The Hippo pathway kinase LATS2 promotes contact inhibition of growth. How LATS2 is activated in response to changes in cell density is unknown. It is found that tight junction protein AMOTL2 is a novel activator of LATS2, raising the possibility that tight junction assembly promotes LATS2-dependent inhibition of cell proliferation.
Abstract
LATS2 kinase functions as part of the Hippo pathway to promote contact inhibition of growth and tumor suppression by phosphorylating and inhibiting the transcriptional coactivator YAP. LATS2 is activated by the MST2 kinase. How LATS2 is activated by MST2 in response to changes in cell density is unknown. Here we identify the angiomotin-family tight junction protein AMOTL2 as a novel activator of LATS2. Like AMOTL2, the other angiomotin-family proteins AMOT and AMOTL1 also activate LATS2 through a novel conserved domain that binds and activates LATS2. AMOTL2 binds MST2, LATS2, and YAP, suggesting that AMOTL2 might serve as a scaffold protein. We show that LATS2, AMOTL2, and YAP all localize to tight junctions, raising the possibility that clustering of Hippo pathway components at tight junctions might function to trigger LATS2 activation and growth inhibition in response to increased cell density.
INTRODUCTION
Studies in both fruit flies and mammalian cells have revealed a central role for the Hippo pathway in contact inhibition of growth, tumor suppression, and stem cell differentiation (Halder and Johnson, 2011). The MST2 and LATS2 kinases, as well as the transcriptional coactivator YAP, form the core of the mammalian Hippo pathway. MST2 activates LATS2, and LATS2 phosphorylates YAP, which inhibits the ability of YAP to promote cell motility and proliferation and maintain stem cell fate. Although many candidate upstream regulators of the Hippo pathway have been identified through genetic screens in flies, it is unclear how and whether these proteins directly affect signaling. A major unanswered question is how upstream signals cause LATS2 activation in response to increased cell density and differentiation signals.
The angiomotin family of proteins localize to tight junctions and regulate cell growth and motility (Patrie, 2005; Sugihara-Mizuno et al., 2007; Ernkvist et al., 2008; Gagne et al., 2009; Zheng et al., 2009; Ranahan et al., 2011). The angiomotin family of proteins has three members, AMOT, AMOTL1, and AMOTL2, with AMOT having both short (AMOT80) and long (AMOT130) isoforms. A previous study showed that the ratio of AMOT80 to AMOT130 expression in endothelial cells regulates a switch from migratory to more stable nonmotile cells (Ernkvist et al., 2008), however the mechanism by which Angiomotin proteins regulate cell migration is not known. Recent studies identified angiomotin family proteins as binding partners and inhibitors of the closely related transcriptional coactivators YAP and TAZ (Chan et al., 2011; Wang et al., 2011; Zhao et al., 2011). These studies proposed that angiomotin proteins regulate the Hippo pathway indirectly by binding and sequestering YAP in the cytoplasm. However, it was not clear from these studies whether angiomotin proteins have a direct role in regulating signaling through the core Hippo pathway kinases MST2 and LATS2.
RESULTS
AMOTL2 binds LATS2 and YAP2 and promotes LATS2 phosphorylation of YAP2
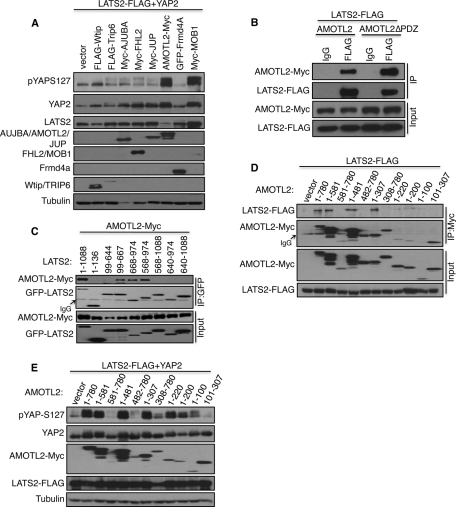
To identify proteins that interact with LATS2, we purified LAP-tagged (Cheeseman and Desai, 2005) LATS2 that was stably expressed in U2OS cells and analyzed the isolated protein complexes using mass spectrometry. This analysis identified known LATS2–binding partners YAP2 (Hao et al., 2008; Oka et al., 2008; Zhang et al., 2008) and the LIM-domain proteins Ajuba and WTIP (Hirota et al., 2000; Abe et al., 2006; Das Thakur et al., 2010), as well as a number of proteins that localize to regions of cell–cell contact (Supplemental Table S1 and Supplemental Figure S1A). Because many upstream regulators of LATS localize to cell–cell junctions (Edgar, 2006), we tested whether overexpression of any of the identified proteins promoted the ability of LATS2 to phosphorylate its target, YAP2. HEK 293 cells were transfected with LATS2, YAP2, and various LATS2-interacting proteins identified in our screen. The levels of LATS2-dependent phosphorylation of YAP2 were measured using phospho-specific antibodies that recognize the LATS2 phosphorylation site (S127) on YAP2 (Zhao et al., 2007). Although expression of most putative LATS2-binding proteins did not affect YAP2 phosphorylation, expression of the AMOTL2 protein caused a major increase in YAP2 phosphorylation, similar to that caused by the known LATS2 activator MOB1 (Figure 1A). AMOTL2 is a member of the angiomotin family of proteins (Bratt et al., 2002). Several recent studies showed that angiomotin proteins bind YAP and were proposed to inhibit YAP by sequestering it to the cytoplasm (Chan et al., 2011; Wang et al., 2011; Zhao et al., 2011). Coimmunoprecipitation experiments confirmed that AMOTL2 bound to LATS2, and the PDZ motif of AMOTL2 is not required for this interaction (Figure 1B). Deletion studies (Supplemental Figure S1, B and C) showed that AMOTL2 binds the kinase domain of LATS2 (amino acids 668–974) and the MOB1-binding region of LATS2 adjacent to the kinase domain (amino acids 644–668; Figure 1C). Surprisingly, several larger LATS2 deletion mutants encompassing the kinase domain and additional adjacent sequences (568–1088, 640–974, 640–1088) did not bind AMOTL2, suggesting that either the constructs did not fold properly or the additional sequences interfered with binding in that context of the deletion mutant. LATS2 bound to the first 307 amino acids of AMOTL2 (Figure 1D). Further deletion analysis of the first 307 amino acids of AMOTL2 showed that although the first 100 amino acids of AMOTL2 are sufficient to promote LATS2 phosphorylation of YAP2 (Figures 1E and 3B), assays for binding of smaller deletions of AMOTL2 to LATS2 gave variable results, perhaps because the smaller fragments have weaker binding interactions that do not survive the immunoprecipitation procedure (Figure 1D).
