The Wee1 kinase inhibits cyclin-dependent kinase 1 (Cdk1) during early mitosis. A low level of Cdk1 activity must escape Wee1 inhibition to initiate early mitotic events, but the underlying mechanisms have remained unknown. In this paper, we show that a specific form of protein phosphatase 2A opposes activation of Wee1, which allows low-level activation of Cdk1 in early mitosis.
Abstract
Entry into mitosis is initiated by synthesis of cyclins, which bind and activate cyclin-dependent kinase 1 (Cdk1). Cyclin synthesis is gradual, yet activation of Cdk1 occurs in a stepwise manner: a low level of Cdk1 activity is initially generated that triggers early mitotic events, which is followed by full activation of Cdk1. Little is known about how stepwise activation of Cdk1 is achieved. A key regulator of Cdk1 is the Wee1 kinase, which phosphorylates and inhibits Cdk1. Wee1 and Cdk1 show mutual regulation: Cdk1 phosphorylates Wee1, which activates Wee1 to inhibit Cdk1. Further phosphorylation events inactivate Wee1. We discovered that a specific form of protein phosphatase 2A (PP2ACdc55) opposes the initial phosphorylation of Wee1 by Cdk1. In vivo analysis, in vitro reconstitution, and mathematical modeling suggest that PP2ACdc55 sets a threshold that limits activation of Wee1, thereby allowing a low constant level of Cdk1 activity to escape Wee1 inhibition in early mitosis. These results define a new role for PP2ACdc55 and reveal a systems-level mechanism by which dynamically opposed kinase and phosphatase activities can modulate signal strength.
INTRODUCTION
Mitosis is an intricate and precisely ordered series of events that results in chromosome segregation and cell division. The key molecular event that initiates mitosis is activation of cyclin-dependent kinase 1 (Cdk1; Morgan, 2007). Cdk1 activation requires binding of mitotic cyclins, which are synthesized anew each cell cycle and accumulate gradually during entry into mitosis. In human somatic cells and Xenopus eggs, activation of Cdk1 occurs in a stepwise manner: a low constant level of Cdk1 activity is initially generated in early mitosis, which is followed by full, switch-like activation of Cdk1 (Solomon et al., 1990; Pomerening et al., 2005; Lindqvist et al., 2007; Deibler and Kirschner, 2010). Thus a gradually increasing signal generated by cyclin synthesis is converted to stepwise activation of Cdk1. This also appears to be true in budding yeast, where a low threshold of Cdk1 activity is required for initiation of mitotic spindle assembly and a higher threshold initiates spindle elongation (Rahal and Amon, 2008). Stepwise activation of Cdk1 helps ensure that mitotic events occur in the proper order (Deibler and Kirschner, 2010). More generally, the molecular mechanisms by which a gradually increasing signal is converted to a step function are of great interest.
The Wee1 kinase and the Cdc25 phosphatase play important roles in modulation of Cdk1 activity during entry into mitosis (Nurse, 1975; Russell and Nurse, 1986, 1987). Wee1 phosphorylates Cdk1 on a conserved tyrosine, thereby inhibiting Cdk1 and delaying entry into mitosis (Gould and Nurse, 1989). Cdc25 promotes entry into mitosis by removing the inhibitory phosphate (Gautier et al., 1991; Kumagai and Dunphy, 1991). Wee1 and Cdc25 are thought to contribute to the abrupt, switch-like activation of Cdk1 via feedback loops in which Cdk1 inhibits Wee1 and activates Cdc25 (Izumi et al., 1992; Kumagai and Dunphy, 1992; Hoffman et al., 1993; Izumi and Maller, 1993; Tang et al., 1993; Mueller et al., 1995). They also enforce cell cycle checkpoints that delay entry into mitosis until previous events have been completed (Rupes, 2002; Kellogg, 2003; Karlsson-Rosenthal and Millar, 2006; Keaton and Lew, 2006).
Recent work established that Wee1 plays an important role in the mechanism that generates a low plateau of Cdk1 activity in early mitosis. In vertebrate cells, low-level activation of Cdk1 in early mitosis occurs in the presence of active Wee1 and is dependent upon Wee1 (Deibler and Kirschner, 2010). These seemingly paradoxical findings suggest that Wee1 does not act in a simple all-or-nothing manner to inhibit Cdk1. Rather, it appears that the activity of Wee1 can be restrained to allow a low level of Cdk1 activity to be generated in early mitosis. A mechanism that restrains Wee1 also appears to operate in budding yeast because two key early mitotic events occur in the presence of active Wee1. First, a low level of Cdk1 activity in early mitosis initiates a positive feedback loop that promotes transcription of mitotic cyclins (Amon et al., 1993). Wee1 is present in excess when mitotic cyclins first accumulate and is capable of inhibiting both nuclear and cytoplasmic pools of Cdk1 (Sreenivasan and Kellogg, 1999; Harvey and Kellogg, 2003; Harvey et al., 2005; Keaton et al., 2008). Thus initiation of the positive feedback loop requires a mechanism by which Cdk1 can be activated in the presence of Wee1. Second, checkpoints that block nuclear division via activation of Wee1 cause an arrest at the short-spindle stage of early mitosis (Carroll et al., 1998; Sreenivasan and Kellogg, 1999; Theesfeld et al., 1999). Since formation of a short spindle requires a low level of mitotic Cdk1 activity, this further suggests that low-level activation of Cdk1 occurs in the presence of active Wee1 (Fitch et al., 1992; Rahal and Amon, 2008).
How is a low level of Cdk1 activity generated in early mitosis in the presence of active Wee1? Also, how is a low constant level of Cdk1 activity maintained in early mitosis as cyclin levels rise? Investigations into the mechanisms that regulate Wee1 and Cdk1 during entry into mitosis have provided clues. In budding yeast, Cdk1 associated with the mitotic cyclin Clb2 directly phosphorylates Wee1, which stimulates Wee1 to bind, phosphorylate, and inhibit Cdk1 (Harvey et al., 2005). Thus Cdk1 activates its own inhibitor. A similar mechanism acts in human cells, which suggests that it is conserved (Deibler and Kirschner, 2010). When Cdk1 activity is inhibited in vivo, Wee1 undergoes rapid, quantitative dephosphorylation, which reveals that phosphorylation of Wee1 by Cdk1 is opposed by a phosphatase (Harvey et al., 2005). After the initial phosphorylation and activation of Wee1 by Cdk1 in early mitosis, further phosphorylation events cause full hyperphosphorylation of Wee1 and dissociation of the Wee1-Cdk1/cyclin complex, which likely represents the inhibition of Wee1 that is necessary for full entry into mitosis (Harvey et al., 2005; Deibler and Kirschner, 2010).
We used these findings as a starting point to investigate how activation of Cdk1 is controlled during entry into mitosis. This led to the discovery that PP2ACdc55 opposes the initial phosphorylation and activation of Wee1 by Cdk1/Clb2 in early mitosis. In vitro reconstitution and mathematical modeling indicate that PP2ACdc55 sets a threshold that limits activation of Wee1 in early mitosis, thereby allowing a low level of Cdk1/Clb2 activity to escape Wee1 inhibition and initiate early mitotic events. Quantitative analysis and mathematical modeling of Wee1 activation further suggest the existence of a systems-level mechanism that keeps the activity of Cdk1/Clb2 at a low constant level as cyclin levels rise during early mitosis.
RESULTS
Cdc28/Clb2 phosphorylates Swe1 on Cdc28 consensus sites
We first carried out an analysis of the mechanism by which mitotic Cdk1/cyclin stimulates activation of Wee1. In budding yeast, Wee1 is referred to as Swe1 and Cdk1 is referred to Cdc28. In previous work, we reconstituted phosphorylation of Swe1 by Cdc28/Clb2 in vitro and used mass spectrometry to map phosphorylation sites (Harvey et al., 2005). We found that Cdc28/Clb2 phosphorylated Swe1 on eight of 13 possible minimal Cdc28 (Cdk1) consensus sites (S/T-P), as well as on 10 nonconsensus sites. Most sites were also identified on Swe1 isolated from cells, suggesting that they are physiologically relevant. A swe1-18A mutant that lacked these sites eliminated Swe1 hyperphosphorylation in vivo and caused premature entry into mitosis. In addition, the mutant protein failed to form a complex with Cdc28/Clb2. These observations suggested a model in which initial phosphorylation of Swe1 by Cdc28/Clb2 leads to formation of a Swe1-Cdc28/Clb2 complex that drives inhibitory phosphorylation of Cdc28/Clb2.
A limitation of these phosphorylation site-mapping experiments is that they were unable to provide information on the stoichiometry of phosphorylation. Thus it was possible that they identified sites phosphorylated at stoichiometries too low to be significant. To determine which of the 18 sites played a significant role in Swe1 activation by Cdc28/Clb2, we used a quantitative method called stable isotope labeling with amino acids in culture (SILAC; Ong et al., 2002). This approach allows estimation of the stoichiometry of phosphorylation via use of heavy isotope–labeled, unphosphorylated reference peptides.
We again reconstituted phosphorylation of Swe1 by Cdc28/Clb2 in vitro (Figure 1A). Unphosphorylated and phosphorylated Swe1 were mixed with isotope-labeled unphosphorylated Swe1, and the accumulation of phosphorylated peptides and depletion of the cognate unphosphorylated peptides were monitored. The relative abundance ratios of the unphosphorylated peptides upon phosphorylation were used to calculate site stoichiometry. We performed two independent analyses using peptides generated by digesting Swe1 with trypsin or lysC.
