Study of the nuclear import behavior of E47 in conjunction with its partner transcription factors shows that although the nuclear import of E47 is importin α dependent, it is capable of accumulating in the nucleus under importin α–blocked conditions by virtue of its interaction with its binding partners NeuroD1 and MyoD.
Abstract
Nuclear import of karyophilic proteins is carried out by a variety of mechanisms. We previously showed that two basic helix-loop-helix proteins, NeuroD1 and E47, synergistically affect each other's nuclear import. In this study, we dissected the molecular pathways underlying nuclear import of the NeuroD1/E47 heterodimer. In vitro nuclear import assays indicated that importin α family members are the major nuclear import receptors for E47. However, inhibition of importin α resulted in cytoplasmic retention of E47 that could be rescued by its binding partner, NeuroD1, through heterodimerization. In addition, nuclear import of NeuroD1 was importin α independent but importin β1 dependent. In primary neurons, localization of endogenous E47 was not affected by importin α inhibition, suggesting that neuronal E47 could be imported into the nucleus as a heterodimer with NeuroD1 by using importin β1 alone. We also found that E47 had similar nuclear import characteristics in C2C12 cells, where E47 heterodimerized with MyoD, another helix-loop-helix protein, suggesting functional conservation within the same family of transcription factors. Collectively, our data reveal that E47 is imported into the nucleus via multiple pathways, depending on the molecular binding mode, establishing a previously uncharacterized cross-talk between two distinct nuclear import pathways.
INTRODUCTION
The regulation of bidirectional movement of macromolecules across the nuclear membrane is critical for gene expression, cell viability, cell cycle progression, differentiation, and development ( Yoneda, 2000; Poon and Jans, 2005; Yasuhara et al., 2009). Various proteins enter or exit the nucleus by using transport receptors called importins or exportins, respectively. Importins identify the nuclear localization signal (NLS) of their cargo to ensure nuclear import, whereas exportins mediate nuclear export of their substrate molecules. The best-characterized protein transport signal is the classic NLS, which consists of either one (monopartite) or two (bipartite) stretches of basic amino acids ( Lange et al., 2007). In the conventional nuclear import pathway, the NLS of the cargo is identified by an adaptor molecule called importin α, which forms a complex with the actual import carrier importin β1. After several interactions with components of the nuclear pore complex during the translocation step, the importin/cargo complex is dissociated by the GTP-bound form of a small Ran GTPase (RanGTP) inside the nucleus ( Imamoto et al., 1995; Cook et al., 2007; Pemberton and Paschal, 2005; Strambio-De-Castillia et al., 2010). The cargo is released to perform its respective function in the nucleus, and the importins are recycled to the cytoplasm for another import cycle.
Importin α–mediated nuclear import is complicated by the fact that, in mammals, there exist five or six isoforms of importin α with differing cargo specificity and tissue-specific expression ( Yoneda, 2000). For example, extracellular, signal-dependent nuclear import of signal transducer and activator of transcription 1 (STAT1) is mediated by importin α5 (NPI-1) via binding with the C-terminal non-armadillo region of importin α5 ( Sekimoto et al., 1997). Alternatively, importin α3 (Qip1) transports DNA helicase Q1-NLS substrates into the nucleus very efficiently ( Miyamoto et al., 1997). It was also found that RanBP3 and RCC1 are imported preferentially by importin α3 ( Köhler et al., 1999; Welch et al., 1999). Furthermore, the physiological significance of differences in importin α isoform expression was shown in a recent report in which switching of the expression of importin α subtypes recapitulated neuronal differentiation in mouse ES cells ( Yasuhara et al., 2007).
In some cases, however, importin β1 directly identifies its cargo molecules and carries out their nuclear import. Although no conserved NLS has been attributed to importin β1, most cargo molecules directly imported by importin β1 have NLSs that are rich in basic amino acids. Examples of proteins that bind importin β1 directly for nuclear import include SREBP-2 ( Nagoshi et al., 1999) and transcription factors important for differentiation, such as SRY, SOX9, Smad, and Snail ( Forwood et al., 2001; Preiss et al., 2001; Xiao et al., 2000; Yamasaki et al., 2005). Although various cargoes bind to the adaptor molecule, importin α, for nuclear import, others directly bind to importin β1; whether and how these two distinct nuclear import pathways cross-talk with each other has remained elusive.
The nuclear accumulation of various proteins is regulated by various means, such as masking of the NLS ( Beg et al., 1992), posttranslational modification ( Kanai et al., 2007), and negative control by import receptors ( Forwood and Jans, 2002). We recently showed that heterodimerization might be another way in which nuclear accumulation of NeuroD1 and E47 proteins might be regulated ( Mehmood et al., 2009). Helix-loop-helix (HLH) transcription factors regulate gene expression to promote cell differentiation ( Murre et al., 1989). NeuroD1 is a type II HLH protein that dimerizes with E47, a type I HLH transcription factor, to transactivate its target genes in neuronal cells, beta pancreatic cells, and enteroendocrine cells ( Naya et al., 1995; Mutoh et al., 1997; Breslin et al., 2003; Liu et al., 2004). The mechanism of nuclear import for this heterodimeric complex has not yet been identified.
In this study, we dissected the nuclear import pathways for NeuroD1 and E47 heterodimer. Using a proteomic approach and in vitro nuclear import assays, we identified distinct nuclear import receptors for both partner proteins. Using primary hippocampal neurons, we showed for the first time cross-talk between two distinct nuclear import pathways. We clearly demonstrated that despite depending on importin α for its nuclear import, E47 is capable of migrating into the nucleus under an importin α–blocked condition via its binding partners. These various modes of nuclear transport and their cross-talk appear to be a strategy used by cells to ensure efficient nuclear import of critical transcription factors.