FIGURE 1:
AMOTL2 interacts with LATS2 and YAP2 and promotes LATS2-mediated YAP phosphorylation. (A) YAP2, LATS2-FLAG, and the indicated plasmids were transfected into HEK293 cells. Cell lysates were analyzed by Western blotting to detect YAP2 phosphorylation levels using anti-pYAP-S127 antibody (top). The blot was then reprobed to obtain the levels of other proteins. (B) HEK293 cells were transfected with LATS2-FLAG along with either AMOTL2 or an AMOTL2 mutant lacking the C-terminal PDZ motif (AMOTL2-ΔPDZ). Cell lysates were subjected to immunoprecipitation (IP) of LATS2 with FLAG antibody or immunoglobulin G (IgG) control. The immunoprecipitates and cell lysates were subjected to immunoblot analysis with anti-Myc and anti-FLAG antibodies. The levels of AMOTL2 and LATS2 proteins in the starting lysates are shown in the bottom half (Input). (C) The AMOTL2-Myc plasmid was transfected along with either full-length GFP-LATS2 (1–1088) or the indicated GFP-LATS2 deletion constructs into HEK293 cells. LATS2 was immunoprecipitated (IP) from cell lysates with anti-GFP antibodies. The immunoprecipitates and cell lysates were subjected to immunoblot with anti-Myc and anti-GFP antibody. The levels of AMOTL2 and LATS2 proteins in the starting lysates are shown in the bottom half (Input). (D) LATS2-FLAG along with full- length AMOTL2-Myc (1–780) or the indicated Myc tagged AMOTL2 deletion constructs were transfected into HEK293 cells. AMOTL2 was immunoprecipitated (IP) from cell lysates using anti-Myc antibody, and the immunoprecipitates and cell lysates were subjected to immunoblot with anti-FLAG and anti-Myc antibodies. (E) YAP2, LATS2-FLAG, and the indicated AMOTL2-Myc deletion constructs were transfected into HEK293 cells, and the resulting cell lysates were subjected to immunoblot analysis with anti-pYAP-S127 antibody (top). The levels of YAP, AMOTL2-Myc, LATS2-FLAG, and tubulin are shown (bottom).
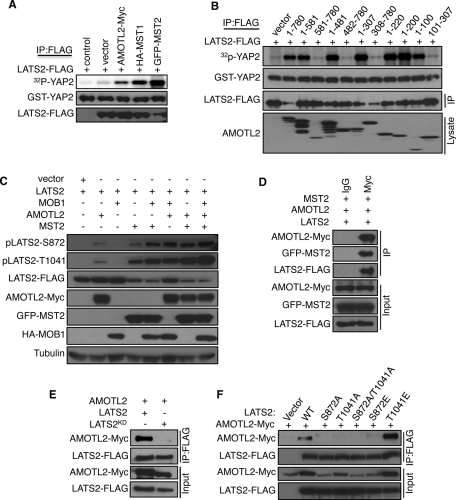
FIGURE 3:
AMOTL2 promotes LATS2 kinase activity and binds to the active form of LATS2. (A) Cell lysates were prepared from HEK293 cells (control) or HEK293 cells transfected with LATS2-FLAG and the indicated plasmids. LATS2-FLAG was immunoprecipitated using anti-FLAG antibodies. The immunoprecipitates were subjected to vitro kinase assays using bacterially expressed GST-YAP2 as a substrate, and LATS2 phosphorylation of GST-YAP2 (top) was measured using a PhosphorImager. Simultaneously, equal volumes of kinase assay samples were subjected to Western blot analysis to detect the levels of GST-YAP2 and LATS2-FLAG. (B) LATS2-FLAG was coexpressed with the indicated AMOTL2 deletion constructs in HEK293 cells and processed for in vitro kinase assays and Western blot analysis as described in A. (C) LATS2-FLAG was coexpressed with indicated plasmids in HEK293, and the lysates were subjected to Western blot with anti-pLATS2-S872, anti-pLATS2-T1041, and other antibodies as marked. (D) AMOTL2-Myc, LATS2-FLAG, and GFP-MST2 plasmids were coexpressed in HEK293 cells, and cell lysates were immunoprecipitated either with anti-Myc or control IgG antibodies. The immunoprecipitates (IP) and cell lysates (Input) were subjected to immunoblot with the indicated antibodies. (E) LATS2-FLAG or kinase-dead LATS2 (LATS2KD-FLAG) were coexpressed with AMOTL2-Myc in HEK293 cells. Cell lysates were immunoprecipitated with anti-FLAG antibodies and immunoprecipitates (IP) and cell lysates (Input) were subjected to immunoblot with the indicated antibodies. (F) AMOTL2-Myc was coexpressed with the indicated phosphorylation-site mutants of LATS2, and cell lysates were processed for immunoprecipitation and Western blot as described in E.