FIGURE 1:
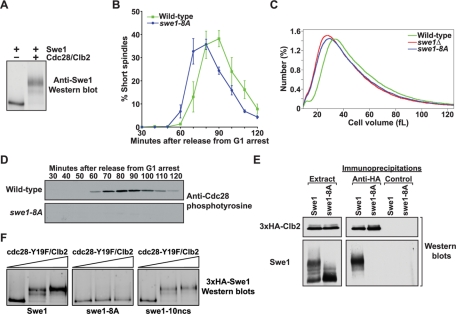
Phosphorylation of Cdc28 consensus sites is necessary for Swe1 activity in vivo. (A) Purified Cdc28/Clb2 quantitatively phosphorylates purified Swe1 in vitro. Phosphorylation of Swe1 causes an electrophoretic mobility shift that is detected by Western blot analysis. (B) Wild-type and swe1-8A cells were released from a G1 arrest, and the percentage of cells with short mitotic spindles was determined at the indicated times and plotted as a function of time. Error bars represent SEM for three independent experiments. (C) Cell size distributions of log-phase cultures of wild-type, swe1Δ, and swe1-8A strains. Each trace is the average of 15 independent cultures. (D) Cells of the indicated genotypes were released from a G1 arrest, and samples were taken at the indicated times. Cdc28 phosphotyrosine was monitored by Western blotting. (E) Wild-type and swe1-8A cells carrying GAL1-3×HA-CLB2 were grown to midlog phase, and expression of 3×HA-Clb2 was induced with 2% galactose for 3 h at 30°C. Extracts were made, and 3×HA-Clb2 was immunoprecipitated with an anti-HA antibody. As a control, identical precipitations were carried out using an anti-GST antibody. Coprecipitation of Swe1 was assayed by Western blotting. The panels labeled “Extract” show Western blots of 3×HA-Clb2 and Swe1 in the crude extracts used for the immunoprecipitations. (F) Purified 3×HA-Swe1, 3×HA-swe1-8A, or 3×HA-swe1-10ncs were incubated with increasing amounts of purified cdc28-Y19F/Clb2-3×HA in the presence of ATP. Phosphorylation of Swe1 was detected as an electrophoretic mobility shift on a Western blot.
Standard site mapping identified many sites, including most of the sites identified in our previous analysis (Supplemental Table S1; Harvey et al., 2005). However, SILAC revealed that the only sites phosphorylated at a measurable stoichiometry were Cdc28 consensus sites (Table 1). An exception was a phosphorylation that occurred within amino acids 159–162, which likely represents Swe1 autophosphorylation on a tyrosine. In two cases, peptides with multiple sites underwent quantitative phosphorylation; however, the SILAC analysis indicates only that these peptides were phosphorylated at high stoichiometry on one or more sites. In these cases, we believe that it is likely that only the Cdc28 consensus sites were phosphorylated at high stoichiometry.
TABLE 1:
Estimated fractional phosphorylation occupancy of key sites in Swe1.a
| Fractional occupancy (%) | |||
|---|---|---|---|
| Site | Local sequence | LysC | Trypsin |
| T45 | IGGSTPTNK | 86 | 88 |
| S111, T121b | IKRWSPFHENESVTTPITK | 95 | 94 |
| S133 | EKTNSPISL | 97 | 97 |
| [159–162] | TSSS[SSYS]VAK | 55 | 62 |
| T196 | RIPETPVKK | 98 | 99 |
| S201 | PVKKSPLVE | 49 | ND |
| S263 | TIDSSPLSE | 97 | 97 |
| T280, S284, [286–297]b | HNNQTNILSP[TNSLVTNSSPQT]LHSN | 96 | 97 |
| T373 | EEISTPTRR | 96 | ND |
| S418 | TDSSSPLNSK | ND | 94 |
aFor peptides with multiple phosphorylation sites, the fractional occupancy indicates the fraction of peptides phosphorylated on one or more sites; the fractional occupancy of individual sites could not be determined. Swe1 was purified from yeast grown in the presence of 13C stably, isotopically labeled lysine (“heavy”) or natural lysine (“light”) and phosphatase treated. Light Swe1 was incubated in a mock reaction without added kinase or with Cdk1/Clb2. The light Swe1 from each reaction was then mixed with heavy Swe1 that had been treated with phosphatase, and the mixtures were resolved by SDS–PAGE. Coomassie Blue–stained bands were excised, proteolysed, and examined by liquid chromatography–mass spectrometry/mass spectrometry on an LTQ-Orbitrap mass spectrometer (Thermo-Fisher Scientific, San Jose, CA), and matched to peptide sequences using SEQUEST software (Thermo-Fisher Scientific). Abundance ratios of unphosphorylated peptides (heavy:light [H:L]) were automatically calculated using Vista (Bakalarski et al., 2008). As peptides became phosphorylated, the pool of light unphosphorylated peptides was depleted and the H:L ratio increased. Using ratios for peptides that lack serine, threonine, and tyrosine as a baseline, we calculated the phosphorylated fraction for each peptide as FPhos = 1 − FUn/FPep, where FPhos = fraction phosphorylated, FUn = average H:L ratio of unphosphorylatable peptides in each experiment, and FPep = average H:L ratio of each unphosphorylated peptide. Phosphorylation sites for each peptide were inferred from the appearance of peptides bearing these sites in the light fraction upon phosphorylation with Cdk1/Clb2. Confidently localized sites are shown bold and underlined. Where we were unable to determine precise site localization, brackets indicate the range of possible assignments. LysC-digested and trypsin-digested samples represent distinct biological replicates of the experiments. ND, not determined.
bThe stoichiometry of these sites could not be assessed independently. Reported data indicate the fraction of peptides phosphorylated on one or more sites.
Phosphorylation of Cdc28 consensus sites is required for Swe1 activity in vivo
SILAC analysis demonstrated that the initial phosphorylation of Swe1 by Cdc28/Clb2 occurs primarily on Cdc28 consensus sites, which suggested that phosphorylation of consensus sites alone drives activation of Swe1 by Cdc28/Clb2. To test whether Cdc28 consensus sites are required for Swe1 activity, we analyzed a mutant version of Swe1 that lacked eight of the Cdc28 consensus sites identified by mass spectrometry (swe1-8A, sites mutated: T45A, S111A, T121A, S133A, T196A, S201A, S263A, and T373A). We previously found that the swe1-8A mutant largely eliminated phosphorylation of Swe1 in vivo (Harvey et al., 2005). Moreover, previous work established that loss of Swe1 causes premature entry into mitosis and reduced cell size (Jorgensen et al., 2002; Harvey and Kellogg, 2003; Harvey et al., 2005; Rahal and Amon, 2008). In this study, we found that swe1-8A caused premature entry into mitosis, as detected by assembly of short mitotic spindles (Figure 1B). The swe1-8A mutant also caused reduced cell size and a complete loss of Cdc28 inhibitory phosphorylation (Figure 1, C– D). Finally, the swe1-8A protein failed to form a complex with Cdc28/Clb2 in crude extracts (Figure 1E). The sites mutated in swe1-8A were located in the N-terminus, far from the kinase domain, and mutation of these sites in the swe1-18A mutant did not affect the intrinsic kinase activity of Swe1 (Harvey et al., 2005). A swe1-13A mutant that lacked all 13 minimal Cdc28 consensus sites also caused premature entry into mitosis and a reduced cell size (unpublished data).
We next tested whether the swe1-8A protein was phosphorylated by Cdc28/Clb2 in vitro. We utilized a mutant form of Cdc28 that cannot be phosphorylated by Swe1 (cdc28-Y19F), which allowed us to carry out reactions without the complication of Cdc28 inhibition by Swe1. The swe1-8A protein was resistant to phosphorylation by purified cdc28-Y19F/Clb2 (Figure 1F). In contrast, a swe1-10ncs protein that lacked the nonconsensus sites identified in our original study was phosphorylated to the same extent as wild-type Swe1, which supported the conclusion that initial phosphorylation of Swe1 by Cdc28/Clb2 occurs on Cdc28 consensus sites (Figure 1F).
These observations are consistent with a model in which Cdc28/Clb2 activates Swe1 and drives formation of the Swe1-Cdc28/Clb2 complex via phosphorylation of Cdc28 consensus sites. Note that only a quantitatively phosphorylated form of Swe1 forms a complex with Cdc28/Clb2 in wild-type cells (Figure 1E and Harvey et al., 2005). Based on its electrophoretic mobility, this form is likely to correspond to the form of Swe1 that has been quantitatively phosphorylated by Cdc28/Clb2 on consensus sites, which suggests that Swe1 must be quantitatively phosphorylated to bind and inhibit Cdc28.
To further investigate the mechanism and function of consensus site phosphorylation, we created two additional phosphorylation site mutants in which a smaller number of sites were mutated. In the first mutant, we mutated two sites that correspond to optimal Cdc28 consensus sites (S/TPXXR/K; swe1-T196A,T373A). In the second mutant, we mutated two sites that correspond to the minimal Cdc28 consensus site (S/TP; swe1-S133A,S263A). In both cases, we chose sites that were quantitatively phosphorylated by Cdc28/Clb2 in the mass spectrometry analysis. There are numerous other phosphorylation site mutants that would be interesting to analyze in this context; these mutants were selected to provide a preliminary analysis of the roles of different classes of sites and to test whether phosphorylation of all sites is necessary for full Swe1 activity. For both mutants, we analyzed cell size, Cdc28 inhibitory phosphorylation, Swe1 phosphorylation, and formation of the Swe1-Cdc28/Clb2 complex (Figure 2).
FIGURE 2:
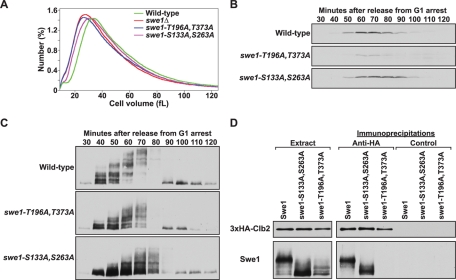
Analysis of Swe1 phosphorylation site mutants. (A) Cell size distributions of log-phase cultures of wild-type, swe1Δ, swe1-T196A,T373A, and swe1-S133A,S263A. Each trace is the average of 15 independent cultures. The wild-type and swe1Δ traces plotted in Figure 1C and panel A are identical. (B and C) Cells of the indicated genotypes were released from a G1 arrest into 30°C YPDA medium and samples were taken at the indicated times. The behavior of (B) Cdc28 phosphotyrosine and (C) Swe1 was assayed by Western blotting. (D) Wild-type, swe1-S133A,S263A, and swe1-T196A,T373A cells carrying GAL1-3×HA-CLB2 were grown to midlog phase and expression of 3×HA-Clb2 was induced with 2% galactose for 3 h at 30°C. Extracts were made and 3×HA-Clb2 was immunoprecipitated with an anti-HA antibody. As a control, identical precipitations were carried out using an anti-GST antibody. Coprecipitation of Swe1 was assayed by Western blotting.