RESULTS
E47 is imported into the nucleus in an importin α–dependent manner, whereas NeuroD1 accumulates in the nucleus under importin α–inhibited conditions
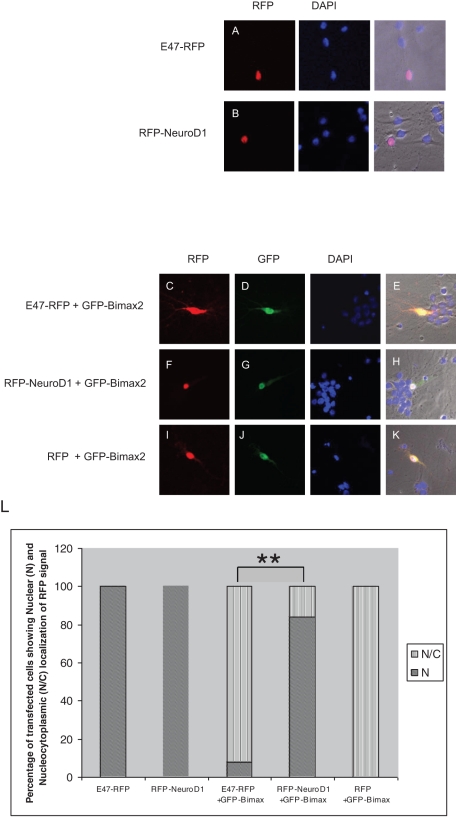
The majority of nuclear proteins studied so far accumulate in the nucleus via a classic nuclear import pathway in which importin α acts as an adaptor molecule between the cargo protein and importin β1 or by binding to importin β1 directly. Unconventional nuclear import pathways have also been described ( Wagstaff and Jans, 2009). Classic NLSs contain a domain of basic amino acids and have been classified as monopartite, with a stretch of consecutive basic amino acids, and bipartite, with two stretches of basic amino acids separated by 10–12 amino acids. Because the NLSs of E47 and NeuroD1 contain basic amino acids, we reasoned that importin α should act as an import receptor for these proteins. We therefore made use of Bimax2, which was reported to be a specific and strong competitive inhibitor of importin α–mediated nuclear transport ( Kosugi et al., 2008). When red fluorescent protein (RFP)–tagged E47 or NeuroD1 alone was transfected into rat embryonic primary hippocampal neurons, their localization was mainly restricted to the nucleus ( Figure 1, A and B). In contrast, upon cotransfection with green fluorescent protein (GFP)–Bimax2, the nuclear accumulation of E47-RFP was significantly hampered ( Figure 1, C–E), whereas that of RFP-NeuroD1 remained unchanged ( Figure 1, F–H). When cotransfected with RFP alone, GFP-Bimax2 did not cause any change in the localization of RFP ( Figure 1, I–K). Figure 1L shows the quantitative analysis of localization of proteins under the importin α–inhibited condition. These results indicate that E47 is imported into the nucleus mainly by the importin α family of proteins, whereas NeuroD1 is imported in an importin α–independent manner.
FIGURE 1:
E47 and NeuroD1 have two distinct nuclear import pathways. Primary cultures of neurons from rat hippocampi were transfected with the indicated expression plasmids. (A, B) RFP-tagged E47 and NeuroD1 were mainly localized to the nuclei of the transfected cells. (C–E) Cotransfection of GFP-Bimax2 with E47-RFP resulted in the cytoplasmic retention of E47. (F–H) The localization of RFP-NeuroD1 remained unaffected when cotransfected with GFP-Bimax2. (I–K) The localization of RFP alone was not affected even when cotransfected with GFP-Bimax2. (L) Quantitative representation of the localization of RFP signal in the cells transfected with indicated plasmids. A two-tailed Fisher's test was performed for statistical analysis using GraphPad software. Significant differences compared with the corresponding control samples are indicated as follows: *p < 0.05, **p < 0.01. The transfections were carried out five times (n = 20). At 24 h posttransfection, the neuronal cells were fixed and observed under a confocal laser scanning microscope attached to an inverted microscope (Zeiss Axiovert 100M) with a ×40/0.75 Plan-Neofluar objective.
Importin α1 and importin α3 are the major import receptors for E47
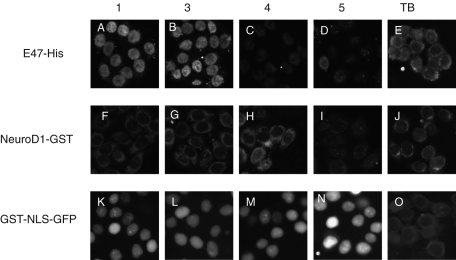
Mammalian importin α consists of five or six family members with tissue-specific expression and cargo specificities. Thus it is imperative to determine whether E47 is imported into the nucleus by specific importin α isoforms. To identify the specific importin α that mediates nuclear import of E47, in vitro nuclear import assays were performed using digitonin-permeabilized HeLa cells. Of the various importin α isoforms tested, only importin α1 (human Rch1) and importin α3 (human Qip1), together with importin β1, efficiently transported E47 into the nucleus ( Figure 2, A–E). In contrast, we observed a very limited nuclear accumulation of NeuroD1 into the nucleus ( Figure 2, F–J) in the presence of importin α and β1. GST-SV40NLS-GFP (referred to as GST-NLS-GFP), which was used as a control, was efficiently imported into the nucleus by all importin α isoforms ( Figure 2, K–O). These data clearly indicate that importin α1 and importin α3 are the major import receptors for E47, consistent with the data in Figure 1, where nuclear import of E47 was inhibited after cotransfection with GFP-Bimax2.
FIGURE 2:
In vitro nuclear import assay for E47 and NeuroD1 using recombinant import factors. Digitonin-permeabilized HeLa cells were incubated with a 10 μl of reaction mixture containing 4 pmol of import substrate, an ATP regeneration system, 6 pmol of importin α, 4 pmol of importin β1, 40 pmol of RanGDP, 3.5 pmol of NTF2, and 0.5 mM GTP. (A, B) E47 was efficiently imported into the nucleus by importin α1 and importin α3, whereas importin α4 and importin α5 did not efficiently import E47 into the nucleus (C, D). (F–I) Nuclear import of NeuroD1 by indicated importin α isoforms. (K–N) GST-NLS-GFP that was used as a positive control was efficiently imported into the nucleus by all importin α isoforms tested. (E, J, O) Only TB and recombinant proteins were added to the import mixture. The cells were fixed as described in Materials and Methods, treated with anti-E47 and anti-NeuroD1, followed by anti–rabbit Alexa 568 and anti–goat Alexa 488, respectively, and visualized under a Zeiss Axiovert 200M microscope.