Of interest, like LATS2 (Oka et al., 2008), AMOTL2 also bound to YAP2 through the two WW domains in YAP2, but the binding did not depend on the PDZ binding motif in either protein (Supplemental Figure S2, A and B). The ability of AMOTL2 to bind both YAP2 and LATS2 suggested that AMOTL2 could promote LATS2 phosphorylation of YAP2 by bringing these two proteins together. However, AMOTL2 was still able to promote LATS2 phosphorylation of a WW-domain mutant in YAP2 that could not bind either LATS2 or AMOTL2 (Supplemental Figure S2C). We also mutated the YAP-binding site in AMOTL2 and found that this mutant (AMOTL2Y213A) promoted phosphorylation just as well as wild-type AMOTL2 (Supplemental Figure S2D), showing that LATS2 and AMOTL2 do not need to bind directly to YAP2 to promote its phosphorylation. The ability of AMOTL2 to promote LATS2 phosphorylation of YAP2 did not require the carboxyl-terminal PDZ binding motifs of YAP2 and AMOTL2 (Supplemental Figure S2, C and D). In addition to YAP2, AMOTL2 was also able to promote phosphorylation of TAZ, another downstream target of LATS2 (Lei et al., 2008; Supplemental Figure S2E).
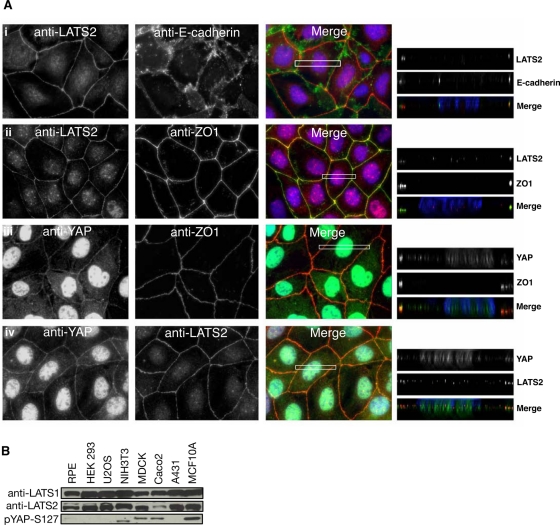
Because AMOTL2 and YAP2 have been shown to localize to tight junctions (Patrie, 2005; Sugihara-Mizuno et al., 2007; Oka et al., 2010), we examined whether LATS2 might also localize to tight junctions in the polarized epithelial cell lines Caco2 (Figure 2A) and MDCK (unpublished data). Of interest, both LATS2 and YAP colocalize with the tight junction marker ZO-1. Moreover, these cell lines, along with the breast epithelial cells MCF10A and NIH3T3 cells, all show significant LATS pathway activity as judged by the level of phosphorylation of the LATS2 target, YAP2 (Figure 2B).
FIGURE 2:
LATS2 and YAP colocalize with the tight junction marker ZO-1 in Caco2 cells. (A) Caco2 cells grown to confluence and processed for indirect immunofluorescence using antibodies against LATS2 (i, ii) with E-cadherin (i) or Zo1 (ii) (top half) or YAP2 (iii, iv) with Zo1 (iii) or LATS2 (iv) (bottom half). The merged green, red, and blue (DAPI/DNA) signals are shown. The boxed region was deconvolved to show LATS2 and YAP2 localization along the apical–basolateral axis (right). (B) Endogenous levels of LATS1, LATS2, and YAP phosphorylation (pYAP-S127) were analyzed by Western blotting of lysates from the indicated cell lines.
AMOTL2 promotes LATS2 kinase activity
To test whether AMOTL2 directly affected LATS2 kinase activity, we measured LATS2 in vitro kinase activity using LATS2 protein immunoprecipitated from HEK293 cells expressing LATS2 with either control plasmid, AMOTL2, or the LATS2-activating kinase MST1 or MST2 (Figure 3A). Expression of AMOTL2 caused a significant increase in LATS2 in vitro kinase activity, although not as high as that caused by MST1 and MST2. As observed in the in vivo assays, the first 100 amino acids of AMOTL2 were sufficient to promote LATS2 in vitro kinase activity (Figure 3B). However, addition of the bacterially expressed AMOTL2 peptides containing the first 100 or 200 amino acids to LATS2 immune complexes was not able to promote LATS2 activity in vitro (Supplemental Figure S2F), suggesting that AMOTL2 needs other proteins or posttranslational modifications to activate LATS2.
LATS2 activity requires phosphorylation on an autophosphorylation site (S872) and phosphorylation on T1041 by the MST1 and MST2 kinases (Chan et al., 2005; Xiao et al., 2011). Therefore we tested whether AMOTL2 expression affected the phosphorylation on either of these sites. AMOTL2 caused an increase in LATS2 phosphorylation on both S872 and T1041 (Figure 3C). Furthermore, AMOTL2 acted synergistically with both MOB1, and MST2 to increase LATS2 phosphorylation on both sites (Figure 3C). MST2 phosphorylates T1041 of LATS2 directly (Chan et al., 2005) and also phosphorylates MOB1A/B (Hirabayashi et al., 2008; Praskova et al., 2008; Bao et al., 2009), which enhances its binding to LATS2 and stimulates LATS2 to autophosphorylate on S872 (Praskova et al., 2008). Therefore AMOTL2 could promote LATS2 phosphorylation by acting as a scaffold to bring together MST2 and LATS2. Consistent with this notion, AMOTL2 immunoprecipitates contained both MST2 and LATS2 (Figure 3D), suggesting that they might form a ternary complex.