Mutation of the optimal Cdc28 consensus sites (swe1-T196A,T373A) caused a small-size phenotype similar to a swe1Δ and a severe reduction in Cdc28 inhibitory phosphorylation, consistent with loss of Swe1 function (Figure 2, A and B). Intermediate phosphorylation forms of the swe1-T196A,T373A protein could be detected, but appeared to be reduced (Figure 2C, compare 50-min time points). The swe1-T196A,T373A protein completely failed to form a complex with Cdc28/Clb2 (Figure 2D).
Mutation of the minimal Cdc28 consensus sites (swe1-S133A,S263A) caused reduced cell size (Figure 2A), as well as delayed Cdc28 inhibitory phosphorylation (Figure 2B; compare 50- to 60-min time points in wild-type cells to the same time points in swe1-S133A,S263A cells). It also appeared to cause a reduction in Swe1 phosphorylation: most of the swe1-S133A,S263A protein was present in a rapidly migrating form at early time points (Figure 2C; compare 50- to 60-min time points). Phosphorylated forms of swe1-S133A, S263A eventually appear, but only at later times, when Cdc28/Clb2 accumulates to higher levels. The swe1-S133A,S263A protein was capable of forming a complex with Cdc28/Clb2.
The behavior of the swe1-S133A,S263A and swe1-T196A,T373A mutants indicates that the consensus sites are not all equivalent in terms of function. A model that could explain the behavior of the mutants is that phosphorylation occurs in an ordered and cooperative manner. In this model, phosphorylation of S133 or S263 would occur first, promoting subsequent phosphorylation events. Phosphorylation of T196 and T373 would occur last, stimulating Swe1 activity by promoting formation of the Swe1-Cdc28/Clb2 complex. This model further postulates that phosphorylation of T196 and T373 eventually occurs in the swe1-S133A,S263A mutant, but inefficiently, and therefore only after Cdc28/Clb2 activity has risen to higher levels. Thus activation of Swe1 would be delayed in the swe1-S133A,S263A mutant, which would explain the observed premature accumulation of active Cdc28 and reduced cell size. This model is consistent with the observation that only the quantitatively phosphorylated form of wild-type Swe1 binds to Cdc28/Clb2 (Figure 1E; Harvey et al., 2005).
Previous work on the fibroblast growth factor receptor found that it undergoes multi-site phosphorylation in a sequential and precisely ordered manner, which provides a precedent for a sequential mechanism of Swe1 phosphorylation (Furdui et al., 2006). However, extensive additional experiments will be required to clearly define the mechanism of multi-site phosphorylation of Swe1. Although the behavior of the Swe1 phosphorylation site mutants suggests the presence of cooperativity, these observations must be interpreted with caution because phosphorylation has an unpredictable effect on gel mobility: multiple phosphorylations may show no gel shift, while a single phosphorylation may show a large one. Thus it is not possible to know definitively which sites or how many sites have been phosphorylated based on the electrophoretic mobility of a protein. Quantitative mass spectrometry will be necessary to fully characterize the effects of the mutations on Swe1 phosphorylation.
PP2ACdc55 opposes phosphorylation of Swe1 by Cdc28 in vivo
We next used a candidate approach to search for the phosphatase that opposes phosphorylation of Swe1 by Cdc28. We reasoned that inactivation of the phosphatase should cause premature phosphorylation of Swe1. PP2ACdc55 was a strong candidate because evidence from diverse systems has indicated it is a key regulator of entry into mitosis and Cdc28 inhibitory phosphorylation. Canonical PP2A is a heterotrimeric complex composed of a catalytic subunit, a scaffolding subunit, and a regulatory subunit. Cells express multiple regulatory subunits that associate with PP2A in a mutually exclusive manner. In budding yeast, PP2A associates primarily with two regulatory subunits called Rts1 and Cdc55, forming distinct complexes called PP2ARts1 and PP2ACdc55, respectively. Inactivation of the catalytic subunit after cells have passed through G1 phase causes defects in entry into mitosis; cells arrest with low levels of mitotic cyclin mRNA and protein and reduced specific activity of mitotic Cdc28 (Lin and Arndt, 1995). Similarly, cdc55Δ causes increased Cdc28 inhibitory phosphorylation, delayed accumulation of mitotic cyclin, and a prolonged delay in early mitosis (Minshull et al., 1996; Yang et al., 2000; Pal et al., 2008). These phenotypes are rescued by swe1Δ or cdc28-Y19F (Yang et al., 2000; Pal et al., 2008). Conversely, PP2ACdc55 mutants are enhanced by inactivation of Mih1, the budding yeast homologue of Cdc25 (Lin and Arndt, 1995; Pal et al., 2008). For example, mih1Δ causes lethality in cdc55Δ cells, which is rescued by swe1Δ. Together, these observations show that PP2ACdc55 promotes entry into mitosis via regulation of Cdc28 inhibitory phosphorylation. Xenopus PP2A also controls entry into mitosis, and is thought to regulate Wee1 and Cdc25 (Félix et al., 1990; Kumagai and Dunphy, 1992; Tang et al., 1993; Izumi and Maller, 1995; Mueller et al., 1995; Castilho et al., 2009; Mochida et al., 2009).
To test whether PP2ACdc55 opposes phosphorylation of Swe1, we assayed Swe1 hyperphosphorylation in synchronized cdc55Δ cdc28-Y19F cells. The cdc28-Y19F allele eliminated the mitotic delay caused by cdc55Δ, which allowed cell cycle synchronization. We found that Swe1 prematurely shifted to a partially phosphorylated form in cdc55Δ cdc28-Y19F cells (Figure 3A). Moreover, there was a loss of hypophosphorylated forms of Swe1 in cdc55Δ cdc28-Y19F cells (Figure 3A). The cdc28-Y19F mutant alone did not cause premature hyperphosphorylation of Swe1 (Figure 3A). In cdc55Δ cells, Swe1 was present at all time points due to poor cell cycle synchrony, and was constitutively in the partially phosphorylated form (Supplemental Figure S1A). Swe1 did not undergo premature hyperphosphorylation in rts1Δ cells, which indicated that PP2ARts1 does not play a role in opposing the initial phosphorylation of Swe1 (Figure S1B). Swe1 failed to undergo full hyperphosphorylation in rts1Δ cells, which suggested that PP2ARts1 plays a role in events required for full hyperphosphorylation of Swe1.
FIGURE 3:
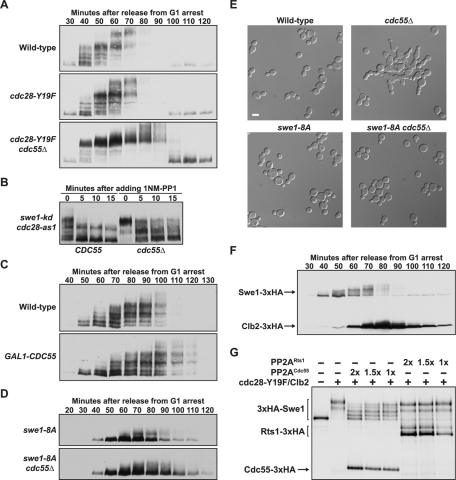
PP2ACdc55 opposes the initial phosphorylation of Swe1 by Cdc28. (A) Cells of the indicated genotypes were released from G1 arrest, and the behavior of Swe1 was assayed by Western blotting. The strains used for this experiment were generated in the A364A background. Similar results were obtained in the W303-1A strain background. (B) 1NM-PP1 was added to cdc28-as1 swe1-kd or cdc28-as1 swe1-kd cdc55Δ cells 65 min after release from G1 arrest. Swe1 phosphorylation was assayed by Western blotting. (C) Cells were grown overnight in YEP medium containing 2% raffinose, and then arrested with α-factor at room temperature. After a 2-h incubation in the presence of α-factor, 2% galactose was added to initiate expression of GAL1-CDC55, and the incubation was continued for an additional 1 h. Cells were released from the arrest into galactose-containing YEP medium, and the behavior of Swe1 was assayed by Western blotting. (D) Cells of the indicated genotypes were released from G1 arrest into 30°C YPD medium, and the behavior of Swe1 was assayed by Western blotting. (E) Cells of the indicated genotypes were grown overnight to log phase at room temperature in YPD medium and imaged by differential interference contrast microscopy. Scale bar: 5 μm. (F) Cells carrying Clb2-3×HA and Swe1-3×HA were released from a G1 arrest into 30°C YPD medium, and the behavior of Clb2 and Swe1 was monitored on the same Western blot. (G) The indicated combinations of purified protein complexes were incubated in the presence of ATP and purified dephosphorylated Swe1 for 20 min, and then analyzed by Western blotting. All of the 3×HA-tagged proteins were detected on the same blot with an anti-HA antibody. Three different concentrations of PP2ACdc55 or PP2ARts1 that varied over a twofold range (indicated as 1×, 1.5×, and 2×) were added to the reactions.
To further investigate the role of PP2ACdc55, we tested whether PP2ACdc55 was required for dephosphorylation of Swe1 in vivo after inhibition of an analogue-sensitive allele of Cdc28 (cdc28-as1). We were unable to recover a cdc28-as1 cdc55Δ double mutant, but we were able to recover the double mutant in the presence of a kinase-dead allele of SWE1 (swe1-kd) We synchronized cdc28-as1 swe1-kd and cdc28-as1 swe1-kd cdc55Δ cells and inhibited cdc28-as1 as cells were entering mitosis. Inhibition of cdc28-as1 in the control cells caused rapid and nearly complete loss of swe1-kd phosphorylation (Figure 3B). Inhibition of cdc28-as1 in cdc55Δ cells caused a partial dephosphorylation of swe1-kd. The partial dephosphorylation of swe1-kd could be due to another phosphatase that makes a minor contribution, or to the nonspecific activity of multiple phosphatases. We further found that overexpression of Cdc55 from the GAL1 promoter caused delayed phosphorylation of Swe1 (Figure 3C, compare 60- to 70-min time points in each strain). These data, combined with the fact that cdc55Δ causes nearly quantitative hyperphosphorylation of Swe1 in vivo, indicate that PP2ACdc55 is the primary phosphatase that acts on Swe1 during early mitosis.
