4, and a final concentration of 1 mg/mL of soluble protein. The reaction was started by the addition of the cell-free extract at 28°C. After addition of 100 μL 1 M HCl to 500-μL samples to stop the reaction, the solution was centrifuged at 12000 rpm for 5 min (Eppendorf Centrifage 5430), and the products were analyzed by HPLC.
The Hmo and DMD assays were performed as described previously [19,20].
Production of R-MA in vitro
In the assay for R-MA production in vitro, the 5-mL reaction mixture contained 200 mM potassium phosphate buffer (pH 7.5), 5 mM S-MA, 2 mM NADH, 40 mM NAD+, and a final concentration of 1 mg/mL of soluble protein. The reaction was started by the addition of the crude extract at 30°C. Following the addition of 100 μL 1 M HCl to 500-μL samples to stop the reaction, the solution was centrifuged at 12000 rpm for 5 min (Eppendorf Centrifage 5430), and the supernatants were analyzed by HPLC.
Production of S- or R-MA in vivo
The engineered strains BCAE containing the plasmid pSUFAAQ or pSUFAAQSD were inoculated into 4 mL LB medium with chloramphenicol (25 mg/L) and incubated at 37°C at 220 rpm overnight. 4 mL of this culture were subsequently added to 250-mL shake flasks containing 50 mL of the fermentation medium with chloramphenicol (25 mg/L) and incubated at 37°C, 220 rpm. After 4~5 h incubation when the OD600 had reached 0.4~0.6, the cells were induced by adding 0.1 mM IPTG, and then the temperature was set to 33°C. Samples were taken at different time points during cultivation and centrifuged at 4°C at 12000 rpm for 5 min (Eppendorf Centrifage 5430). Then the supernatant was frozen at -20°C and prepared for analysis by HPLC, GC, and GC-MS. Cell densities were determined by spectrophotometer OD600 (Beckman coulter DU730, USA). Dry cell weight (g/L) was calculated using a conversion coefficient of 0.375 g/L/OD600.
Phenylpyruvate feeding experiments
To test HmaS conversion of phenylpyruvate to S-MA in vivo, feeding experiments were performed. The concentration gradient was as follows: 0 mM, 1 mM, 2 mM, 4 mM, and 8 mM. The phenylpyruvate was fed when the OD600 reached 0.4~0.6, meanwhile 0.1 mM IPTG was added to the cultures inducing expression of HmaS, and the fermentation temperature was adjusted from 37°C to 33°C for 24 h. Culture samples were analyzed by HPLC.
HPLC analysis
L-phenylalanine, phenylpyruvate, and S- or R-MA were quantified using a high-performance liquid chromatography system (Agilent Technologies 1,100 series) equipped with a Zorbax Eclipse XDB-C8 column (Agilent, 4.6 mm × 150 mm, 5 μm) at 40°C. The mobile phase consisted of 50 mM phosphate sodium buffer (pH 6.5) and methanol at a volume ratio of 95:5, at a flow rate of 1 mL/min, with UV detection was 215 nm. The pure substances were used as standards.
The S- or R-MA was identified by HPLC with a CHIRALCEL OD-H column (4.6 mm × 250 mm, 5 μm; Daicel Co., Japan) at 30°C. The mobile phase consisted of hexane, isopropanol, and trifluoroacetic acid in a volume ratio of 94:6:0.2, at a flow rate of 1.2 mL/min, and the UV detection wavelength was 228 nm. The pH of samples was adjusted to 1~2 using HCl, then S- or R-MA in the broth was extracted using an equal volume of ethyl acetate.
Glucose concentrations in the broth were determined by HPLC (Model 1200, Agilent) using a Sugar-Pak TM I column (Waters Corp., MA, USA), and the refractive index detector was used [36].
Standard samples were racemic mandelic acid ordered from Fluka, L-phenylalanine from Tokyo Chemical Industry (TCI, Shanghai, China), and phenylpyruvate (sodium), S-MA, and R-MA from Sigma.