Of interest, AMOTL2 could not bind to inactive versions of LATS2 such as the kinase-dead allele (Figure 3E) or LATS2 mutants with the two activating phosphorylation sites (S872 and T1041) mutated to alanines (Figure 3F), suggesting that AMOTL2 might be able to amplify weak LATS2 signaling by binding activated LATS2 and keeping it together with its activator MST2. Consistent with this idea, AMOTL2 bound strongly to LATS2 bearing the phosphomimetic T1041E mutation (Figure 3F). AMOTL2 did not bind the LATS2-S872E mutant, most likely because this mutation does not effectively mimic phosphorylation and is inactivating like the S872A mutant (Figure 3F; Millward et al., 1999; Chan et al., 2005). LATS2 may also have an effect on AMOTL2 levels since the AMOTL2 levels were highest when wild type or the T1041E mutant LATS2 were expressed, suggesting that the active versions of LATS2 are stabilizing the protein. This could be either because AMOTL2 binds only the active versions of LATS2 and having a binding partner stabilizes the overexpressed AMOTL2 protein, or because active LATS2 promotes AMOTL2 stability through some other mechanism. Similarly, AMOTL2 levels are decreased when kinase-dead LATS2 is expressed compared with wild-type LATS2 (Figure 3E). Together these data show that AMOTL2 binds active forms of LATS2 and enhances their ability to promote YAP2 phosphorylation, and LATS2 might in turn promote AMOTL2 stability.
AMOT family proteins are both positive and negative regulators of LATS2
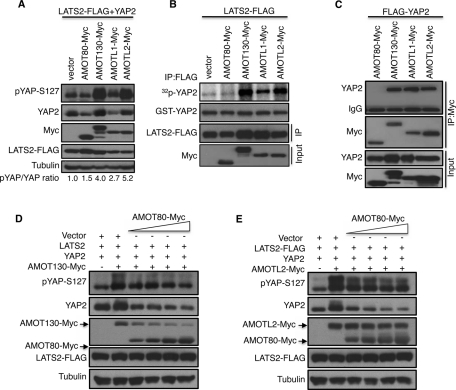
We next addressed whether the two other angiomotin family proteins, AMOTL1 and AMOT, which comes in 80-kDa (AMOT80) and 130-kDa (AMOT130) splice variants (Bratt et al., 2005), also regulate LATS2 activity. Of interest, overexpression of AMOTL1 and AMOT130, but not AMOT80, promoted LATS2 phosphorylation of YAP2 in vivo and vitro (Figure 4, A and B). This is not surprising since AMOT130 and AMOTL1, but not AMOT80, have N-terminal sequences (Supplemental Figure S3, A and B) homologous to the domain of AMOTL2 that binds and activates LATS2, and this domain of AMOT130 is sufficient to activate LATS2 (Supplemental Figure S3C). In addition, AMOT130, AMOTL1, and AMOTL2, but not AMOT80, were able to bind YAP2 (Figure 4C).
FIGURE 4:
AMOT family proteins are both positive and negative regulators of LATS2 signaling. (A) YAP2 was coexpressed with LATS2-FLAG and the indicated Myc-tagged AMOT proteins in HEK293 cells. Cell lysates were processed and subjected to immunoblot detection of YAP2 phosphorylation (pYAP-S127). The levels of AMOT proteins (Myc), YAP2, LATS2, and tubulin in the lysates are shown. The numbers under the blots show the ratio of the pixel densities of phospho-YAP to YAP. (B) LATS2-FLAG was coexpressed with Myc-tagged AMOT proteins as indicated, and the cell lysates were processed for LATS2 in vitro kinase assays and Western blot analysis as described in Figure 3A. (C) FLAG-YAP2 was transfected with Myc-tagged AMOT plasmids in HEK293 cells as indicated. Cell lysates were subjected to immunoprecipitation of AMOT proteins with anti-Myc antibodies, and the immunoprecipitates and lysates were probed with FLAG and Myc antibodies. (D) AMOT130-Myc, YAP2, and LATS2-FLAG were coexpressed with various amounts of AMOT80 as indicated in HEK293 cells, and the level of YAP2 phosphorylation was analyzed by Western blot. The levels of YAP2, AMOT130, AMOT80, LATS2, and tubulin are shown. (E) AMOTL2-Myc, YAP2, and LATS2-FLAG were coexpressed with various amounts of AMOT80 as indicated in HEK293 cells, and the level of YAP2 phosphorylation was analyzed by Western blot. The levels of YAP2, AMOTL2, AMOT80, LATS2, and tubulin are shown.
A previous study showed that the AMOT80 and AMOT130 have opposing effects on cell motility, with AMOT80 promoting cell motility and AMOT130 inhibiting cell motility (Ernkvist et al., 2008). Of interest, when both forms of AMOT were expressed, the ability of the AMOT80 to promote cell motility was dominant over the inhibitory effects of AMOT130 (Ernkvist et al., 2008). At least some of these observations could be explained if AMOT80 antagonizes the ability of AMOT130 to promote LATS2 signaling, since inactivation of LATS2 promotes YAP-mediated cell motility (Overholtzer et al., 2006; Wang et al., 2011). To see whether AMOT80 inhibits the ability of AMOT130 and AMOTL2 to promote LATS2 signaling, we coexpressed AMOT130 or AMOTL2 with increasing amounts of AMOT80. Although AMOT80 inhibited the ability of AMOT130 and AMOTL2 to promote YAP2 hyperphosphorylation (Figure 4, D and E), the effects on AMOT130 were much stronger. Expression of AMOT80 caused a reduction in the levels of AMOT130, perhaps explaining its antagonism. The levels of AMOTL2 were not affected by AMOT80 expression. These results reveal complex interactions among angiomotin family proteins and show that differential regulation of angiomotin isoforms can lead to enhancement or inhibition of Hippo pathway signaling.