The swe1-8A mutant failed to undergo premature hyperphosphorylation in cdc55Δ cells, which demonstrated that PP2ACdc55 opposes phosphorylation of the Cdc28 consensus sites (Figure 3D). Moreover, swe1-8A rescued the elongated cell morphology caused by cdc55Δ, providing further evidence that PP2ACdc55 targets the consensus sites (Figure 3E; 62% of cdc55Δ cells were elongated, whereas no elongated cells were detected in cdc55Δ swe1-8A cells). Full hyperphosphorylation of Swe1 was delayed in cdc55Δ cdc28-Y19F cells, which indicated that PP2ACdc55 controls additional events required for full hyperphosphorylation of Swe1 later in mitosis (Figure 3A).
PP2ACdc55 can effectively oppose phosphorylation of Swe1 by Cdc28/Clb2 in vitro
To determine whether PP2ACdc55 is likely to act directly on Swe1, we used in vitro reconstitution to test whether PP2ACdc55 can effectively oppose phosphorylation of Swe1 by Cdc28/Clb2. We used immunoaffinity chromatography with specific peptide elution to purify triple hemagglutinin-Swe1 (3×HA-Swe1), cdc28-Y19F/Clb2-3×HA, and PP2ACdc55-3×HA. Purifications were carried out in the presence of 1 M KCl to remove peripherally associated proteins. Examples of the purified proteins are shown in Figure S2. We used the cdc28-Y19F mutant to create a simplified system in which Swe1 cannot phosphorylate and inhibit Cdc28, which allowed us to focus on phosphorylation of Swe1.
To ensure that reactions recapitulated conditions during early mitosis, we used Western blotting to assess relative levels of Clb2 and Swe1 during the cell cycle. Clb2 and Swe1 were tagged with 3×HA at their endogenous loci in the same strain, and levels of both proteins were assessed simultaneously in synchronized cells (Figure 3F). This revealed that Swe1 is present in excess during early mitosis, when Clb2 is first accumulating, and that Clb2 levels eventually exceed Swe1 levels. Note that the partially hyperphosphorylated form of Swe1 appears at low Clb2 levels in early mitosis, and that full hyperphosphorylation of Swe1 only occurs when Clb2 levels exceed Swe1 levels. Samples were electrophoresed for a short time to keep both proteins on the gel, so the resolution of Swe1 isoforms was reduced compared with Figure 3A. A similar approach revealed that Cdc55 is present in excess of both Swe1 and Clb2 (unpublished data).
Reactions were initiated that contained Swe1, cdc28-Y19F/Clb2, and increasing amounts of PP2ACdc55. The reactions were allowed to proceed for 20 min and an anti-HA antibody was used to detect phosphorylated forms of Swe1 by shifts in electrophoretic mobility (Figure 3G). The antibody also detected Clb2-3×HA and Cdc55-3×HA, which allowed assessment of the relative amounts of each protein. Clb2-3×HA levels were just below detection, and Swe1 was present in excess of Clb2, as in early mitosis. In the absence of PP2ACdc55, Swe1 was quantitatively phosphorylated by cdc28-Y19F/Clb2. Added PP2ACdc55 effectively opposed phosphorylation of Swe1. Note that under these conditions, with very low concentrations of cdc28-Y19F/Clb2 and excess Swe1, the extensive, full hyperphosphorylation of Swe1 by cdc28-Y19F/Clb2 we previously reported was not observed (Harvey et al., 2005). PP2ARts1 purified under identical conditions failed to oppose phosphorylation of Swe1 by Cdc28/Clb2, which suggests Cdc55 confers specific functions upon PP2A (Figure 3G).
Inactivation of PP2ACdc55 causes a Swe1-dependent delay in accumulation of mitotic cyclin
We carried out in vivo experiments to further analyze the role of PP2ACdc55 in entry into mitosis. Previous work found that cdc55Δ caused a large increase in Cdc28 inhibitory phosphorylation and delayed accumulation of Clb2 (Minshull et al., 1996; Yang et al., 2000; Pal et al., 2008). However, these studies were carried out using cdc55Δ cells. In contrast to the cdc55Δ cdc28-Y19F cells used for the experiments in Figure 3, cdc55Δ cells have significant cell cycle defects and rapidly accumulate suppressors, which makes it difficult to clearly define the immediate effects of Cdc55 inactivation. We therefore generated a temperature-sensitive allele of CDC55 (cdc55-4), which allowed us to determine the immediate effects of inactivating Cdc55. DNA sequencing identified a single point mutation (C875T) in the cdc55-4 sequence compared with the wild-type CDC55 sequence, which results in conversion of threonine 292 to methionine. Wild-type, cdc55-4, and cdc55-4 swe1Δ cells were released from a G1 arrest at the restrictive temperature, and formation of mitotic spindles was assayed, as were levels of Clb2 protein (Figure 4, A and B). The cdc55-4 mutant caused a prolonged delay in accumulation of Clb2 and formation of short mitotic spindles. The delay was eliminated by swe1Δ, which indicated the delay was due to a failure to properly regulate Cdc28 inhibitory phosphorylation. The cdc55-4 mutant caused a 10-min delay in bud emergence, but this could not account for the 30-min delay in formation of short mitotic spindles (Figure S3).
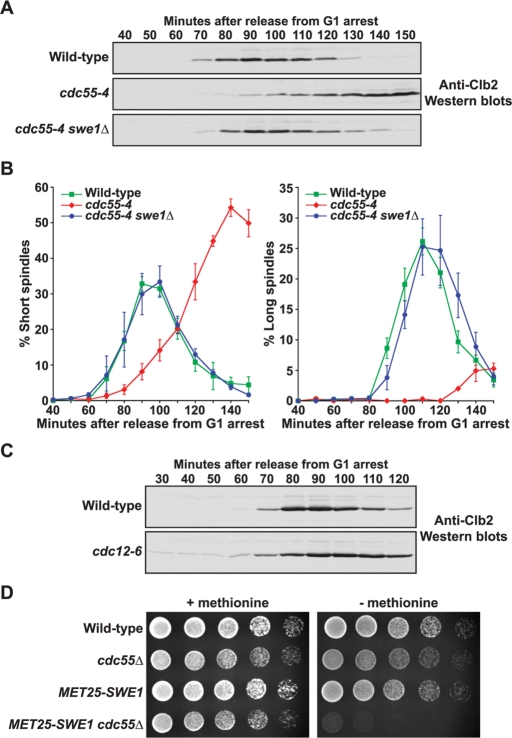
FIGURE 4:
Inactivation of PP2ACdc55 causes a Swe1-dependent delay in Clb2 accumulation. (A and B) Cells of the indicated genotypes were released from G1 arrest into 35°C YPD medium. Samples were taken every 10 min at the indicated times. (A) Clb2 protein levels were monitored by Western blotting. (B) The percentage of cells with short or long spindles was determined and plotted as a function of time. Error bars represent SEM for three independent experiments. (C) Cells of the indicated genotypes were grown overnight to log phase, and then arrested with α-factor for 4 h at 20°C. Cells were released from the G1 arrest into 37°C YPD medium. Samples were taken every 10 min at the indicated times, and Clb2 protein levels were monitored by Western blotting. (D) Fivefold serial dilutions of cells of the indicated genotypes were grown on minimal media plates containing 2% dextrose in the presence or absence of 20 μg/ml l-methionine at 30°C. The MET25 promoter is induced in the absence of l-methionine.
The cdc55-4 mutant also caused a delay in formation of long mitotic spindles that was much greater than the delay in formation of short spindles. The delay was largely eliminated by swe1Δ, which demonstrated that PP2ACdc55-dependent regulation of Cdc28 inhibitory phosphorylation also plays a role in events required for the normal timing of spindle elongation.
A potential explanation for the delay in early mitotic events is that inactivation of Cdc55 causes defects in morphogenesis that activate a Swe1-dependent checkpoint. However, previous work suggested that activation of the morphogenesis checkpoint does not cause a significant delay in accumulation of Clb2 (Carroll et al., 1998). To further test this, we used a conditional septin mutant (cdc12-6) to activate the morphogenesis checkpoint. We detected a slight delay in accumulation of Clb2 protein in cdc12-6 mutants; however, the delay was not comparable to the prolonged delay caused by cdc55-4 (Figure 4C).
A threshold model for the role of PP2ACdc55 in regulation of Swe1 and Cdc28
In previous work, it was hypothesized that inactivation of PP2A catalytic subunits causes reduced specific activity of Cdc28/Clb2, thereby disrupting a positive feedback loop in which Cdc28/Clb2 promotes Clb2 transcription (Amon et al., 1993; Lin and Arndt, 1995). Our results provide strong support for this hypothesis: inactivation of Cdc55 caused a prolonged delay in accumulation of Clb2. Moreover, Clb2 levels rose abruptly in wild-type cells but more gradually in cdc55-4 cells, consistent with a failure in the positive feedback loop (Figure 4A). Our analysis of Swe1 regulation suggests an explanation for these observations. We found that Cdc28/Clb2 phosphorylates and activates Swe1, and that activation of Swe1 is opposed by PP2ACdc55. Thus inactivation of PP2ACdc55 would be expected to result in hyperactive Swe1, which would phosphorylate and inhibit Cdc28/Clb2, leading to reduced specific activity of Cdc28/Clb2 and a failure in the positive feedback loop. In this model, PP2ACdc55 creates a threshold that limits activation of Swe1 by Cdc28/Clb2, thereby allowing a low level of Cdc28/Clb2 activity to accumulate in early mitosis. The low level of Cdc28/Clb2 activity triggers the positive feedback loop that promotes Clb2 transcription without triggering full entry into mitosis. When Cdc28/Clb2 activity rises above the threshold, Swe1 would be activated to bring Cdc28/Clb2 activity back below the threshold, thus maintaining a low level of Cdc28/Clb2 activity in early mitosis. Swe1 is present in excess when mitotic cyclins first accumulate and is capable of inhibiting both nuclear and cytoplasmic pools of Cdc28 (Keaton et al., 2008). Thus a mechanism for restraining Swe1 is necessary for initiation of the positive feedback loop. Cdc55 is localized to the nucleus and the cytoplasm, so it is present where activation of Swe1 must be restrained (Gentry and Hallberg, 2002).