GC analysis
The concentration of acetic acid in the supernatant was determined using gas chromatography (GC) (7890A, Agilent, Wilmington, DE, USA)[36].
GC-MS analysis
S- or R-MA was further identified by gas chromatography-mass spectrometry (GC-MS). Lyophilized samples were oximated with 20 mg/mL methoxyamine hydrochloride in pyridine at 30°C for 60 min. Then 80 μL pyridine and 20 μL N-methyl-N-[tert-butyldimethylsilyl] trifluoroacetamide were added to the samples for derivatization at 70°C for 30 min. Then 3 μL of derivatized sample were injected for GC-MS analysis after filtration. The GC-MS system (Agilent 6890-5973) was equipped with an HP-5MS column (30 m, 0.25 mm, 0.25 μm), and the electron impact (EI) mode of the mass spectrometer was set to 70 eV. The GC oven temperature was initially held at 60°C and raised with a gradient of 5°C per minute until it reached 180°C. It was then raised with a gradient of 10°C per minute until it reached 260°C. The flow rate of the mobile phase was set to 1 mL/min. All reagents used in this assay were ordered from Sigma.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SY conceived and supervised the research and revised the manuscript. ZTS designed and performed main experiments, carried out the data analysis and drafted the manuscript. YYN and YML participated in plasmid construction. LXL performed the GC-MS experiments. BBS contributed to enzyme assays. CY contributed to the revision of the manuscript and GC-MS data analysis. WHJ participated in the coordination of this study. All authors read and approved the final manuscript.
Supplementary Material
Chiral chromatography of the MA produced in vitro. (a) racemic mixture standards; (b) BL21(DE3) with pET24a as negative control; (c) HmaS stereoconversion of phenylpyruvate to S-MA; (d) BL21(DE3) with pTrc99a as negative control; (e) Hmo and DMD together transform S-MA to R-MA.
Chiral chromatography of the MA produced in vivo. (f) racemic mixture standards; (g) the fermentation broth of BCAE with pSUFAQ as negative control; (h) S-MA synthesized by strains BCAE harboring pSUFAAQ; (i) R-MA synthesized by strains BCAE containing pSUFAAQSD.
GC-MS analysis of the S-MA produced in vivo. S-MA in the broth was further verified by GC-MS: (a) the total ion chromatogram of S-MA; (b) the m/z profile of S-MA; (c) the standard m/z profile of MA from the GC-MS library.
GC-MS analysis of the R-MA produced in vivo. R-MA in the broth was further verified by GC-MS: (a) the total ion chromatogram of R-MA; (b) the m/z profile of R-MA; (c) the standard m/z profile of MA from the GC-MS library.
Cell densities, acetate and S-MA concentrations in LB medium with glucose. All single gene mutants and the control strain (BCAE) were cultured in LB medium supplemented with 20 g/L glucose for 48 h. All of the strains harbor the plasmid pSUFAAQ. ND: not detected.
Cell densities, acetate and S-MA concentrations in fermentation medium. The triple genes mutant and control strain (BCAE) were grown in fermentation medium for 48 h. Both of the strains harbor the plasmid pSUFAAQ. Data shown are means ± standard deviations, calculated from triplicate individual experiments.
Contributor Information
Zhoutong Sun, Email: ztsun@sibs.ac.cn.
Yuanyuan Ning, Email: yyning@cibt.ac.cn.
Lixia Liu, Email: lxliu@sibs.ac.cn.
Yingmiao Liu, Email: ymliu@cibt.ac.cn.
Bingbing Sun, Email: bbsun@cibt.ac.cn.
Weihong Jiang, Email: whjiang@sibs.ac.cn.
Chen Yang, Email: chenyang@sibs.ac.cn.
Sheng Yang, Email: syang@sibs.ac.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Funded No. 31170102) and the National Basic Research Program of China (973: 2011CBA00806).