Under importin α–inhibited conditions, NeuroD1 facilitates translocation of E47 to the nucleus
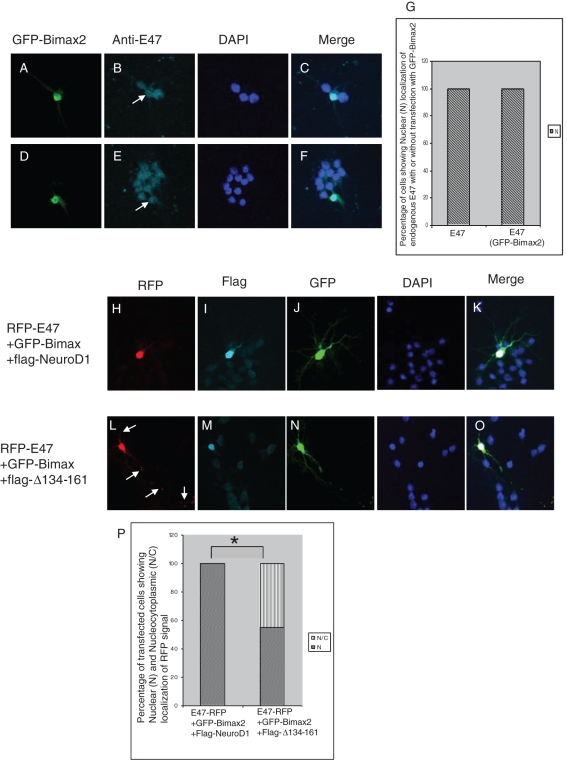
In primary hippocampal neurons, where importin α3 is strongly expressed and importin α1 is not expressed (Supplementary Figure S1), overexpressed E47-RFP was clearly mislocalized to the cytoplasm under importin α–inhibited conditions ( Figure 1), suggesting that E47 is primarily imported by importin α3 with importin β1. Next, to determine the impact of Bimax2 on the localization of endogenous E47 in primary hippocampal neurons, we transfected cultured cells with GFP-Bimax2 and checked the localization of endogenous E47. Surprisingly, we found that there was no apparent change in the localization of endogenous E47 in the transfected cells compared with controls ( Figure 3, A–G).
FIGURE 3:
NeuroD1 redirects the nuclear import of E47 under importin α–inhibited conditions. Primary cultures of neurons from rat hippocampi were transfected with the indicated expression vectors and observed under a microscope 24 h posttransfection. (A–F) Transfection of cells with GFP-Bimax2 and immunostaining with anti-E47. (G) Quantitative comparison of localization of endogenous E47 with and without GFP-Bimax2 transfection (n = 26). (H–K) FLAG-NeuroD1 redirects E47-RFP into the nucleus in the presence of GFP-Bimax2. (L–O) NeuroD1-assisted nuclear redirection of E47-RFP was lost upon cotransfection with E47-RFP, GFP-Bimax2, and FLAG-Δ134-161 plasmids, indicating the specificity of the interaction with HLH domains. (P) Percentage of cells with nuclear (N) and nucleocytoplasmic (N/C) localization of E47-RFP in the presence of GFP-Bimax2 and FLAG-NeuroD1 or FLAG-Δ134-161. A two-tailed Fisher's test was performed for statistical analysis using GraphPad software. Significant differences compared with the corresponding control samples are indicated as follows: *p < 0.05. Transfection was carried out at least five times. Cells cotransfected with all the three constructs with nuclear FLAG staining were analyzed.
We previously demonstrated that E47 and its binding partner, NeuroD1, synergistically support each other's nuclear import ( Mehmood et al., 2009). In addition, we found that NeuroD1 is imported into the nucleus through a mechanism that is distinct from that of E47 ( Figure 1). These results raise the possibility that endogenous NeuroD1 can direct endogenous E47, but not excess amounts of the overexpressed E47-RFP ( Figure 1, C–E), to the nucleus in primary hippocampal neurons. To investigate this possibility, we next tested whether overexpressed flag-NeuroD1 with nuclear localization could rescue the nuclear import of overexpressed E47-RFP under importin α–blocked conditions. We cotransfected E47-RFP with GFP-Bimax2 and FLAG-NeuroD1 in primary hippocampal neurons and found that even in the presence of GFP-Bimax2, overexpressed flag-NeuroD1 efficiently redirected E47-RFP to the nucleus ( Figure 3, H–K), demonstrating interplay of two distinct nuclear import pathways for E47.
To confirm the specificity of this heterodimerization-dependent nuclear import of E47, we constructed a mutant of NeuroD1 that lacks amino acids 134–161(FLAG-Δ134-161) and cannot interact with E47 through the helix-loop-helix domain. This mutant was mainly localized to the nucleus, although a little cytoplasmic retention was observed compared with wild-type NeuroD1 (Supplementary Figure S2). The mutant, which was localized to the nucleus, was inefficient to redirect E47-RFP to the nucleus in the presence of GFP-Bimax2 ( Figure 3, L–O) compared with wild-type NeuroD1 ( Figure 3P). These data show that NeuroD1 specifically binds E47 and supports its nuclear import under conditions where the importin α–mediated nuclear accumulation of E47 is inhibited. Therefore, we conclude these two different nuclear import pathways, one importin α dependent and one importin α independent, cross-talk during the nuclear import of the NeuroD1/E47 heterodimer.
Importin β1 binds to NeuroD1 in a Ran-dependent manner and preferentially carries out its nuclear import in heterodimeric form
To identify the import receptor for NeuroD1, we initially microinjected recombinant NeuroD1-GFP with or without the Ran mutant RanQ69L-GTP, which is devoid of GTP hydrolysis, into the cytoplasm of HeLa and NIH3T3 cells because RanGTP releases the import receptors from the bound cargo molecules by binding to the N-terminal Ran-binding domain of importin β family molecules ( Görlich et al., 1996). Although the microinjected NeuroD1-GFP mainly accumulated in the nucleus, the coinjection of NeuroD1-GFP with RanQ69L-GTP led to cytoplasmic retention of NeuroD1-GFP (Supplementary Figure S3), suggesting the involvement of importin β family members in the nuclear transport of NeuroD1.