Swe1 is hyperactive in cells that lack PP2ACdc55
We used a combination of in vivo analysis, quantitative analysis, and mathematical modeling to test the threshold model for PP2ACdc55 function. If PP2ACdc55 creates a threshold that limits activation of Swe1 by Cdc28/Clb2, one would predict that Swe1 is hyperactive in cells that lack PP2ACdc55. We tested this by determining whether cdc55Δ cells are sensitive to increased levels of Swe1. To do this, we induced overexpression of Swe1 from the MET25 promoter in wild-type and cdc55Δ cells. Expression of Swe1 in wild-type cells did not affect the rate of colony formation. In contrast, expression of Swe1 in cdc55Δ cells caused lethality, consistent with the idea that Swe1 is hyperactive in the absence of PP2ACdc55, which leads to mitotic arrest (Figure 4D).
The phosphorylation state of Swe1 represents a steady state
We next carried out further analysis of the reconstituted system. The threshold model suggests that phosphorylation of Swe1 will approach a steady state determined by the relative activities of Cdc28/Clb2 and PP2ACdc55. In vivo analysis is consistent with this: acute inhibition of Cdc28 causes rapid dephosphorylation of Swe1, and inactivation of PP2ACdc55 causes hyperphosphorylation of Swe1 (Figure 3 and Harvey et al., 2005). To determine whether the reconstituted system recapitulates steady-state regulation of Swe1, we set up reactions containing Swe1, PP2ACdc55, and two different concentrations of cdc28-Y19F/Clb2. The reactions were initiated by addition of ATP, and samples were taken at intervals to determine the time required to approach steady state (Figure 5A). Swe1 phosphorylation approached steady state within 10–15 min in the presence of both low and high levels of cdc28-Y19F/Clb2. This is a biologically relevant timescale, because Clb2 accumulation in vivo occurs over a period of 40 min at the temperature used to carry out these reactions.
FIGURE 5:
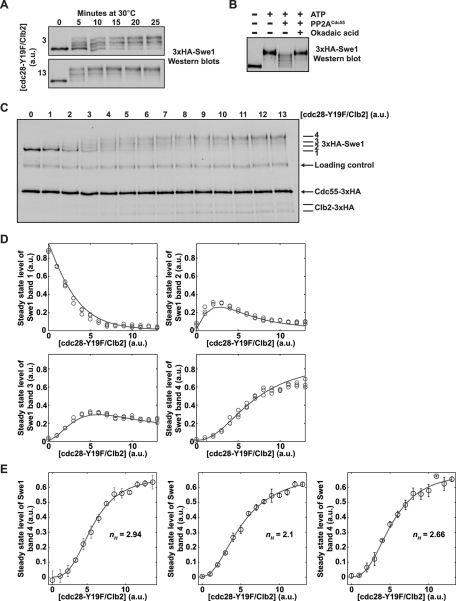
The response of Swe1 to rising levels of cdc28-Y19F/Clb2 is ultrasensitive. (A) Reactions containing Swe1, PP2ACdc55, and two concentrations of cdc28-Y19F/Clb2 were initiated by addition of ATP. Samples were taken at the indicated times, and the phosphorylation state of Swe1 was assayed by Western blotting. The amounts of cdc28-Y19F/Clb2 were chosen to demonstrate that the reactions carried out in (C) approach steady state within the reaction time at both low and high concentrations of cdc28-Y19F/Clb2 (a.u., arbitrary units). (B) Reactions containing Swe1 and cdc28-Y19F/Clb2 were incubated for 15 min in the presence or absence of ATP and PP2ACdc55 (lanes 1–3). Samples were taken from each reaction, and okadaic acid was then added to the reaction in lane 3, which was incubated for an additional 10 min before taking a sample (lane 4). Swe1 phosphorylation was assayed by Western blotting. (C) Phosphorylation of Swe1 (bands labeled 1–4) as a function of cdc28-Y19F/Clb2 concentration in the presence of a constant amount of PP2ACdc55. Reactions were allowed to proceed for 20 min. Alexa Fluor 647–labeled phosphorylase B was used as a loading control. Proteins were assayed by quantitative Western blotting using a monoclonal HA antibody to detect each 3×HA-tagged protein. (D) Quantitation of the amount of each Swe1 band generated in the reactions shown in (C). Reactions were analyzed in triplicate; circles represent data from each of the three replicates. Each graph also shows a line that represents the solution of the pipeline model described in the Supplemental Information. (E) Swe1 shows an ultrasensitive response to rising levels of cdc28-Y19F/Clb2. We repeated the analysis using three independent preparations of purified cdc28-Y19F/Clb2 and two independent preparations of PP2ACdc55 to create three biological replicates of the steady-state analysis, and each biological replicate was measured in triplicate. The graphs show the average quantitation of the amount of Swe1 band 4 generated as a function of cdc28-Y19F/Clb2 concentration for each biological replicate. Error bars represent SD. In each graph, a Hill function was fit through least-squares approximation, and the Hill coefficients (nH) are displayed.
To further establish that the reactions were in dynamic opposition, a reaction was allowed to proceed for 15 min and okadaic acid was then added to inhibit PP2ACdc55. This caused rapid hyperphosphorylation of Swe1, which confirmed that phosphorylation and dephosphorylation were occurring in dynamic opposition (Figure 5B).
Quantitative analysis reveals an ultrasensitive response of Swe1 to rising levels of Cdc28/Clb2
Diverse experimental observations support the threshold model. However, a system that includes opposing kinase and phosphatase activities and multi-site phosphorylation can exhibit complex behaviors, and multiple distinct behaviors are possible. To determine which of the possible behaviors is exhibited by the system, one must use quantitative analysis and fitting of experimental data to mathematical models. We therefore analyzed the reconstituted system, as well as the individual phosphorylation and dephosphorylation reactions, with the goal of determining whether the system was capable of generating the behavior postulated by the model.
We first analyzed phosphorylation of Swe1 in steady-state reactions containing Swe1, PP2ACdc55, and increasing amounts of cdc28-Y19F/Clb2. We used fluorescent dye–based Western blotting with an anti-HA antibody to detect phosphoforms of Swe1, as well as the HA-tagged Cdc55 and Clb2 added to the reactions (Figure 5C). The amounts of added Cdc28/Clb2 were set to be very low, and Swe1 levels were set to be in excess, to mimic the conditions during early mitosis. Comparison of the Clb2 signal in the in vitro reactions with the Clb2 signal in crude extracts indicated the amount of cdc28-Y19F/Clb2 added to the reactions was significantly lower than the peak level of Cdc28/Clb2 reached in vivo.
Four forms of Swe1 could be detected in the presence of increasing amounts of cdc28-Y19F/Clb2. These are labeled in Figure 5C as bands 1–4. Based on its electrophoretic mobility, band 4 corresponds to the form of Swe1 that is quantitatively phosphorylated on Cdc28 consensus sites. Because phosphorylation has an unpredictable effect on gel mobility, each of the lower bands may correspond to multiple forms of phosphorylated Swe1. However, the bands showed decreased electrophoretic mobility with increasing Cdc28/Clb2, indicating that they corresponded to increasingly phosphorylated forms of Swe1.
All four forms of Swe1 were quantitated in samples run in triplicate and plotted as a function of increasing amounts of cdc28-Y19F/Clb2 (Figure 5D, circles show data from three experimental replicates). Analysis of the data for three independent experiments utilizing different preparations of cdc28-Y19F/Clb2 and PP2ACdc55 revealed that formation of the most phosphorylated form of Swe1 (band 4) at steady state was nonlinear with respect to increasing amounts of cdc28-Y19F/Clb2. This kind of nonlinearity is also referred to as ultrasensitivity, which can be quantified by fitting the data to a Hill function. The corresponding Hill coefficient provides a quantitative measure of the ultrasensitivity. The Hill coefficient is sensitive to noise in the data, so we repeated the analysis using three independent preparations of purified cdc28-Y19F/Clb2 and two independent preparations of PP2ACdc55 and found the average Hill coefficient to be 2.56 ± 0.43, which indicates an ultrasensitive response (Figure 5E; each panel shows the results of an independent experiment).
We also carried out experiments to analyze phosphorylation of Swe1 as a function of increasing cdc28-Y19F/Clb2 in the absence of PP2ACdc55. Reactions without PP2ACdc55 will go to completion (i.e., complete phosphorylation of Swe1), and therefore do not represent a steady-state system. Thus we stopped the reactions after an arbitrary time to capture Swe1 in a range of intermediate phosphorylation states. We carried out three independent repetitions of the experiment, using the same protein preparations used for the experiments in Figure 5E, and measured Hill coefficients of 1.9, 2.0, and 2.3 (Figure S4). The fact that these reactions produced a Hill coefficient of greater than 1 suggests that there may be ultrasensitivity that is intrinsic to phosphorylation of Swe1 by Cdc28. However, these results must be treated with caution for several reasons. First, if one repeated the experiment and stopped the reactions after different amounts of time, the Swe1 dose–response would look different. Therefore the ultrasensitivity of the dose–response in the absence of PP2ACdc55 is transient and can vary over time. In particular, as time increases sufficiently for the phosphorylation reaction to go to completion, the dose–response becomes constant, and the Hill coefficient loses its practical meaning. Since the ultrasensitivity observed in the absence of PP2ACdc55 is transient, it is a different type of phenomenon and cannot be compared with that observed for steady-state dose–responses in the presence of PP2ACdc55. A related consideration is that PP2ACdc55 may enforce a different order of phosphorylation if it dephosphorylates different sites on Swe1 with different efficiencies. In this case, phosphorylation of Swe1 in the presence or absence of PP2ACdc55 would represent fundamentally different reactions.