The authors wish to express thanks to Professor Jianhe Xu, Dr. Jianping Wu and Dr. Bo Wang for their advices, and also thanks to Dr. Yinhua Lu for his improvement of the manuscripts, finally, thanks to Mr. Jun Chen, Mr. Zhijun Zhang, Mr. Liuyang Diao, Dr. Yuanheng Cai, and Dr. Dalong Zhang for their advices and assistances.
References
- Furlemmeier A, Quitt P, Vogler K, Lanz P. 6-Acyl derivatives of aminopenicillanic acid. US Patent. 1976;3:957–758. [Google Scholar]
- Chang TL, Teleshova N, Rapista A, Paluch M, Anderson RA, Waller DP, Zaneveld LJD, Granelli-Piperno A, Klotman ME. SAMMA, a mandelic acid condensation polymer, inhibits dendritic cell-mediated HIV transmission. FEBS Lett. 2007;581(24):4596–4602. doi: 10.1016/j.febslet.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M, Yu B, Wyatt V, Griffith J, Craft T, Neurath AR, Strick N, Li Y-Y, Wertz DL, Pojman JA, Lowe AB. Anti-HIV-1 Activity of Poly(mandelic acid) Derivatives. Biomacromolecules. 2007;8(11):3308–3316. doi: 10.1021/bm070221y. [DOI] [PubMed] [Google Scholar]
- Saravanan P, Singh VK. An efficient synthesis of chiral nonracemic diamines: application in asymmetric synthesis. Tetrahedron Lett. 1998;39:167–170. doi: 10.1016/S0040-4039(97)10578-0. [DOI] [Google Scholar]
- Bhushan R, Agarwal C. Direct enantiomeric TLC resolution of dl-penicillamine using (R)-mandelic acid and l-tartaric acid as chiral impregnating reagents and as chiral mobile phase additive. Biomed Chromatogr. 2008;22(11):1237–1242. doi: 10.1002/bmc.1052. [DOI] [PubMed] [Google Scholar]
- Whitesell JK, Reynolds D. Resolution of chiral alcohols with mandelic acid. J Org Chem. 1983;48:3548–3551. doi: 10.1021/jo00168a035. [DOI] [Google Scholar]
- Brittain HG. Analytical Profiles of Drug Substances and Excipients. Vol. 29. Academic Press; 2002. Mandelic acid; pp. 179–211. [Google Scholar]
- Gathergood N, Zhuang W, Jorgensen KA. Catalytic enantioselective Friedel-Crafts reactions of aromatic compounds with glyoxylate: a simple procedure for the synthesis of optically active aromatic mandelic acid esters. J Am Chem Soc. 2000;122(50):12517–12522. doi: 10.1021/ja002593j. [DOI] [Google Scholar]
- Nishiguchi N, Moritoki M, Shinohara T, Toyokura M. Separation of L-mandelic acid from asymmetric mixtures by means of high-pressure crystallization. JP Patent. 1997. p. 09066202.
