Abstract
Hibernation is an energy-saving strategy used by diverse species of mammals to survive winter. It is characterized by cycles between multi-day periods of torpor with low body temperature (Tb), and short periods of rapid, spontaneous rewarming. The ability to retain cellular integrity and function throughout torpor and rewarming is a key attribute of hibernation. Livers from winter hibernators are resistant to cellular damage induced by cold storage followed by warm reperfusion. Identifying proteins that differ between the summer-sensitive and winter-protected phenotypic states is one useful approach that may elucidate the molecular mechanisms that underlie this protection. Here we employ a novel quantitative proteomics screening strategy whereby a newly-weaned 13-lined ground squirrel was metabolically labeled by ingesting heavy-isotope substituted (15N) Spirulina. The liver protein extract from this animal provided a common reference for quantitative evaluation of protein differences by its addition to extracts from pooled samples of summer active (SA) or winter entrance (Ent) phase hibernating ground squirrels. We identified 61 significantly different proteins between the two groups and compared them to proteins identified previously in the same samples using 2D gels. Of the 20 proteins common to the two datasets, the direction and magnitude of their differences were perfectly concordant for 18, providing confidence that both sets of altered proteins reflect bona fide differences between the two physiological states. Furthermore, the 41 novel proteins recovered in this study included many new enzymes in pathways identified previously: specifically, additional enzymes belonging to the urea cycle, amino acid and carbohydrate degradation, and lipid biosynthetic pathways were decreased, whereas enzymes involved in ketone body synthesis, fatty acid utilization, protein synthesis and gluconeogenesis were increased in the samples from entrance hibernators compared to summer active animals, providing additional specific evidence for the importance of these pathways in the hibernating phenotype.
Keywords: heavy isotope, hibernation, Ictidomys tridecemlineatus
1. Introduction
In contrast to most mammals, 13-lined ground squirrels and other hibernators become heterothermic in winter. Heterothermy consists of repeated cycling between multiday periods of torpor, during which core Tb drops to near freezing, and short (10–12 hr) periods of euthermia, known as interbout arousal (IBA, Fig. 1). Dramatic depressions of metabolic, heart and respiratory rates are also characteristic of torpor. At the molecular level, basic cellular processes such as DNA replication, transcription, and protein synthesis slow or cease. All of these depressed molecular and physiological processes are fully restored, however, during each IBA (Carey et al., 2003; van Breukelen and Martin, 2002). Tissues able to sustain these dramatic swings in metabolism, oxygen delivery and temperature are expected to be more resistant to damage resulting from conditions of low oxygen delivery. Consistent with this expectation, there are numerous reports of resistance to experimentally-induced ischemia-reperfusion injuries in various organs and tissues from hibernators (Dave et al., 2006; Frerichs and Hallenbeck, 1998; Kurtz et al., 2006; Lindell et al., 2005; Ma et al., 2005).
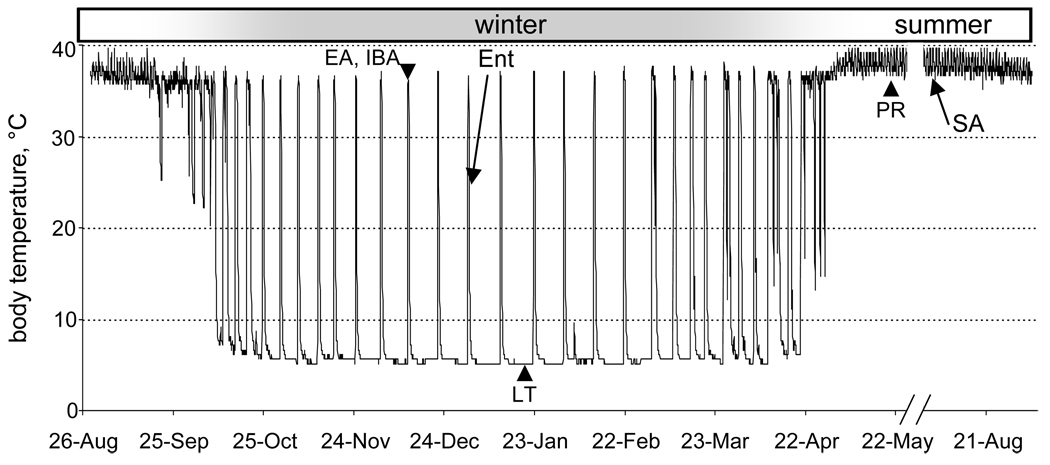
Fig. 1.