The ultrasensitive response of Swe1 to rising levels of Cdc28/Clb2 may be due to multi-step effects
Multiple mechanisms can generate ultrasensitivity, including zero-order effects, substrate competition, and multi-step effects (Goldbeter and Koshland, 1981; Gunawardena, 2005; Kim and Ferrell, 2007). Multi-step effects can arise when a kinase or phosphatase acts distributively to modify multiple sites. An enzyme that acts distributively requires more “steps” to complete the task, which leads to multi-step effects that can give rise to limited ultrasensitivity (Gunawardena, 2005). Note that distributivity says nothing about the order in which an enzyme modifies sites; a distributive enzyme may modify sites in a specific order or in a random order.
The presence of four Swe1 bands in steady-state reactions indicated that at least four distinct phosphoforms of Swe1 were generated by largely distributive phosphorylation, since a completely processive mechanism would generate only two bands (i.e., unphosphorylated and fully phosphorylated Swe1). This suggested that the observed ultrasensitivity may arise predominantly from multi-step effects. To test this, we modeled the system as a phosphorylation pipeline with four stages corresponding to the four bands in the gel, and assumed purely linear rates for phosphorylation and dephosphorylation (details provided in the Supplemental Information). To constrain the data further, we combined the steady-state data in Figure 5, C and D, with data obtained by assaying the rate of phosphorylation of Swe1 by cdc28-Y19F/Clb2 in the absence of PP2ACdc55, and the rate of dephosphorylation of phosphorylated Swe1 by PP2ACdc55 in the absence of cdc28-Y19F/Clb2 activity, using the same preparations of cdc28-Y19F/Clb2 and PP2ACdc55 that were used for the steady-state reactions shown in Figure 5, C and D (Figure S5, A and B, circles represent data from three replicates). We found linear rate constants that provided an excellent fit to the steady-state data (represented by lines in the graphs in Figure 5D). Since we did not assume any nonlinearity in the model, which could give rise to zero-order effects or other forms of amplification, this suggests that the observed ultrasensitivity could be accounted for entirely by multi-step effects. The data for the individual phosphorylation and dephosphorylation reactions were also fitted (Figure S5, A and B, lines represent the behavior predicted by the model). These data were also reasonably well reproduced by the model, although there was a slight systematic error in at least one of the graphs, which points to the limitations of describing this enzymatic system with a simple linear-rate model involving only six variables. We conclude nevertheless that the model is a surprisingly accurate qualitative and quantitative representation of Swe1 phosphorylation. The linear pipeline model therefore provides a simple abstraction that largely accounts for the data, as well as a useful framework for carrying out further experiments to investigate the mechanisms underlying Swe1 phosphorylation. We emphasize that the model is not intended to provide a final or conclusive description of the mechanism of Swe1 phosphorylation. Moreover, our analysis thus far does not rule out the possibility that additional factors, such as cooperative phosphorylation of Swe1, contribute to the observed ultrasensitivity.
Interestingly, the model predicts that the threshold value (EC50) of Cdc28/Clb2 increases proportionally as the concentration of PP2ACdc55 is increased (Figure S6 and Supplemental Information). With higher concentrations of PP2ACdc55, a higher level of active Cdc28/Clb2 is needed to counter it, which suggests that PP2ACdc55 can tune the level of Swe1 activation by Cdc28/Clb2.
Ultrasensitive activation of Swe1 allows generation of a stable low level of Cdc28 activity in early mitosis
How does ultrasensitivity contribute to the mechanisms that regulate Swe1 and Cdc28? In principle, the system could have evolved to generate a more simple linear response of Swe1 to increasing Cdc28/Clb2 activity. Thus it seemed likely that the ultrasensitivity plays an important role in control of early mitotic events. We therefore used modeling to test how ultrasensitivity could affect the behavior of the system in its biological context. To do this, we followed a strategy widely adopted in engineering of “opening the loop” and considering separately the steady-state response of Cdc28/Clb2 to Swe1 and the steady-state response of Swe1 to Cdc28/Clb2 (details provided in the Supplemental Information). We then put the separate responses back together, thereby closing the loop, and in this way determined the system behavior.
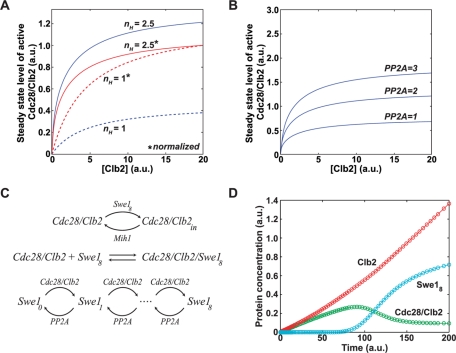
This analysis suggested that an ultrasensitive response of Swe1 to Cdc28/Clb2 helps generate a stable level of active Cdc28/Clb2 during early mitosis, despite rising levels of Clb2. This can be seen in Figure 6A, in which we plot steady-state levels of Cdc28/Clb2 activity as a function of Clb2 levels. The red lines show plots in which the final level of Cdc28/Clb2 activity was normalized to 1 to emphasize the different shapes of the curves. The blue lines show plots that were not normalized, which indicate the different levels of Cdc28/Clb2 activity achieved under each condition. In the absence of ultrasensitivity (nH = 1), the levels of Cdc28/Clb2 activity rise in a nearly linear manner. In contrast, an ultrasensitive response of Swe1 to Cdc28/Clb2 produces an initial rapid rise in Cdc28/Clb2 activity that then begins to level off (nH = 2.5). Thus the ultrasensitivity may allow the system to rapidly establish and maintain a low level of Cdc28/Clb2 activity that can initiate early mitotic events, while preventing a further rise in Cdc28/Clb2 activity that would trigger full entry into mitosis.
FIGURE 6:
Systems-level regulation of Cdc28 and Swe1 by PP2ACdc55 in early mitosis. (A) The steady-state activity of Cdc28/Clb2 was calculated by intersecting the dose–response curves in Figure S7, A–B, and the resulting values were plotted for different Clb2 concentrations. The solid and dotted lines were determined using the solid and dotted lines in Figure S7A (see Supplemental Information for details, including parameter values; a.u., arbitrary units). The red lines show plots in which the final level of Cdc28/Clb2 activity was normalized to 1 to emphasize the different shapes of the curves. The blue lines show plots that were not normalized, which indicate the different levels of Cdc28/Clb2 activity achieved under each condition. (B) Using the response curve for nH = 2.5 in (A), the same graph as in (A) is plotted for increasing PP2ACdc55 concentrations. (C and D) A detailed quantitative model of the interactions among Swe1, Clb2, Cdc28, and PP2ACdc55 during entry into mitosis. (C) The chemical reactions that define the network, using mass-action terms for each interaction. (D) A sample time course of the model, set in motion by a steadily increasing Clb2 concentration. The red line shows rising Clb2 levels, the green line shows Cdc28/Clb2 activity, and the blue line shows Swe1 activity.
We also carried out an analysis of the effects of varying PP2ACdc55 activity on the system. This showed that the level of active Cdc28/Clb2 is strongly influenced by levels of PP2ACdc55 activity, which provided further evidence that PP2ACdc55 can tune the system to control the level of Cdc28/Clb2 activity (Figure 6B). This implies that regulation of PP2ACdc55 could be an important mechanism for controlling levels of active Cdc28/Clb2 during early mitosis.
A quantitative model for systems-level regulation of Cdc28 and Swe1
We next generated a mechanistically detailed mathematical model to test whether signaling interactions known to occur during mitosis are capable of generating a system that exhibits the proposed threshold behavior. We based the model on the chemical reactions described schematically in Figure 6C. Details of the model are provided in the Supplemental Information.
A sample simulation of the model, which is set in motion by the introduction of steadily increasing levels of Clb2, is shown in Figure 6D. The system has several dozen variables; however, for simplicity, only three key variables are displayed in the simulation: levels of Clb2, active Cdc28/Clb2, and active Swe1. Initially, Cdc28/Clb2 is activated since active Swe1 is unavailable. After a delay in which the intermediate states of Swe1 are being formed, active Swe1 is produced, which then inactivates Cdc28/Clb2 through phosphorylation, leading to a decrease in Cdc28/Clb2 activity. At this time, a basal level of Cdc28/Clb2 activity is reached and maintained. At steady state, the Cdc28/Clb2 dimer is undergoing cycles of inhibition by Swe1 and activation by Mih1, which leads to a low basal level of Cdc28/Clb2 activity. Previous work has shown that a low basal activity of Mih1/Cdc25 is present during interphase and early mitosis, which would allow for the reversibility of Cdc28 inhibition (Kumagai and Dunphy, 1992; Harvey et al., 2005). In Xenopus, the low basal activity of Cdc25 is stimulated fivefold during entry into mitosis, which is thought to help trigger full activation of Cdc28 (Kumagai and Dunphy, 1992).
Importantly, the model shows that the opposing activity of PP2ACdc55 can act as a threshold to generate a dynamic steady state in which a low level of Cdc28/Clb2 activity is generated and maintained in early mitosis in the presence of Swe1, despite rising Clb2 levels.
DISCUSSION
Measurement of mitotic Cdk1 activity in vertebrate cells has shown that a low plateau level of Cdk1 activity is generated in early mitosis, which is followed by switch-like, full activation of Cdk1 later in mitosis (Pomerening et al., 2005; Lindqvist et al., 2007; Deibler and Kirschner, 2010). In budding yeast, low-level activation of Cdc28 in early mitosis initiates mitotic spindle assembly, as well as a feedback loop that promotes transcription of mitotic cyclins (Amon et al., 1993; Rahal and Amon, 2008). Low-level activation of Cdk1 in early mitosis therefore appears to be a conserved mitotic control mechanism. A mechanism that keeps Cdk1 activity low as cyclin levels rise would allow the cell to accumulate inactive Cdk1/cyclin complexes, which could be rapidly activated via posttranslational modifications to trigger full entry into mitosis. Low-level activation of Cdk1 in the presence of Wee1 would also allow the cell to become poised for cell division once checkpoint conditions have been satisfied. Here we define a systems-level mechanism by which a low level of Cdk1 activity can be generated and maintained in early mitosis, despite the presence of the Wee1 inhibitor and constantly rising cyclin levels.