- Mateo C, Chmura A, Rustler S, van Rantwijk F, Stolz A, Sheldon RA. Synthesis of enantiomerically pure (S)-mandelic acid using an oxynitrilase-nitrilase bienzymatic cascade: a nitrilase surprisingly shows nitrile hydratase activity. Tetrahedron: Asymmetry. 2006;17(3):320–323. doi: 10.1016/j.tetasy.2006.01.020. [DOI] [Google Scholar]
- Yamamoto K, Oishi K, Fujimatsu I, Komatsu K. Production of R-(-)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl Environ Microbiol. 1991;57(10):3028–3032. doi: 10.1128/aem.57.10.3028-3032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustler S, Motejadded H, Altenbuchner J, Stolz A. Simultaneous expression of an arylacetonitrilase from Pseudomonas fluorescens and a (S)-oxynitrilase from Manihot esculenta in Pichia pastoris for the synthesis of (S)-mandelic acid. Appl Microbiol Biotechnol. 2008;80(1):87–97. doi: 10.1007/s00253-008-1531-1. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Kaul P, Banerjee U. Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol. 2006;72(1):77–87. doi: 10.1007/s00253-005-0255-8. [DOI] [PubMed] [Google Scholar]
- He Y-C, Xu J-H, Pan J, Ouyang L-M, Xu Y. Preparation of (R)-(-)-mandelic acid and its derivatives from racemates by enantioselective degradation with a newly isolated bacterial strain Alcaligenes sp. ECU0401. Bioprocess Biosyst Eng. 2008;31(5):445–451. doi: 10.1007/s00449-007-0181-5. [DOI] [PubMed] [Google Scholar]
- Huang H-R, Xu J-H. Preparation of (S)-mandelic acid from racemate using growing cells of Pseudomonas putida ECU1009 with (R)-mandelate degradation activity. Biochem Eng J. 2006;30(1):11–15. doi: 10.1016/j.bej.2006.01.010. [DOI] [Google Scholar]
- Takahashi E, Nakamichi K, Furui M, Mori T. R-(-)-mandelic acid production from racemic mandelic acids by Pseudomonas polycolor with asymmetric degrading activity. J Ferment Bioeng. 1995;79(5):439–442. doi: 10.1016/0922-338X(95)91258-7. [DOI] [Google Scholar]
- Takahashi E, Nakamichi K, Furui M. R-(-)-mandelic acid production from racemic mandelic acids using Pseudomonas polycolor IFO 3918 and Micrococcus freudenreichii FERM-P 13221. J Ferment Bioeng. 1995;80(3):247–250. doi: 10.1016/0922-338X(95)90824-J. [DOI] [Google Scholar]
- Tsou AY, Ransom SC, Gerlt JA, Buechter DD, Babbitt PC, Kenyon GL. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990;29(42):9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- Muller U, van Assema F, Gunsior M, Orf S, Kremer S, Schipper D, Wagemans A, Townsend CA, Sonke T, Bovenberg R, Wubbolts M. Metabolic engineering of the E-coli L-phenylalanine pathway for the production of D-phenylglycine (D-Phg) Metab Eng. 2006;8(3):196–208. doi: 10.1016/j.ymben.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Rosli MI, Graem AR, Stephen KC, Charles AF, John SM. Molecular Cloning and Sequencing of D-mandelate Dehydrogenase Gene from Rhodotorula graminis. Pakistan J Biol Sci. 2002;5(8):871–877. [Google Scholar]
- Bongaerts J, Kramer M, Muller U, Raeven L, Wubbolts M. Metabolic Engineering for Microbial Production of Aromatic Amino Acids and Derived Compounds. Metab Eng. 2001;3(4):289–300. doi: 10.1006/mben.2001.0196. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Herrmann KM. Cloning of an aroF allele encoding a tyrosine-insensitive 3-deoxy-d-arabino-heptusonolate-7-phosphate synthase. J Bacteriol. 1990;172:6581–6584. doi: 10.1128/jb.172.11.6581-6584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms J, Edwards RM, Warwick J, Fotheringham I. Novel mutations in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Appl Environ Microbiol. 1992;58(8):2592–2598. doi: 10.1128/aem.58.8.2592-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey M, De Maeseneire S, Soetaert W, Vandamme E. Minimizing acetate formation in E-coli fermentations. J Ind Microbiol Biot. 2007;34(11):689–700. doi: 10.1007/s10295-007-0244-2. [DOI] [PubMed] [Google Scholar]