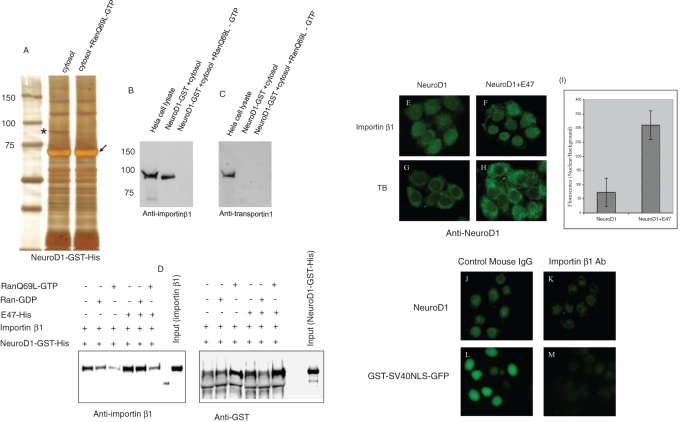
On the basis of observation that NeuroD1 accumulates in the nucleus in a Ran-dependent manner in live HeLa cells, we carried out GST pull-down assays using HeLa cell cytosol and recombinant NeuroD1-GST in the absence or presence of RanQ69L-GTP. As shown in Figure 4A, a band of ∼95 kDa was repeatedly associated with GST-NeuroD1; this band disappeared in the presence of RanQ69L-GTP. Mass spectrometric analysis revealed that the band contained importin β1. Ran-dependent binding of importin β1 with NeuroD1 was further confirmed by Western blotting using a specific anti–importin β1 antibody ( Figure 4B). Although transportin1 is another importin β family member with a similar molecular size ( Arnold et al., 2006), specific antibodies against transportin1 did not detect any Ran-dependent binding of transportin1 to NeuroD1 ( Figure 4C). We also detected Ran-independent binding of RanBP5 and RanBP7 with NeuroD1 (Supplementary Figure S4). However, as the nuclear import of NeuroD1 was Ran dependent, we characterized importin β1, which binds NeuroD1 in a Ran-dependent manner. To see whether the interaction between NeuroD1 and importin β1 is direct, we carried out in vitro binding assays using recombinant proteins. NeuroD1–glutathione S-transferase (GST)–His was immobilized on GST beads, and recombinant importin β1 was added to it. As can be seen in Figure 4D, NeuroD1 bound directly to importin β1, and this binding was disrupted with the addition of RanQ69L-GTP to the complex. Moreover, the addition of recombinant E47-His stabilized the binding between NeuroD1 and importin β1, which was also disrupted in the presence of RanQ69L-GTP ( Figure 4D).
FIGURE 4:
Importin β1 binds NeuroD1 in a Ran-dependent manner and carries out its nuclear import. (A) GST pull-down assay. Glutathione–Sepharose prebound with NeuroD1-GST was incubated with the clear HeLa cell cytosol with or without RanQ69L-GTP for 4 h at 4°C. After five washings, the bound proteins were eluted by sample buffer, resolved by SDS–PAGE, and stained by silver staining. The band marked with the asterisk was put to mass spectrometric analysis and was identified to be importin β1. The experiment was repeated three times, and every time the marked band appeared in the gel. The resolved proteins were immunoblotted using anti–importin β1 (B) and anti–transportin1 (C). (D) In vitro binding assay using recombinant NeuroD1-GST-His, importin β1, E47-His, and RanQ69L-GTP, or RanGDP. GST beads were incubated with the indicated recombinant proteins in TB containing 4% BSA. Eluted samples were immunoblotted using anti-GST and anti–importin β1. (E–H) In vitro nuclear import assay. (E) NeuroD1 was very weakly imported into the nucleus by importin β1. (F) Robust nuclear accumulation of NeuroD1 when it was added to the import mixture together with E47. (G, H) In vitro import assay using TB alone with the indicated substrates. (I) Quantitative comparison of the florescence intensity (nuclear/background) when NeuroD1 was added alone to the import mixture or together with E47. Experiment was performed three times (n = 30). (J–M) In vitro nuclear import assay using Ehrlich cell lysate after importin β1 depletion. Anti–importin β1 was added to the Ehrlich cell lysate, followed by in vitro import assay.
Importin β1 was shown to directly interact with the NLS of a number of cargo molecules ( Harel and Forbes, 2004). To see whether importin β1 is a direct nuclear import receptor for NeuroD1, we carried out the in vitro nuclear import assay using importin β1 as the only import factor. As shown in Figure 4E, there was only a limited nuclear accumulation of NeuroD1 with importin β1. Addition of importin α had no major effect on this weak nuclear import of NeuroD1 (Supplementary Figure S5). In contrast, we observed a robust nuclear accumulation of NeuroD1 with the addition of E47 to the import mixture ( Figure 4, F and I). These results indicate that NeuroD1 binds importin β1 in a Ran-dependent manner and is preferentially imported into the nucleus in heterodimeric form with E47. Finally, to confirm that importin β1 is the major nuclear import receptor for NeuroD1, we immunodepleted importin β1 from Ehrlich cell lysate using anti–importin β1 and carried out nuclear import assay ( Figure 4, J–M). As can be seen, compared with control mouse immunoglobulin G (IgG), the nuclear accumulation of NeuroD1 was negatively affected in the presence of anti–importin β1. This result shows that importin β1 is the major nuclear import receptor for NeuroD1. This also explains the nuclear accumulation of E47 upon importin α inhibition in primary neurons, where it is imported into the nucleus through heterodimeric complex formation with NeuroD1 and robustly expresses endogenous importin β1 (Supplementary Figure S1).
The cross-talk between two distinct nuclear import pathways is conserved for bHLH proteins
Type II bHLH transcriptional regulators are involved in a variety of differentiation programs in various tissues. One founding member of the type II bHLH proteins, MyoD, is specifically expressed in myoblasts and is essential for skeletal muscle differentiation. MyoD also heterodimerizes with E47 to transactivate its target genes during muscle differentiation ( Lassar et al., 1991). Thus, to identify functional conservation among members of the same family of transcription factors, we focused on the nuclear import of MyoD and E47 in C2C12 cells, which are typically used as an in vitro model system to study myogenesis.