Reduction in AMOT130 and AMOTL2 levels inhibits LATS2 signaling
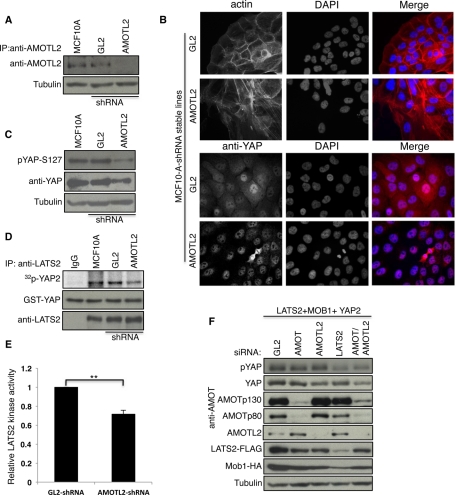
To test the effect of down-regulation of AMOTL2 on LATS2 activity, we generated MCF10A breast epithelial cells stably expressing a Lentivirus short hairpin RNA (shRNA) construct specific to AMOTL2 (Figure 5A). In comparison to control cells, AMOTL2 knockdown cells showed a striking change in actin organization from normal epithelial morphology to more spindle-shaped fibroblast morphology as observed previously (Wang et al., 2011; Figure 5B, top). Of interest, AMOTL2 knockdown cells displayed increased nuclear localization of YAP2 (Figure 5B, bottom) and a marked reduction in YAP phosphorylation (Figure 5C). Furthermore, LATS2 kinase activity was reduced in AMOTL2 knockdown cells, consistent with the observed decrease in YAP phosphorylation (Figure 5, D and E).
FIGURE 5:
Reduction in AMOT130 and AMOTL2 levels inhibits LATS2 signaling. (A) AMOTL2 was immunoprecipitated from lysates of MCF10A cells or MCF10A cells stably expressing luciferase control (GL2) or AMOTL2 shRNA vectors, and then the levels of AMOTL2 were analyzed using anti-AMOTL2 antibodies. (B) MCF10A cells stably expressing the indicated shRNA constructs described in A were processed for immunofluorescence for F-actin (top) and anti-YAP (bottom). Nuclei were stained with DAPI. (C) Lysates of MCF10A cells or MCF10A cells stably expressing the indicated shRNA constructs (see A) were subjected to Western blot analysis for detection of endogenous YAP phosphorylation using anti–pYAP-S127, and the blot was subsequently reprobed for YAP and tubulin. (D) Lysates of MCF10A cells alone or MCF10A cells stably expressing the indicated shRNA vectors (see A) were subjected to immunoprecipitation of endogenous LATS2 using anti-LATS2 antibodies and processed for in vitro kinase assays and Western blot analysis as described in Figure 3A. IgG immunoprecipitates from MCF10A cells (IgG) were used as a control. (E) The levels of kinase activity of LATS2 from the indicated MCF10A lines were quantified using ImageJ and normalized to the amount of LATS2. The values are the average of three independent experiments. Error bars, SD of the relative kinase activity; **p < 0.0003 (based on p value calculation by t-test analysis). (F) AMOTL2-, AMOT-, LATS2-, or luciferase (GL2)-specific siRNA pools were transfected into HEK293 cells along with LATS2-FLAG, HA-MOB1, and YAP2 as indicated. Cell lysates were prepared 72 h after transfection and analyzed by Western blot for detection of pYAP-S127 and other proteins as marked.
The importance of individual angiomotin family members for LATS2 signaling may vary, depending on the cell type. For example, HEK293 cells express high levels of AMOT130 in addition to AMOTL2 (Aase et al., 2007). To test whether AMOT130 might contribute to LATS2 signaling, AMOTL2, AMOT, and LATS2 were knocked down in HEK293 cells. Note that the small interfering RNA (siRNA) pool used to knock down AMOT targets both the AMOT80 and the AMOT130 isoforms (Figure 5F). Although knockdown of LATS2 caused a reduction in YAP2 phosphorylation as expected, knockdown of AMOTL2 or AMOT individually had only mild effects. In contrast, knockdown of both AMOTL2 and AMOT together caused a reduction in YAP2 phosphorylation similar to that caused by LATS2 knockdown (Figure 5F). Together these studies identify AMOT family proteins as novel activators of LATS2 and the Hippo pathway in mammalian cells.
DISCUSSION
The Hippo/LATS pathway is emerging as a key regulator of organ size, contact inhibition of growth, and stem cell differentiation (Zhao et al., 2008). All components of the mammalian pathway were first identified in genetic screens in Drosophila melanogaster. Of interest, the AMOT family of proteins does not appear to exist in flies and thus represents the first mammalian components of the pathway not identified based on their homology to proteins in the fly pathway. Given that AMOT family members have both positive and negative roles in Hippo/LATS signaling, the angiomotin family provides another layer of control and could have allowed the pathway to take on new functions in mammalian cells. We showed that all three angiomotin family members are capable of promoting LATS2 activation, raising the possibility that angiomotin family members act redundantly, as we observed in HEK293 cells, or are used in a tissue-specific manner to control Hippo pathway signaling. It is also not completely resolved from our studies how AMOTL2 activates LATS2. Addition of bacterially expressed AMOTL2 was not able to activate LATS2 in vitro, suggesting that AMOTL2 requires additional proteins or secondary modifications to promote LATS2 activation. Our favored model is that AMOTL2 activates LATS2 by acting as a scaffold protein to bring LATS2 together with its activating kinase, MST2.