- He P, Conrad JA, Moran GR. The Rate-Limiting Catalytic Steps of Hydroxymandelate Synthase from Amycolatopsis orientalis. Biochemistry. 2010;49(9):1998–2007. doi: 10.1021/bi901674q. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kuramitsu S, Aki K, Watanabe Y, Takagi T, Nishigai M, Ikai A, Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: overproduction and properties. J Biochem. 1988;104(5):777–784. doi: 10.1093/oxfordjournals.jbchem.a122549. [DOI] [PubMed] [Google Scholar]
- Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24(11):530–536. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wong MS, Wu S, Causey TB, Bennett GN, San K-Y. Reduction of acetate accumulation in Escherichia coli cultures for increased recombinant protein production. Metab Eng. 2008;10(2):97–108. doi: 10.1016/j.ymben.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Mey MD, Lequeux GJ, Beauprez JJ, Maertens J, Horen EV, Soetaert WK, Vanrolleghem PA, Vandamme EJ. Comparison of Different Strategies to Reduce Acetate Formation in Escherichia coli. Biotechnol Prog. 2007;23(5):1053–1063. doi: 10.1021/bp070170g. [DOI] [PubMed] [Google Scholar]
- Ojima Y, Komaki M, Nishioka M, Iwatani S, Tsujimoto N, Taya M. Introduction of a stress-responsive gene, yggG, enhances the yield of l -phenylalanine with decreased acetic acid production in a recombinant Escherichia coli. Biotechnol Lett. 2009;31(4):525–530. doi: 10.1007/s10529-008-9906-z. [DOI] [PubMed] [Google Scholar]
- Kramer M, Bongaerts J, Bovenberg R, Kremer S, Muller U, Orf S, Wubbolts M, Raeven L. Metabolic engineering for microbial production of shikimic acid. Metab Eng. 2003;5(4):277–283. doi: 10.1016/j.ymben.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Kieser T, Chater KF. PCR targeting system in Streptomyces coelicolor A3(2) John Innes Centre. 2002.
- Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007;Chapter 1:Unit 1.17.11–11.17.18. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Ren C, Gu Y, Hu SY, Wu Y, Wang P, Yang YL, Yang C, Yang S, Jiang WH. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metabolic Engineering. 2010;12(5):446–454. doi: 10.1016/j.ymben.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Jia S-J, Chen D-j, Xu W-S. Development of DNA Transformation System for Amycolatopsis orientalis. Chin J Pharm. 2005;36(6):332–335. [Google Scholar]
- Kieser T, Bibb MJ, Chater K, Hopwood D. Norwich. The John Innes Foundation; 2000. Practical Streptomyces Genetics. [Google Scholar]
- Martinez E, Bartolomé B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68(1):159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chiral chromatography of the MA produced in vitro. (a) racemic mixture standards; (b) BL21(DE3) with pET24a as negative control; (c) HmaS stereoconversion of phenylpyruvate to S-MA; (d) BL21(DE3) with pTrc99a as negative control; (e) Hmo and DMD together transform S-MA to R-MA.
Chiral chromatography of the MA produced in vivo. (f) racemic mixture standards; (g) the fermentation broth of BCAE with pSUFAQ as negative control; (h) S-MA synthesized by strains BCAE harboring pSUFAAQ; (i) R-MA synthesized by strains BCAE containing pSUFAAQSD.
GC-MS analysis of the S-MA produced in vivo. S-MA in the broth was further verified by GC-MS: (a) the total ion chromatogram of S-MA; (b) the m/z profile of S-MA; (c) the standard m/z profile of MA from the GC-MS library.
GC-MS analysis of the R-MA produced in vivo. R-MA in the broth was further verified by GC-MS: (a) the total ion chromatogram of R-MA; (b) the m/z profile of R-MA; (c) the standard m/z profile of MA from the GC-MS library.
Cell densities, acetate and S-MA concentrations in LB medium with glucose. All single gene mutants and the control strain (BCAE) were cultured in LB medium supplemented with 20 g/L glucose for 48 h. All of the strains harbor the plasmid pSUFAAQ. ND: not detected.
Cell densities, acetate and S-MA concentrations in fermentation medium. The triple genes mutant and control strain (BCAE) were grown in fermentation medium for 48 h. Both of the strains harbor the plasmid pSUFAAQ. Data shown are means ± standard deviations, calculated from triplicate individual experiments.