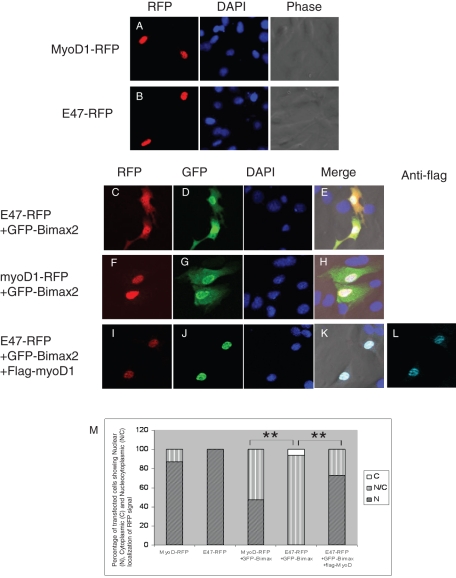
RFP-tagged MyoD or E47 cells alone were transfected into C2C12 cells, and the intracellular localization was examined 20 h posttransfection. Both of the transcription factors localized to the nucleus ( Figure 5, A and B). Cotransfection of GFP-Bimax2 with E47-RFP led to the cytoplasmic retention of E47-RFP ( Figure 5, C–E). Alternatively, there was no change in the localization of MyoD-RFP when cotransfected with GFP-Bimax2 ( Figure 5, F–H). To explore the potential of MyoD to redirect E47 into the nucleus, three constructs including E47-RFP, GFP-Bimax2, and flag-MyoD were simultaneously transfected into C2C12 cells. As shown in Figure 5, I–M, a clear nuclear redirection of E47-RFP was observed upon cotransfection with FLAG-MyoD, even in the presence of GFP-Bimax2. This clearly indicates that, like NeuroD1, MyoD is imported into the nucleus independent of importin α and is competent to redirect E47 into the nucleus under importin α–inhibited conditions. These data support the notion that, in spite of varying differentiation programs, members of the same protein family share common functional features regarding nuclear localization.
FIGURE 5:
MyoD is also capable of redirecting E47 into the nucleus in C2C12 cells. C2C12 cells were transfected with the indicated expression vectors and observed under a microscope 20 h posttransfection. RFP-tagged MyoD (A) and E47 (B) were mainly localized to the nuclei of C2C12 cells. (C–E) The nuclear import of E47-RFP was hampered when cotransfected with GFP-Bimax2. (F–H) The nuclear accumulation of MyoD remained unchanged when cotransfected with GFP-Bimax2. (I–L) FLAG-MyoD rescued the GFP-Bimax2–mediated cytoplasmic retention of E47-RFP. (M) Quantitative analysis showing the localization of RFP-myoD and E47-RFP when transfected alone or cotransfected with GFP-Bimax2. A two-tailed Fisher's test was performed for statistical analysis using GraphPad software. Significant differences compared with the corresponding control samples are indicated as follows: *p < 0.05, **p < 0.01. The transfection was carried out at least three times independently (n = 30).
DISCUSSION
In recent years, many papers have reported on importin α/β1- ( Lange et al., 2007) or importin α-independent/importin β1-dependent nuclear import pathways ( Fried and Kutay, 2003; Harel and Forbes, 2004), but cross-talk between these two nuclear import pathways was not clearly shown. In this study, using the NeuroD/E47 heterodimer as a model, we showed that there is cross-talk between these two distinct nuclear import pathways. High levels of cytoplasmically retained E47-RFP can act in a dominant-negative manner and retain NeuroD1 in the cytoplasm ( Mehmood et al., 2009); however, corresponding overexpressed NeuroD1 not only can migrate into the nucleus, but it can also redirect E47-RFP into the nucleus in the presence of Bimax2 ( Figure 3). E47 was specifically imported by importin α1 and importin α3, but not importin α4 or importin α5, in conjunction with importin β1, which corroborates previous results showing that substrate specificity within importin α family members is an important determinant of the regulatory nuclear import of transcriptional regulators ( Yasuhara et al., 2007). The most striking observation of this study is that through the process of heterodimerization with partner proteins, the nuclear import of E47 is safeguarded even under conditions in which its inherent nuclear import is suppressed, which means that E47 can accumulate and function in the nucleus of cells in which only importin α4 or importin α5 is expressed among importin α family members. A recent report identified RanBP17 and RanBP16, both having structural similarity with importin β superfamily members, as novel E12/E47–binding proteins ( Lee et al., 2010). Although these proteins had no influence on the nuclear accumulation of E12/E47, their overexpression led to increased expression of the E47 target gene, suggesting novel functions for import receptors ( Lee et al., 2010). We also found Ran-independent binding of NeuroD1 with RanBP5 and RanBP7. These observations may suggest import independent roles of some of the importin family members.
Importin β1 is one of the most versatile karyopherins, as it can identify a variety of cargo molecules mainly by virtue of its inherent molecular flexibility ( Lee et al., 2003). Although no consensus NLS has been attributed as a target of importin β1, basic amino acids are rich in the NLSs of most of its substrates, including NeuroD1. Our pull-down in vitro nuclear import assays indicated that importin β1 is the major import receptor that binds NeuroD1 in a Ran-dependent manner. Efficient importin β1–mediated nuclear import of NeuroD1 in the presence of E47 might indicate conformation changes taking place when NeuroD1 is complexed with its partner E47.
Another interesting aspect of this study is the role of MyoD in redirecting E47 into the nucleus under importin α–inhibited conditions. E12 and E47 were shown to regulate not only the degradation of MyoD, but also its nuclear localization ( Lingbeck et al., 2005). However, it was also reported that the NLS-mutant of E47 was not redirected into the nucleus by wild-type MyoD ( Lingbeck et al., 2005). In contrast, in our assay we used wild-type E47 that was forcibly retained in the cytoplasm. The nuclear import of cytoplasmically retained wild-type E47 was rescued by MyoD, indicating the dominant-positive role of MyoD for the nuclear redirection of E47 and suggesting that the higher-order configuration of wild-type proteins is an important determinant of the nuclear import property of MyoD. Thus NeuroD1 and MyoD, both of which belong to the same family of proteins, share conserved features: their nuclear import is independent of importin α, and they mediate nuclear redirection of E47 through heterodimerization, although their nuclear localization signals vary greatly, and it remains unknown whether these proteins are imported into the nucleus by the same set of importin β family members.