Cdc28-dependent regulation of Swe1 during early mitosis
Quantitative mass spectrometry revealed that Cdc28/Clb2 phosphorylates Swe1 on a set of minimal Cdc28 consensus sites. Analysis of a swe1-8A mutant that lacked consensus sites showed that they are required for Swe1 activity in vivo and for formation of a Swe1-Cdc28/Clb2 complex that likely maintains Cdc28 in the inhibited state in vivo. Together, these observations suggest that phosphorylation of Cdc28 consensus sites alone drives stimulation of Swe1 activity.
Standard mass spectrometry showed that Cdc28/Clb2 phosphorylated Swe1 on both nonconsensus sites and consensus sites. However, the nonconsensus sites were phosphorylated at stoichiometries that were too low to be measured. Phosphorylation of these sites likely represents inefficient phosphorylation events that occur before Swe1 inhibits Cdc28/Clb2. The detection of these events emphasizes the high sensitivity of mass spectrometry, as well as the importance of using quantitative approaches to reveal which sites are phosphorylated at significant stoichiometries.
PP2ACdc55 restrains activation of Swe1 by Cdc28
Previous studies found that inactivation of budding yeast PP2ACdc55 causes increased inhibitory phosphorylation of Cdc28 and delayed mitotic progression; however, the precise roles of PP2ACdc55 were unclear (Lin and Arndt, 1995; Minshull et al., 1996; Yang et al., 2000; Pal et al., 2008). We discovered that an important function of PP2ACdc55 in budding yeast is to oppose phosphorylation of Swe1 by Cdc28/Clb2. Since initial phosphorylation of Swe1 by Cdc28/Clb2 activates Swe1, we propose that PP2ACdc55 sets a threshold that limits activation of Swe1 during early mitosis, thereby allowing accumulation of a low level of active Cdc28/Clb2 that can initiate the positive feedback loop that promotes transcription of mitotic cyclins (Amon et al., 1993). Diverse in vivo experiments support this model. For example, inactivation of Cdc28 in vivo caused rapid dephosphorylation of Swe1 (Harvey et al., 2005). Conversely, inactivation of PP2ACdc55 caused premature phosphorylation of Swe1, and a mutant form of Swe1 that lacked Cdc28 consensus sites did not undergo premature phosphorylation when PP2ACdc55 was inactivated. Overexpression of Cdc55 caused delayed phosphorylation of Swe1. Together, these observations demonstrate that PP2ACdc55 opposes the initial activating phosphorylation of Swe1 that occurs on consensus sites. Mild overexpression of Swe1 was lethal in cdc55Δ cells, consistent with the idea that PP2ACdc55 opposes activation of Swe1. In addition, inactivation of Cdc55 caused a prolonged delay in accumulation of Clb2, which is consistent with a failure in the positive feedback loop that promotes transcription of CLB2. Deletion of SWE1 rescued the delay, which established that it was due to inappropriate inhibition of Cdc28/Clb2 by Swe1. Cells that lack PP2ACdc55 may eventually enter mitosis when enough Clb2 accumulates via basal transcription to overcome the inhibitory activity of Swe1 without the help of PP2ACdc55.
In vitro reconstitution provided further support for the model. Cdc28/Clb2 was able to efficiently phosphorylate Swe1 under the conditions that prevail in early mitosis, when there are low levels of Cdc28/Clb2 and Swe1 is in excess. Moreover, purified PP2ACdc55 could effectively oppose phosphorylation of Swe1 by Cdc28/Clb2. Swe1 phosphorylation in the reconstituted system approached steady state on a timescale that was relevant in vivo.
In addition to causing defects in early mitotic events, inactivation of the cdc55-4 allele caused a prolonged delay in spindle elongation. Thus PP2ACdc55 must also control events occurring later in mitosis that are required for the normal timing of spindle elongation. The nature of the these PP2ACdc55-dependent events is unknown; however, the fact that swe1Δ almost completely eliminated the delay in spindle elongation indicates that PP2ACdc55 exerts its effects on mitotic progression primarily via control of Cdc28 inhibitory phosphorylation. Since a high level of Cdc28 activity is required for spindle elongation, the delay may be due to delayed accumulation of active Cdc28 in the presence of hyperactive Swe1 (Rahal and Amon, 2008). Previous work found that inactivation of PP2ACdc55 causes premature exit from a mitotic arrest. This phenotype is independent of Cdc28 inhibitory phosphorylation, which indicates an additional role for PP2ACdc55 in late mitosis (Minshull et al., 1996; Queralt et al., 2006; Tang and Wang, 2006; Chiroli et al., 2007).
In previous work, we discovered that Mih1 is dephosphorylated during entry into mitosis by a form of PP2ACdc55 that is tightly associated with a pair of redundant regulatory subunits called Zds1 and Zds2 (Pal et al., 2008; Wicky et al., 2010). The Zds1/2 proteins are not required for dephosphorylation of Swe1, which suggests they specifically target PP2ACdc55 to Mih1. Although the functional significance of Mih1 dephosphorylation is unclear, these results, when combined with the results reported here, indicate that PP2ACdc55 regulates the phosphorylation states of both Swe1 and Mih1. Importantly, cdc55Δ is lethal in mih1Δ cells and the lethality is rescued by swe1Δ (Pal et al., 2008). Since the effects of deleting CDC55 in mih1Δ cells cannot be due to misregulation of Mih1, this observation argues that PP2ACdc55 plays a direct role in inhibition of Swe1.
Systems-level control of Cdc28 activity during entry into mitosis
We used quantitative analysis and mathematical modeling to test the PP2ACdc55 threshold model and to investigate the systems-level mechanisms by which PP2ACdc55 controls Swe1 and Cdc28. These approaches revealed that the most phosphorylated form of Swe1 was generated in an ultrasensitive manner in response to rising levels of cdc28-Y19F/Clb2 in the reconstituted system. Systems-level modeling further suggested that the biological function of the ultrasensitivity in the system is to allow a relatively stable level of Cdc28/Clb2 activity to be rapidly generated and maintained in early mitosis despite rising Clb2 levels. The data from the reconstituted system could be fitted to a simple mathematical model, in which Cdc28/Clb2 and PP2ACdc55 act in the linear range to move Swe1 through a series of isoforms in a phosphorylation “pipeline.” The linear pipeline model provides what is perhaps the simplest abstract description of the system that could account for the data and is intended as a guide for future investigations into the underlying mechanisms, rather than as a definitive description of the system. Although the model demonstrated that ultrasensitivity could be accounted for by multi-step effects caused by multi-site phosphorylation of Swe1, our results thus far do not rule out the possibility that cooperativity or other factors contribute to the observed ultrasensitivity in multi-site Swe1 phosphorylation.
We also generated a mechanistically detailed systems-level model to test whether a system based on known signaling interactions in mitosis can generate the behavior postulated by the threshold model. This analysis provided an important proof-of-principle test of the PP2ACdc55 threshold model.
Quantitative analysis, modeling, and analysis of phosphorylation site mutants suggest that multi-site phosphorylation of Swe1 by Cdc28/Clb2 may occur in an ordered and distributive manner. However, rigorous experimental verification of an ordered phosphorylation mechanism will require direct analysis of the phosphoforms that are produced at specific times during reactions, which may be achievable in the future via mass spectrometry. Combined experimental and mathematical analysis is an iterative process. Our analysis thus far provides a first iteration and suggests new experimental tests that will lead to a more refined understanding of the system.
Numerous studies in vertebrate cells and yeast have detected stepwise activation of Cdk1 during entry into mitosis (Pomerening et al., 2005; Lindqvist et al., 2007; Rahal and Amon, 2008; Deibler and Kirschner, 2010). In contrast, a recent study utilizing an in vivo biosensor detected a steady rise in mitotic Cdk1 activity in vertebrate cells (Gavet and Pines, 2010). The biosensor measured Cdk1 activity via phosphorylation of a Cdk1 consensus site in an engineered protein, which triggered a phosphorylation-dependent binding event that resulted in a fluorescence resonance energy transfer signal. Chemical inhibition of Cdk1 caused a slow loss of the biosensor signal over a period of 25 min, which indicated that the biosensor has low temporal resolution. It is unclear whether a phosphatase acts efficiently on the biosensor phosphosite, so the slow loss of the signal could be due to cyclin destruction. The low temporal resolution of the biosensor may preclude detection of dynamic Cdk1 regulation. For example, if Cdk1 phosphorylates the biosensor and is then inhibited by Wee1, the low temporal resolution of the biosensor likely means the biosensor signal would persist long after Cdk1 is inhibited by Wee1. Thus the biosensor may not detect dynamic Wee1-dependent mechanisms that keep Cdk1 activity low in early mitosis.
In vertebrate cells, it has been proposed that Cdk1/cyclin A turns down Wee1 activity to allow low-level activation of Cdk1/cyclin B in early mitosis (Deibler and Kirschner, 2010). In budding yeast, the Clb3 cyclin appears before Clb2 during entry into mitosis, which suggests that it could turn down Swe1 activity. If this were true, one would predict that deletion of the CLB3 gene should cause a phenotype consistent with hyperactivity of Swe1. However, deletion of the CLB3 gene causes no obvious phenotype (Fitch et al., 1992). We therefore favor the PP2ACdc55 threshold model as a mechanism to restrain Swe1 activity in early mitosis. PP2A also plays a crucial role in controlling mitosis in vertebrate cells, so it could play a conserved role in restraining the activity of Wee1 family members. However, additional experiments will be necessary to test the cyclin A hypothesis and the role of PP2A in controlling Wee1 activity in vertebrates.