It will be interesting to determine which previously described functions of angiomotin proteins are mediated by effects on Hippo/LATS signaling. For example, AMOT80 and AMOT130 were previously shown to have positive and negative effects, respectively, on cell motility (Ernkvist et al., 2008). These results could be explained by effects on YAP phosphorylation since nonphosphorylated YAP promotes cell motility (Overholtzer et al., 2006) and we showed that YAP phosphorylation is inhibited by AMOT80 and promoted AMOT130. In addition, AMOT80 was recently shown to promote cell proliferation through enhanced ERK1/2 signaling (Ranahan et al., 2011). Another study showed that Amot promotes ERK1/2 activity in a direct pathway through the Rac1 GTPase (Yi et al., 2011). However it is possible that AMOT80 expression could also affect ERK1/2 activity through YAP-dependent transcription since YAP promotes transcription of amphiregulin (Zhang et al., 2009; Dong et al., 2011), a growth-promoting epidermal growth factor receptor ligand, which would lead to ERK1/2 activation. In the future these issues could be resolved using alleles of AMOT that specifically disrupt its ability affect Hippo/YAP pathway signaling.
A key question is how LATS2 becomes activated in response to increased cell density. Pertinent to this issue might be the fact that many upstream activators of LATS2 localize to tight junctions (Halder and Johnson, 2011). Because tight junctions only form when cells come into close contact, tight junction assembly could serve as a signal to trigger LATS2 activation. LATS2, YAP, and angiomotin family proteins all localize to tight junctions (this study; Patrie, 2005; Sugihara-Mizuno et al., 2007; Chan et al., 2011; Wang et al., 2011; Zhao et al., 2011). One model could be that tight junctions serve as scaffolds to concentrate Hippo pathway components to promote signal propagation. Tight junctions could also serve to bring LATS2 together with angiomotin proteins. Because AMOTL2 can bind both LATS2 and its activator MST2, AMOTL2 might act as an adaptor protein to promote MST2 activation of LATS2. It will be interesting to determine whether the angiomotin family and other tight junction proteins have a role in cell types such as NIH3T3 that do not have tight junctions yet require Hippo pathway to mediate contact inhibition of growth (Zhao et al., 2007). In conclusion, we expect that study of the angiomotin family of proteins will shed important light on mechanisms governing regulation of the Hippo/LATS pathway in the processes of cell proliferation, motility, and differentiation.
MATERIALS AND METHODS
Cell culture
Human HEK293, 293T, and U2OS cell lines were grown in DMEM supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin/streptomycin (Invitrogen, Carlsbad, CA). Caco2 cells were maintained in MEM Alpha supplemented with 20% FBS and 1% (vol/vol) penicillin/streptomycin. Human mammary epithelial MCF10A cells were cultured in DMEM/F12 complemented with 5% horse serum (Invitrogen), 10 mg/ml insulin (Sigma-Aldrich, St. Louis, MO), 100 ng/ml cholera toxin (Sigma-Aldrich), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 20 ng/ml epidermal growth factor (PeproTech, Rocky Hill, NJ), and 1% of penicillin and streptomycin (Invitrogen). All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
Plasmids
pcDNA3-LATS2-FLAG (wild type and kinase dead) plasmids were a kind gift of Tadashi Yamamoto (University of Tokyo, Tokyo, Japan). The LATS2 deletion constructs were cloned into the pC113-GFP-LAP plasmid (GFP, green fluorescent protein; a gift from Iain Cheeseman, Broad Institute, Cambridge, MA). pcDNA4-AMOTL2-Myc and pcDNA4-AMOTL2-ΔPDZ-Myc (lacking the C-terminal PDZ motif) were kindly provided by Anming Meng (Tsinghua University, Beijing, China), and AMOTL2 deletion constructs were PCR amplified using Herculase Tag polymerase (Stratagene, Santa Clara, CA) and cloned in pcDNA4/myc plasmid using standard procedures. All clones were verified by DNA sequencing. The pCS-GFP-AMOTL1 plasmid came from the Fernandes lab stock, and the pBABE-AMOT80 and pBABE-AMOT130 plasmids were provided by Lars Holmgren (Karolinska Institute) and were subcloned into the pcDNA4/myc plasmid. pcDNA4-YAP2, pcDNA4-YAP2WW, pcDNA4-YAP2-ΔPDZ, p2xFLAG-YAP2, and EGFP-MST2 (plasmids 19060, 19000, 21125, 19045, and 19050 [Addgene, Cambridge, MA], submitted by Marius Sudol, Weis Center for Research, Danville, PA), 3x-FLAG-pCMV5-TOPO TAZ and 3x-FLAG-pCMV5-TOPO TAZ (S89A) (plasmids 24809 and 24815 [Addgene], submitted by Jeff Warner) were obtained from Addgene. HA-MOB1 plasmid DNA was a kind gift of Thanos Halazonetis (University of Geneva, Geneva, Switzerland). FLAG-Trip6 and FLAG-Wtip plasmids were kindly provided by Mary Beckerle (University of Utah, Salt Lake City, Utah) and Sigmar Stricker (Max-Planck-Institute, Berlin, Germany), respectively. The cDNA clones for JUP, Frmd4a, PEZ, and AJUBA were obtained from Open Biosystems (Thermo Biosystems, Huntsville, AL) and cloned in pcDNA3.1 with a Myc epitope tag at the N-terminus of the proteins or in the pC113-GFP vector. YAP2 was PCR amplified and cloned into the GST vector pGEX-5×2. The lentiviral shRNA plasmid pLKO.1 and the packaging plasmids psPAX.2 and pMD2.G were obtained from Addgene. The point mutations in LATS2 (S872A, S872E, T1041A, T1041E) were made using the Quick-Change Site Mutagenesis Kit (Stratagene). All PCR-generated clones were verified by DNA sequencing.