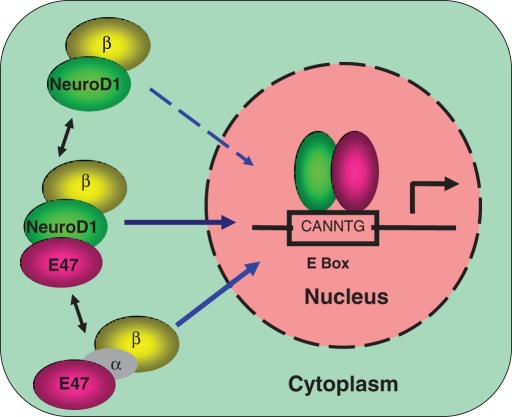
On the basis of our findings, we propose a model for the nuclear transport of the NeuroD1/E47 heterodimer ( Figure 6) that underscores the significance of the existence of parallel pathways that support nuclear import of various interacting proteins under various physiological conditions. Nuclear import of E47 depends on importin α1/β1 or α3/β1, whereas import of NeuroD1 is independent of importin α but depends on importin β1. Although both proteins can be imported independently (although with variable efficiencies), there is significant cross-talk between the two nuclear import pathways for E47 and its partner protein NeuroD1. It has been shown that the high-mobility group box-containing transcription factors SRY and SOX9 are imported into the nucleus by two distinct mechanisms; one uses importin β1 and the other involves calmodulin, although the two pathways work under different physiological conditions (low vs. high calcium levels) so as to maintain a threshold level of SRY and SOX9 ( Harley et al., 1996; Forwood et al., 2001; Argentaro et al., 2003). Thus cross-talk might safeguard the efficient nuclear import of the heterodimer for transactivation of target genes under certain physiological conditions or in certain cell types. For example, in cells in which only importin α4 or importin α5 is expressed, E47 is likely not to be efficiently transported into the nucleus in an importin α/β1–dependent manner. The presence of cross-talk between nuclear import pathways ensures efficient and timely nuclear import of certain transcriptional regulators. Cellular environment inside a neuronal cell keeps on changing during development and neuronal activity. Unweaving the relationship between nuclear import pathways and changing cellular environment is an exciting research area for future research.
FIGURE 6:
Proposed model for the nuclear import of the NeuroD1/E47 heterodimer. Details are explained in the text.
MATERIALS AND METHODS
Cell culture
NIH3T3 and HeLa cells, both of which do not express endogenous NeuroD1, were cultured in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with heat-inactivated 10% fetal bovine serum (FBS) at 37°C in 10% CO2. C2C12 cells that endogenously express MyoD and are used as a model for muscle differentiation were also maintained in DMEM containing 10% FBS.
Construction of mammalian expression vectors
E47-RFP was previously described ( Mehmood et al., 2009). RFP-NeuroD1 was constructed by subcloning NeuroD1 between the EcoRI and SmaI sites of a pmRFP-C1 vector. FLAG-NeuroD1 and FLAG-Δ134-161 were cloned between the HindIII and EcoRI sites of a pFLAG-CMV-2 expression vector (Sigma-Aldrich). GFP-Bimax2 was constructed using the EcoRI and ApaI sites of a pEGFP-C2 vector. MyoD was amplified from C2C12 cDNA and cloned between the HindIII and SacII sites in a pmRFP-N1 vector. FLAG-MyoD was constructed by subcloning MyoD from MyoD-RFP with HindIII and SmaI restriction enzymes into the pFLAG-CMV-2 vector.
Bacterial expression vectors and purified recombinant proteins
E47-pET21d and NeuroD1-GST vectors were previously described ( Mehmood et al., 2009). NeuroD1-EGFP was constructed by swapping GST from the NeuroD1-GST vector with EGFP using the XhoI restriction enzyme.
NeuroD1-GST and NeuroD1-EGFP were transformed into the BL21 strain of Escherichia coli. Expression of the fusion proteins was induced with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) at 20°C for 16 h. E47-pET21d was transformed into another E. coli strain, Rosetta. Expression of the fusion protein was induced with 0.1 mM IPTG at 37°C for 5 h. Bacterial cultures were harvested, lysed in lysis buffer (50 mM Tris-HCl, pH 8.3, 500 mM NaCl, 1 mM EDTA), gently mixed with lysozyme, treated with liquid nitrogen, and stored at –80°C overnight. After thawing and sonication, the cleared lysates of GST-tagged proteins were incubated with glutathione–Sepharose 4B (Amersham Biosciences, Piscataway, NJ), and the lysates of His-tagged proteins were incubated with Ni-NTA agarose (Qiagen, Valencia, CA) according to the manufacturer's instructions. Bacterially expressed recombinant importin α1, α3, α4, α5, and β1, NTF2, and RanQ69L-GTP were purified as previously described ( Sekimoto et al., 1996). The purified proteins were diluted in transport buffer (TB; 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.3, 110 mM CH3COOK, 5 mM CH3COONa, 2 mM (CH3COO)2Mg, 0.5 mM ethylene glycol tetraacetic acid [EGTA], 2 mM dithiothreitol [DTT], and 1 μg/ml each aprotinin, pepstatin, and leupeptin) using a PD-10 column (GE Healthcare, Piscataway, NJ).