Body temperature of a 13-lined ground squirrel over a year. Note summer homeothermy and winter heterothermy where animals cycle between torpor and spontaneous arousals to euthermia (interbout arousal, IBA). Animals in this study (arrows) were re-entering torpor (Ent) after an IBA, or SA in August. Other stages mentioned in the text are noted with arrowheads including three unique to Shao et al.(2010): PR, post-reproductive; LT, late torpor; and EA, early in the interbout arousal.
Livers taken from hibernating 13-lined ground squirrels are resistant to damage caused by cold storage and reperfusion. This protection occurs throughout the hibernation season and is independent of torpor status; i.e., livers taken from both interbout aroused and torpid ground squirrels are more resistant to cold storage and reperfusion injury than livers from summer-active ground squirrels or rats (Lindell et al., 2005). The mechanism of resistance has not been fully established. We hypothesize that the reprogramming of gene expression that occurs during winter hibernation in ground squirrel liver at the mRNA (Epperson and Martin, 2002; Williams et al., 2005; Yan et al., 2008) and protein (Epperson et al., 2004; 2010; Shao et al., 2010) levels plays a role in protecting the hibernating liver from damage, and that these differences will be reflected by differences between the liver proteomes of summer-active (SA) ground squirrels and those re-entering torpor (Ent) following an IBA.
Our previous work provided information about liver protein differences between SA and Ent ground squirrels (Epperson et al., 2004; 2010). Entrance into torpor (Fig. 1) was used because this timepoint appears to re-establish a proteome with adequate integrity for quantitative 2D gel electrophoresis and analysis (Epperson et al., 2004). Although 2D gels offer a robust method for quantification of protein differences between two states, they have the disadvantage of excluding most membrane proteins, a group that is likely to be important for maintaining homeostasis in hibernation. Membrane proteins are notoriously difficult to solubilize in the first dimension isoelectric focusing buffer (Santoni et al., 2000). In addition, many membrane proteins are removed by the chloroform:methanol extraction that has proven critical to obtain high-quality resolution in the isoelectric focusing dimension. A method was developed for the successful analysis of membrane proteins together with soluble components in large-scale proteomic screens using MS/MS (Wu et al., 2003); however, this method was not quantitative and could not be used to compare the proteome from different physiological states.
Here, we applied a method to metabolically label all proteins in vivo with 15N (Wu et al., 2004) by feeding newly-weaned ground squirrels 15N-labeled Spirulina. Proteins extracted from the livers of these 15N-labeled animals were then used as an internal reference standard for comparison of pooled samples from untreated SA and Ent animals, in order to quantify protein differences between these two states of the circannual hibernation cycle without the use of 2D gels. The same Ent and SA liver extracts were used previously in a proteomics screen employing a 2D gel-based technique (Epperson et al., 2010), allowing direct comparison of the results obtained using the two methods. Although the present approach only modestly increased the number of membrane proteins recovered we observed excellent concordance of the direction and magnitude of protein differences between SA and Ent livers for proteins recovered by both methodologies. In addition, many previously unreported proteins were found to differ seasonally. Metabolic enzymes involved in the urea cycle, amino acid and carbohydrate degradation and lipid biosynthesis decreased in Ent compared to SA, whereas enzymes involved with ketone body synthesis, fatty acid metabolism and gluconeogenesis increased in Ent compared to SA, along with several proteins involved in protein synthesis, folding and stability. Hence, the use of the 15N internal standard confirmed and substantially extended the findings reported previously for liver protein differences between these two states.