Antibodies
The rabbit anti-LATS2 antibody used for immunofluorescence (Abe et al., 2006) was kindly provided by Tadashi Yamamoto (University of Tokyo). The rabbit anti-LATS2 antibody (A300-479A) used for immunoprecipitation and Western blot analysis was obtained from Bethyl Laboratories (Montgomery, TX). Mouse anti-tubulin and mouse anti-FLAG (M2) were purchased from Sigma-Aldrich. Mouse anti-HA (16B12) was obtained from Covance (Berkeley, CA). The rabbit anti-YAP (sc15407), mouse anti-YAP (sc10199), mouse anti-GFP (9996), rabbit anti-Myc (sc789), mouse anti-Myc 9E10 (sc46), and goat anti-AMOTL2 (82501) were obtained from Santa Cruz Biotechnology. The rabbit anti-pYAP-S127 (4911S), anti–rabbit pLATS2-S872 (9157S), and rabbit pLATS2-T1041 (9159S) were purchased from Cell Signaling Technology (Beverly, MA). The rabbit anti-AMOT antibody was generated by the Fernandes lab (Université Laval, CHUQ-CHUL Research Centre).
LAP purification and mass spectrometry
U2OS cells were transiently transfected with LAP-tagged pC113-GFP or pC113-LATS2 plasmids using Lipofectamine 2000 (Invitrogen). Stable clones were selected with 800 μg/ml G418 (Invitrogen) in the medium, and pooled cells were subjected to flow sorting to obtain a single positive line that stably expressed the full-length recombinant protein. A large-scale purification of GFP (as control) or the LATS2 protein complex was carried out as described (Cheeseman and Desai, 2005), and resultant protein complexes were subjected to mass spectrometry analysis.
In vivo YAP phosphorylation assay and Western blotting
For detection of LATS2-mediated phosphorylation of YAP (Ser127), HEK293 cells were transfected in 12-well plates with empty vector, YAP2, and LATS2 with different upstream regulators as indicated, using Lipofectamine 2000. Forty hours after transfection, cells were lysed in immunoprecipitation buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.0% Nonidet P-40, 2% glycerol) supplemented with 1× protease inhibitor cocktail (Sigma-Aldrich), 100 nM sodium vanadate (Sigma-Aldrich), and 50 mM sodium fluoride (Sigma-Aldrich), and lysates were cleared by centrifugation at 13,000 rpm for 10 min at 4°C. After determining the protein concentration by bicinchoninic acid assay (Pierce, Thermo Fisher Scientific, Rockford, IL), 25 μg of protein lysate was mixed with 2× SDS sample buffer, boiled for 5 min at 100°C, and subjected to electrophoresis (SDS–PAGE) and transferred to polyvinylidene fluoride membrane (Millipore, Billerica, MA). Membranes were blocked using 5% dry milk (Bio-Rad, Hercules, CA) or bovine serum albumin in phosphate-buffered saline (PBS) containing 0.1% Tween 20. Subsequently, the membranes were probed with pYAP-S127 (1:3000) or appropriate antibodies for 1 h to overnight, followed by incubation with horseradish peroxidase– or alkaline phosphatase (AP)–conjugated secondary antibodies and detected with enhanced chemiluminescence or AP substrates, respectively. Blots were stripped using Western blot strip buffer (Pierce) and then reprobed as needed.
Immunoprecipitation
For immunoprecipitation, plasmids were transfected into HEK293 cells using Lipofectamine 2000. Forty hours later, cells were collected and lysates were processed in immunoprecipitation buffer as mentioned previously. An amount of 500 μg of total protein lysate was precleared with Protein G agarose beads (Pierce) for 1 h at 4°C. Then, the lysates were incubated with appropriate primary antibodies for 2 h at 4°C on a rotating platform. Subsequently, 20 μl of Protein G agarose was added to each mixture, and the samples were incubated for an additional hour. The beads were washed five times with immunoprecipitation buffer. After the final wash, residual buffer was removed with a 1-cc syringe. Beads were then mixed with 2× SDS sample buffer, boiled for 5 min at 100°C, and subjected to SDS–PAGE and Western blot analysis as described previously. To pull down endogenous proteins from MCF10A cells, 600–800 μg of protein lysate was mixed with 1 μg of goat anti-AMOTL2 or rabbit anti-LATS2 antibodies.
siRNA/shRNA transfection
The siRNA sequence corresponding to firefly luciferase 5′ CGUACGCGGAAUACUUCGA 3′ (used as a control and referred to as GL2) and human-LATS2 5′ TCAACGTGGACCTGTATGA 3′ (Aylon et al., 2006) were synthesized at Dharmacon (Lafayette, CO). SMARTpool siRNA, consisting of four siRNAs targeting multiple sites on AMOTL2 (AMOTL2-siRNA), and AMOT (targeting both AMOT80 and AMOT130) were obtained from Dharmacon. The sequences for AMOTL2 siRNAs are 5′ GAAAGCAGGUUAAAGGUGC 3′, 5′ CAAGGGCUCUCUUCUAGUG 3′, 5′ GAACGGCUCCUUCAGUUGU 3′, and 5′ CAGAACAACUGCGAGAGAA 3′; and the sequences for AMOT are 5′ CCACAUCGUUUGUCUAUAC 3′, 5′ GACCUGCAAUCCAGACAAA 3′, 5′ CCAAAGACGACACAUCGAA 3′, and 5′ CAAGCUAGAGGGCGAGAUU 3′. For the production of lentiviral shRNA particles the sequences specific to AMOTL2 (Wang et al., 2011) and GL2 were cloned in pLKO.1 vector
For knockdown experiments in HEK293 cells, 100–200 nM of each siRNA were mixed with LATS2-FLAG, pcDNA4-YAP2, and HA-MOB1 plasmids and transfected using Lipofectamine 2000. Cells were incubated for 48–72 h and analyzed for protein expression using immunoblotting with specific antibodies. For lentiviral shRNA production, 1 d before transfection 5 × 105 293T cells were plated on a 60-mm plate and incubated at 37°C overnight. Next morning, 4 μg of pLKO.1-GL2 or pLKO1-AMOTL2 shRNA transfer vector were mixed with 3 μg of PAX (packing), 1 μg of MD2G (envelope) plasmids, and 16 μl of Lipofectamine 2000. After incubating for 20 min at room temperature, the mixture was added dropwise into each plate. Twenty hours later, the media was replaced with 3 ml of OPTI-MEM. The media containing Lentivirus particles were collected 48 h later and passed through a 0.45-μm filter and directly added to MCF10A cells with 6 μg/ml Polybrene. Subsequently, the stable pools of cells were selected for 5 d in growth medium containing 2 μg/ml puromycin.