In vitro nuclear import assay and immunofluorescence
Digitonin-permeabilized HeLa cells were prepared as previously described ( Adam et al., 1990). Permeabilized cells were incubated at room temperature for 10 min in TB and washed twice with TB to minimize residual proteins in the cytoplasm. The cells were incubated at 37°C for 50 min in TB containing 4 pmol of transport substrates, 6 pmol of importin α, 4 pmol of importin β1, 40 pmol of RanGDP, 3.5 pmol of nuclear transport factor 2 (NTF2), 0.5 mM of GTP, and an ATP regeneration system (0.5 mM ATP, 20 U/ml creatine phosphokinase, and 5 mM creatine phosphate; Sigma-Aldrich) for a total volume of 10 μl per sample. A nuclear import assay involving a NeuroD1/E47 heterodimer was carried out similarly, except that equimolar quantities of NeuroD1 and E47 were incubated at 37°C for 30 min before being added to the import mixture. After incubation, the cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) followed by permeabilization of the nuclear membrane with 0.5% Triton X-100 in PBS. Immunostaining of NeuroD1 was performed with a primary anti-NeuroD1 polyclonal antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) and a secondary Alexa 488–labeled anti–goat IgG antibody (1:100; Molecular Probes, Invitrogen, Carlsbad, CA). For detection of E47, primary anti-E47 antibody (1:100; Santa Cruz Biotechnology) was used, followed by Alexa 568–labeled anti–rabbit IgG antibody (1:100; Molecular Probes).
Transfection assays
The transfection of C2C12 cells expressing endogenous MyoD was carried out using Effectene (Qiagen) according to the manufacturer's protocol. C2C12 cells were grown in 12-well plates (Nalge Nunc International, Rochester, NY). For each expression vector, 300 ng of plasmid was transfected per well, and the cells were incubated for 20 h before observation with a confocal laser scanning microscope attached to an inverted microscope (Axiovert 100M; Carl Zeiss, Jena, Germany) with a ×40/0.75 Plan-Neofluar objective.
Primary neuronal culture and transfection
Primary hippocampal neurons that express endogenous NeuroD1 were cultured as previously described ( Takano et al., 2007). The hippocampi from embryonic day 18 rats were isolated. Following dissociation with trypsin, cells were plated in Neurobasal Media (Invitrogen) containing 2.5 mM l-glutamine (Invitrogen), B-27 (used at 1:50 dilution; Invitrogen), and antibiotics/antimycotics (used at 1:100 dilution; Invitrogen) in 12-well plates on polyethyleneimine (P2636, Sigma-Aldrich)–coated coverslips. The transfection was carried out 8–12 d after plating using a TransMessenger Transfection Kit (Qiagen) with 1.6 μg of expression vectors. Anti-FLAG (F7425, 1:200; Sigma-Aldrich) was used to detect FLAG-tagged proteins followed by goat polyclonal antibody to rabbit IgG DyLight 649 (1:100; Abcam, Cambridge, MA). Fluorescence images of the neurons were taken using a confocal laser scanning microscope attached to an inverted microscope (Axiovert 100M; Carl Zeiss) with a ×40/0.75 Plan-Neofluar objective.
GST pull-down assay, silver staining, and Western blotting
HeLa cells were prepared and lysed as described previously ( Kehlenbach et al., 1998). The cells were grown to confluency in five 20-cm dishes, collected by trypsinization, washed with PBS and then washing buffer (10 mM HEPES-KOH, pH 7.3, 110 mM CH3COOK, 2 mM (CH3COO)2Mg, 2 mM DTT, protease inhibitors), incubated in lysis buffer (5 mM HEPES-KOH, pH 7.3, 10 mM CH3COOK, 2 mM (CH3COO)2Mg, 2 mM DTT, protease inhibitors) for 10 min on ice, and lysed with five strokes of a stainless steel Dounce homogenizer. After centrifugation at 1500 × g for 15 min, the supernatant was ultracentrifuged at 30,000 rpm for 1 h. The clear cytosol was collected and dialyzed with binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 5% glycerol). Glutathione–Sepharose beads prebound with NeuroD1-GST were incubated with the clear cytosol, with or without RanQ69L-GTP, for 4 h at 4°C. Beads were washed with the binding buffer five times. Interacting proteins bound to the beads were eluted in the sample buffer (2% SDS, 10% glycerol, 60 mM Tris-HCl, pH 6.8, 5% β-mercaptoethanol, and 0.01% bromophenol blue) and resolved by 12.5% SDS–PAGE. Silver staining was carried out using the SilverQuest Kit (Invitrogen). For Western blotting, the eluted proteins were resolved by SDS–PAGE and transferred onto a nitrocellulose transfer membrane (Whatman, Piscataway, NJ). The blots were first incubated in blocking buffer (5% [wt/vol] nonfat dry milk in PBS plus 0.05% Tween 20) overnight at 4°C. The blots were then incubated with primary antibody in dilution buffer (1% [wt/vol] nonfat dry milk in PBS plus 0.05% Tween 20) for 1 h at room temperature. Anti–importin β1 (1:1000) antibody was from BD Biosciences (San Diego, CA), and anti–importin α3 (1:1000) was from Santa Cruz Biotechnology, whereas rat monoclonal antibodies specific for transportin1 and importin α1 were generated as previously described ( Kamikubo et al., 2004). After four washings, the blots were incubated with horseradish peroxidase–conjugated secondary antibodies in dilution buffer for 1 h at room temperature. Antigen–antibody complexes were visualized using enhanced chemiluminescence (GE Healthcare).
In vitro binding assay
In vitro binding assay was performed in TB with 4% bovine serum albumin. NeuroD1-GST (2 μg), importin β1 (4 μg), E47-His (2 μg), and RanQ69L-GTP (4 μg) or Ran-GDP (4 μg) were mixed and added to the GST beads to a final volume of 50 μl. The mixture was incubated at 37°C for 1 h with tapping every 10 min. Beads were washed with TB, treated with sample buffer, and immunoblotted using anti–importin β1 and anti-GST antibodies (Santa Cruz Biotechnology).
Mass spectrometry
The samples were separated by electrophoresis on 10% bis-Tris SDS–PAGE gels (Wako Pure Chemical, Osaka, Japan) that were silver stained using a SilverQuest Kit. The unique bands were excised and digested with trypsin ( Shevchenko et al., 1996). Samples subjected to analysis by matrix-assisted laser desorption ionization mass spectrometry on an Ultraflex TOF/TOF time-of-flight mass spectrometer (Bruker Daltonics, Billerica, MA) were first desalted using C18 ZipTips (Millipore, Billerica, MA) and eluted with 0.5 μl of α-cyano-4-hydroxy cinnamic acid and then allowed to dry at room temperature. The monoisotopic peptide mass fingerprinting (PMF) data obtained from mass spectrometry were used to automatically search for proteins identified using Mascot 2.2.01 (http://www.matrixscience.com). For PMF analysis against the National Center for Biotechnology Information nonredundant databases permutations, the following search parameters were considered: trypsin missed one cleavage, mass spectrometry tolerance 0.5 Da, fixed and variable modifications (Cys as an S-carbamidomethyl derivate and Met as oxidized methionine), with the taxonomy Homo sapiens (human). Protein identification with a statistically significant Mascot identity score (p < 0.05) was accepted.