Phosphorylation of Wee1 has also been analyzed in Xenopus extracts (Kim and Ferrell, 2007). Increasing amounts of Cdk1/cyclin B1 were added to interphase egg extracts, which were incubated for 1 h to allow the system to reach steady state. Phosphorylation of Wee1 was measured using a phosphospecific antibody that recognized a single Cdk1 consensus site. These experiments measured a Hill coefficient of 3.5 for phosphorylation of the site in crude extracts. In contrast, the Hill coefficient for phosphorylation of the site by purified cdk1-AF/cyclin B1 showed a lower Hill coefficient of 1.5. Further work suggested a model in which high concentrations of Cdk1 substrates act as competitive inhibitors in vivo to restrain phosphorylation of Wee1 by Cdk1 and to generate ultrasensitivity. An important difference between Xenopus egg Wee1 and Swe1 is that phosphorylation of Cdk1 consensus sites on Wee1 is thought to inactivate Wee1, although it has not yet been possible to replace endogenous Xenopus egg Wee1 with a phosphorylation site mutant to test this model (Kim et al., 2005). The difference could be due to the presence of unique phosphorylation sites on Swe1 that function to stimulate Swe1 activity. A similar module could be present in human somatic Wee1, since it is also thought to undergo initial activation by Cdk1 (Deibler and Kirschner, 2010). Currently, it is difficult to compare regulation of Wee1 in yeast and Xenopus, due to their evolutionary divergence and the significant technical differences in how experimental analysis has been carried out.
Theoretical analysis has shown that opposing kinase and phosphatase activities could play an important role in generating diverse and powerful signaling behaviors (Goldbeter and Koshland, 1981, 1982, 1984; Ferrell, 1996; Thomson and Gunawardena, 2009). In a number of cases, experimental analysis has provided support for this idea (LaPorte and Koshland, 1983; Nakajima et al., 2005). Often, however, phosphatases are thought of as playing relatively simple roles, such as serving to reverse the activated state of a signaling pathway once the original stimulus has been removed, or blocking phosphorylation of proteins to inhibit signal transduction. Analysis of the mechanisms that control Cdc28 activity in early mitosis suggests that opposing kinase and phosphatase activities are capable of generating a dynamic systems-level mechanism for modulation of signal strength. Similar mechanisms may be used to modulate signal strength in other signaling networks.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions
The strains, plasmids, and culture conditions used for this study are listed in the Supplemental Information. For time-course experiments, cells were grown in YPD or YPDA medium (1% yeast extract, 2% peptone, 2% dextrose, with or without 0.004% adenine sulfate) overnight at room temperature prior to synchronization. For induced expression experiments, cells were grown overnight in YEP medium (1% yeast extract, 2% peptone, 0.004% adenine sulfate) containing 2% glycerol/2% ethanol or 2% raffinose and then induced with 2% galactose.
Western blotting
For quantitative Western blotting, proteins were transferred to Immobilon-FL (Millipore, Billerica, MA) polyvinylidene fluoride membranes and probed with mouse monoclonal HA antibody followed by Alexa Fluor 647 donkey anti–mouse secondary antibody. Blots were scanned using a Typhoon 9410 Variable Mode Imager (Amersham Biosciences, GE Healthcare, Waukesha, WI), and the results were quantified with Odyssey version 2.1.12 (LI-COR Biosciences, Lincoln, NE) and MATLAB software (Mathworks, Natick, MA).
Protein purifications
Immunoaffinity purifications of Cdc28/Clb2-3×HA, cdc28-Y19F/Clb2-3×HA, 3×HA-Swe1, 3×HA-swe1-8A, 3×HA-swe1-10ncs, PP2ACdc55-3×HA, and PP2ARts1-3×HA were carried out in the presence of 1 M KCl as previously described, with some modifications (Harvey et al., 2005). Detailed methods for purification of proteins, including growth conditions for SILAC, are provided in the Supplemental Information.
SILAC analysis
Swe1 labeled with heavy isotope was made by expressing 3×HA-Swe1 from the GAL1 promoter in lys1Δ cells in medium containing l-lysine-13C6,15N2 hydrochloride as the sole source of lysine. The protein was purified by immunoaffinity chromatography and treated with λ phosphatase to create an unphosphorylated reference protein. The isotope-labeled 3×HA-Swe1 was mixed with unphosphorylated or phosphorylated 3×HA-Swe1, and the stoichiometry of phosphorylation was determined by monitoring the depletion of unphosphorylated protein. Standard site mapping was also carried out to identify all phosphorylated sites, including those not phosphorylated at significant stoichiometries.
Coimmunoprecipitation
Association of Cdc28/3×HA-Clb2 and Swe1, swe1-8A, swe1-S133A,S263A, or swe1-T196A,T373A in crude extracts was assayed as described previously (Harvey et al., 2005).
In vitro assays
See Supplemental Information for detailed reaction conditions.
Generation of Cdc55 temperature-sensitive alleles
Error-prone PCR was used to generate temperature-sensitive alleles of CDC55. DNA sequencing identified a single point mutation (C875T) in the cdc55-4 sequence that converts threonine 292 to methionine.
Supplementary Material
Acknowledgments
We thank David Morgan, Mart Loog, Seth Rubin, Eva Murdock, Adam Rudner, Derek McCusker, Christopher Carroll, and Needhi Bhalla for critical reading of the manuscript and for helpful discussion. We thank Tracy MacDonough for help with strain construction and the members of the laboratory for their generous support and many helpful discussions. This work was supported by a grant from the National Institutes of Health (GM-069602).
Abbreviations used:
- Cdk1
cyclin-dependent kinase 1
- HA
hemagglutinin
- PP2A
protein phosphatase 2A
- SILAC
stable isotope labeling with amino acids in culture
- YEP
yeast extract, peptone, adenine sulfate
- YPD/A
yeast extract, peptone, dextrose with/without adenine sulfate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0340) on August 17, 2011.
REFERENCES
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Bakalarski CE, Elias JE, Villén J, Haas W, Gerber SA, Everley PA, Gygi SP. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7:4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Altman R, Schieltz D, Yates J, Kellogg DR. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho PV, Williams BC, Mochida S, Zhao Y, Goldberg ML. The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55δ, a phosphatase directed against CDK phosphosites. Mol Biol Cell. 2009;20:4777–4789. doi: 10.1091/mbc.E09-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiroli E, Rossio V, Lucchini G, Piatti S. The budding yeast PP2ACdc55 protein phosphatase prevents the onset of anaphase in response to morphogenetic defects. J Cell Biol. 2007;177:599–611. doi: 10.1083/jcb.200609088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Cohen P, Karsenti E. Cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: evidence from the effects of okadaic acid. EMBO J. 1990;9:675–683. doi: 10.1002/j.1460-2075.1990.tb08159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE Jr. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Hallberg RL. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol Biol Cell. 2002;13:3477–3492. doi: 10.1091/mbc.02-05-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr. An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr. Sensitivity amplification in biochemical systems. Q Rev Biophys. 1982;15:555–591. doi: 10.1017/s0033583500003449. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr. Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J Biol Chem. 1984;259:14441–14447. [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Gunawardena J. Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc Natl Acad Sci USA. 2005;102:14617–14622. doi: 10.1073/pnas.0507322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Harvey SL, Kellogg DR. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr Biol. 2003;13:264–275. doi: 10.1016/s0960-9822(03)00049-6. [DOI] [PubMed] [Google Scholar]
- Hoffman I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Maller JL. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell. 1993;12:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Maller JL. Phosphorylation and activation of the Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol Biol Cell. 1995;6:215–216. doi: 10.1091/mbc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Walker DH, Maller JL. Periodic changes in the phosphorylation of the Xenopus Cdc25 phosphatase regulate its activity. Mol Biol Cell. 1992;3:927–939. doi: 10.1091/mbc.3.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Karlsson-Rosenthal C, Millar JB. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16:285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Keaton MA, Lew DJ. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr Opin Microbiol. 2006;9:540–546. doi: 10.1016/j.mib.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Keaton MA, Szkotnicki L, Marquitz AR, Harrison J, Zyla TR, Lew DJ. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol Biol Cell. 2008;19:4016–4018. doi: 10.1091/mbc.E08-03-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J Cell Sci. 2003;116:4883–4890. doi: 10.1242/jcs.00908. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ferrell JE Jr. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Kim SY, Song EJ, Lee KJ, Ferrell JE Jr. Multisite M-phase phosphorylation of Xenopus Wee1A. Mol Cell Biol. 2005;25:10580–10590. doi: 10.1128/MCB.25.23.10580-10590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell. 1991;64:903–914. doi: 10.1016/0092-8674(91)90315-p. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- LaPorte DC, Koshland DE Jr. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature. 1983;305:286–290. doi: 10.1038/305286a0. [DOI] [PubMed] [Google Scholar]
- Lin FC, Arndt KT. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 1995;14:2745–2759. doi: 10.1002/j.1460-2075.1995.tb07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Straight A, Rudner AD, Dernburg AF, Belmont A, Murray AW. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. New York: Oxford University Press; 2007. [Google Scholar]
- Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pal G, Paraz MT, Kellogg DR. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J Cell Biol. 2008;180:931–945. doi: 10.1083/jcb.200711014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening JR, Kim SY, Ferrell JE Jr. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Rahal R, Amon A. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 2008;22:1534–1548. doi: 10.1101/gad.1638308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes I. Checking cell size in yeast. Trends Genet. 2002;18:479–485. doi: 10.1016/s0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- Sreenivasan A, Kellogg D. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang Y. Pds1/Esp1-dependent and -independent sister chromatid separation in mutants defective for protein phosphatase 2A. Proc Natl Acad Sci USA. 2006;103:16290–16295. doi: 10.1073/pnas.0607856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Coleman TR, Dunphy WG. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. EMBO J. 1993;12:3427–3436. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld CL, Irazoqui JE, Bloom K, Lew DJ. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S, Tjandra H, Schieltz D, Yates J III, Kellogg DR. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol Biol Cell. 2010;22:20–32. doi: 10.1091/mbc.E10-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jiang W, Gentry M, Hallberg RL. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol Cell Biol. 2000;20:8143–8156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.