Kinase assays
For in vitro kinase assays, HEK293 cells were transfected with either wild-type LATS2-FLAG or active-site mutants of LATS2-FLAG as indicated. Forty hours after transfection, cells were lysed in immunoprecipitation buffer, and 300 μg of protein lysate was processed for immunoprecipitation of LATS2 as described earlier. Samples were processed for kinase assays as described earlier (Yang et al., 2004), with some modification. The immunoprecipitates were washed three times in immunoprecipitation buffer; once with immunoprecipitation buffer containing 500 mM NaCl and 1 mM dithiothreitol (DTT; rotation for 2 min at 4°C); twice with kinase assay buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 1 mM DTT, and 1× phosphatase inhibitor. Beads were then mixed with 20 μl of kinase assay mix containing 2 μg of GST-YAP2, 10 μM ATP, and 5 μCi of [γ-32P]ATP and incubated at 30°C for 30 min. The reaction was stopped by adding 5× SDS sample dye, boiled at 100°C for 5 min, and subjected to SDS–PAGE. After electrophoresis, gels were dried, and exposed to a PhosphorImager screen for 1–5 h, scanned on a Storm PhosphorImager (GE Healthcare, Piscataway, NJ), and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD).
Immunocytochemistry
Caco2, U2OS, and MCF01A cells cultured in coverslips were fixed with 4% paraformaldehyde for 10 min and permeabilized/blocked with 0.5% Triton X-100 and 5% normal goat serum for 20 min. Cells were subsequently incubated with appropriate primary antibodies overnight at 4°C. They were washed three times in PBS and incubated with Alexa Fluor–conjugated secondary antibodies (Molecular Probes, Invitrogen) for 1 h at room temperature. After three washes with PBS, nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The coverslips were mounted on slides using ProLong Gold antifade reagent (Invitrogen) and viewed using fluorescence microscopy (DMR; Leica Microsystems, Wetzlar, Germany). Images were acquired and processed using Slidebook software 5.0 (Intelligent Imaging Innovations, Denver, CO) and Photoshop (Adobe, San Jose, CA).
Supplementary Material
Acknowledgments
We are grateful to T. Yamamoto, A. Meng, L. Holmgren, T. Halazonetis, M. Beckerle, and S. Stricker for plasmids and antibodies. We also thank Clark Wells for helpful discussions. M.J.G.F. was funded by The Arthritis Society and the Canadian Institutes of Health Research. This work was supported by a National Institutes of Health grant to D. McCollum (GM058406-12).
Abbreviations used:
- BSA
bovine serum albumen
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- IgG
immunoglobulin G
- PBS
phosphate-buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0300) on August 10, 2011.
REFERENCES
- Aase K, et al. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–2068. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Sumita K, Kudo T, Withanage K, Nakagawa K, Ikeda M, Ohno K, Wang Y, Hata Y. Roles of mammalian sterile 20-like kinase 2-dependent phosphorylations of Mps one binder 1B in the activation of nuclear Dbf2-related kinases. Genes Cells. 2009;14:1369–1381. doi: 10.1111/j.1365-2443.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, Ernkvist M, Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- Bratt A, Wilson WJ, Troyanovsky B, Aase K, Kessler R, Van Meir EG, Holmgren L. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298:69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Gupta A, Pai RK, Tun M, Lowe AW. The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through Hippo pathway co-activator YAP1 activation. J Biol Chem. 2011;124:267–273. doi: 10.1074/jbc.M110.215707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;286:18301–18310. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Birot O, Sinha I, Veitonmaki N, Nystrom S, Aase K, Holmgren L. Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim Biophys Acta. 2008;1783:429–437. doi: 10.1016/j.bbamcr.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Gagne V, Moreau J, Plourde M, Lapointe M, Lord M, Gagnon E, Fernandes MJ. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil Cytoskeleton. 2009;66:754–768. doi: 10.1002/cm.20405. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Hirabayashi S, Nakagawa K, Sumita K, Hidaka S, Kawai T, Ikeda M, Kawata A, Ohno K, Hata Y. Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene. 2008;27:4281–4292. doi: 10.1038/onc.2008.66. [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward TA, Hess D, Hemmings BA. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem. 1999;274:33847–33850. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- Oka T, et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrie KM. Identification and characterization of a novel tight junction-associated family of proteins that interacts with a WW domain of MAGI-1. Biochim Biophys Acta. 2005;1745:131–144. doi: 10.1016/j.bbamcr.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranahan WP, Han Z, Smith-Kinnaman W, Nabinger SC, Heller B, Herbert BS, Chan R, Wells CD. The adaptor protein AMOT promotes the proliferation of mammary epithelial cells via the prolonged activation of the extracellular signal-regulated kinases. Cancer Res. 2011;71:2203–2211. doi: 10.1158/0008-5472.CAN-10-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, Furuse M, Tsukita S. Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells. 2007;12:473–486. doi: 10.1111/j.1365-2443.2007.01066.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates the Hippo signaling activity via interactions with Lats kinases. J Biol Chem. 2011;286:7788–7796. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2004;6:609–617. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- Yi C, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Vertuani S, Nystrom S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlen P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105:260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.