Microinjection
NIH3T3 and HeLa cells on a 35-mm culture dish were cytoplasmically microinjected using a micromanipulator system as previously described ( Kotera et al., 2005). Samples were prepared in TB and filtered through a 0.22-mm polyvinylidene fluoride filter (Millipore) prior to injection. Alexa 568–labeled anti–rat IgG was used as an injection marker. Live-cell imaging was carried out 1 h postmicroinjection using a fluorescence microscope.
Supplementary Material
Acknowledgments
We thank the members of the Yoneda lab for discussions. We also thank David Jans and Olivier Raineteau for helpful discussions. This work was supported in part by the Japanese Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science, and the Takeda Science Foundation.
Abbreviations used:
- HLH
helix loop helix
- NLS
nuclear localizing signal
- TB
transport buffer
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0809) on August 10, 2011.
REFERENCES
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentaro A, Sim H, Kelly S, Preiss S, Clayton A, Jans DA, Harley VR. A SOX9 defect of calmodulin-dependent nuclear import in campomelic dysplasia/autosomal sex reversal. J Biol Chem. 2003;278:33839–33847. doi: 10.1074/jbc.M302078200. [DOI] [PubMed] [Google Scholar]
- Arnold M, Nath A, Wohlwend D, Kehlenbach RH. Transportin is a major nuclear import receptor for c-Fos: a novel mode of cargo interaction. J Biol Chem. 2006;281:5492–5499. doi: 10.1074/jbc.M513281200. [DOI] [PubMed] [Google Scholar]
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- Breslin MB,, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Forwood JK, Jans DA. Nuclear import pathway of the telomere elongation suppressor TRF1: inhibition by importin alpha. Biochemistry. 2002;41:9333–9340. doi: 10.1021/bi025548s. [DOI] [PubMed] [Google Scholar]
- Forwood JK, Harley V, Jans DA. The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin beta 1. J Biol Chem. 2001;276:46575–46582. doi: 10.1074/jbc.M101668200. [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN, Hextall PJ. The HMG box of SRY is a calmodulin binding domain. FEBS Lett. 1996;391:24–28. doi: 10.1016/0014-5793(96)00694-1. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- Kamikubo Y, Sakaguchi N, Shikata K, Furuta M, Miyamoto Y, Imamoto N, Yoneda Y, Ogino K, Tachibana T. Specific monoclonal antibody against nuclear import factor, importin α 1/Rch1. Hybrid Hybridomics. 2004;23:301–304. doi: 10.1089/hyb.2004.23.301. [DOI] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Görlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Kotera I, Sekimoto T, Miyamoto Y, Saiwaki T, Nagoshi E, Sakagami H, Kondo H, Yoneda Y. Importinα transports CaMKIV to the nucleus without utilizing importin β. EMBO J. 2005;24:942–951. doi: 10.1038/sj.emboj.7600587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lee SJ, et al. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- Lee JH, Zhou S, Smas CM. Identification of RANBP16 and RANBP17 as novel interaction partners for the bHLH transcription factor E12. J Cell Biochem. 2010;111:195–206. doi: 10.1002/jcb.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingbeck JM, Trausch-Azar JS, Ciechanover A, Schwartz AL. E12 and E47 modulate cellular localization and proteasome-mediated degradation of MyoD and Id1. Oncogene. 2005;24:6376–6384. doi: 10.1038/sj.onc.1208789. [DOI] [PubMed] [Google Scholar]
- Liu Y, Encinas M, Comella JX, Aldea M, Gallego C. Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004;24:2662–2672. doi: 10.1128/MCB.24.7.2662-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood R, Yasuhara N, Oe S, Nagai M, Yoneda Y. Synergistic nuclear import of NeuroD1 and its partner transcription factor, E47, via heterodimerization. Exp Cell Res. 2009;315:1639–1652. doi: 10.1016/j.yexcr.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Mühlhäusser P, Müller EC, Otto A, Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Mutoh H,, Fung BP, Naya FJ, Tsai MJ, Nishitani J, Leiter AB. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Imamoto N, Sato R, Yoneda Y. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol Biol Cell. 1999;10:2221–2233. doi: 10.1091/mbc.10.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Preiss S, Argentaro A, Clayton A, John A, Jans DA, Ogata T,, Nagai T, Barroso I, Schafer AJ, Harley VR. Compound effects of point mutations causing campomelic dysplasia/autosomal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. J Biol Chem. 2001;276:27864–27872. doi: 10.1074/jbc.M101278200. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of STAT1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Nakajima K, Tachibana T, Hirano T, Yoneda Y. Interferon-gamma-dependent nuclear import of STAT1 is mediated by the GTPase activity of Ran/TC4. J Biol Chem. 1996;271:31017–31020. doi: 10.1074/jbc.271.49.31017. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Takano K, Miki T, Katahira J, Yoneda Y. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res. 2007;35:2513–2521. doi: 10.1093/nar/gkm125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff KM, Jans DA. Importins and beyond: non-conventional nuclear transport mechanisms. Traffic. 2009;10:1188–1198. doi: 10.1111/j.1600-0854.2009.00937.x. [DOI] [PubMed] [Google Scholar]
- Welch K, Franke J, Köhler M, Macara IG. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-alpha3. Mol Cell Biol. 1999;19:8400–8411. doi: 10.1128/mcb.19.12.8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad3. J Biol Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sekimoto T, Ohkubo T, Douchi T, Nagata Y, Ozawa M, Yoneda Y. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 2005;10:455–464. doi: 10.1111/j.1365-2443.2005.00850.x. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Oka M, Yoneda Y. The role of nuclear transport system in cell differentiation. Semin Cell Dev Biol. 2009;20:590–599. doi: 10.1016/j.semcdb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5:777–787. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.