2. Materials and methods
2.1. Animals
13-lined ground squirrels (Ictidomys tridecemlineatus) were trapped in July or August in the vicinity of Madison, WI, USA. Twelve squirrels were housed individually under a 12:12 h light:dark cycle with food (Purina rat chow supplemented with sunflower seeds) and water ad libitum at 22 °C. Livers were harvested from six summer animals killed by decapitation in early to mid-August, after they had been laboratory-housed for at least 3 weeks. Six additional animals were surgically implanted with radiotelemeters (VMFH disks, Minimitter, Bend, OR, USA) in late August or September to allow undisturbed monitoring of Tb, and hence torpor and arousal cycles, as reported previously (Serkova et al., 2007). Hibernators in the entrance phase of the torpor-arousal cycle were used for this study. Animals were killed when Tb<27 °C but ≥21 °C following an interbout arousal after exhibiting regular torpor-arousal cycles for 1–5 months. Three additional ground squirrels were fed a special diet of protein-free rat chow (Harlan) supplemented with the blue green algae, Spirulina platensis (Spectra Stable Isotopes), as the sole source of nitrogen from weaning until killed for tissue harvest at 12 weeks. For two of these three animals, the Spirulina was >99% 15N-labeled, whereas the third had natural abundance isotopes. All livers were removed, flash-frozen in liquid nitrogen, and then stored at −80 °C. The University of Wisconsin Institutional Animal Care and Use Committee approved all animal procedures.
2.2. Protein extracts
Frozen liver tissue (0.1–0.18 g) from each individual was homogenized in 100 mM potassium phosphate, 0.5 M sucrose, 5 mM MgCl2, 1 mM PMSF, and 10 µg/mL mammalian protease inhibitor cocktail (Sigma P8340) using a 7 mm generator probe attached to a Brinkmann Kinematica Polytron homogenizer. This homogenate was passed 10 times through a 25 gauge needle and then centrifuged for 10 min at 500 g at 4 °C. Aliquots (30 µL) of the supernatant were snap frozen in liquid N2; each of these was thawed and used only once. The protein concentration of each liver extract was determined by comparison with a BSA standard using the Pierce Micro BCA Protein Assay Kit following the manufacturer’s recommendations. The 15N-enrichment in hydrolyzed liver proteins from animals fed the 15N-substituted diet was determined using gas chromatography/mass spectrometry as described (Wu et al., 2004).
2.3. Protein and peptide fractionation
Protein (100 µg) from six each of Ent and SA liver extracts were individually pooled to make the Ent and SA samples, respectively. 400 µg of the 15N-labeled liver protein extract was mixed into each pool and 125 µg of each mixture were separated by SDS-PAGE (17.5”× 16”× 0.1”, 8–16% gradient polyacrylamide, 1:37.5 bis:acrylamide). The gels were stained with SyproRUBY (Bio-Rad), destained, and the image captured on a Typhoon 9400. Each protein-containing lane was excised under UV illumination, and then cut into 2 mm slices. Each slice, containing a mixture of proteins in a narrow molecular weight window, was divided into thirds for protease treatment; proteins were digested to peptides in situ as described previously (Epperson et al., 2004), except that subtilisin (Sigma) and elastase (Roche) were used in addition to trypsin in three separate reactions. All proteolytic enzymes were used at the same concentration (0.1–0.2 µg/µL) and the peptide-containing supernatants from the extractions were combined. The pooled peptides extracted from each slice were resuspended in 10–20 µL of 5% formic acid and applied to a nanocolumn (Supelco 0.1 mm ID fused silica pulled to a 5 µm-diameter tip, packed with 7 cm of Aqua reverse-phase C18 followed by 5 cm of SCX Partisphere) using a pressure bomb. After washing, peptides were eluted directly into an LCQ-Deca ion trap mass spectrometer using a seven-step MuDPIT. The first six steps began with a three-minute flow of 0, 10, 20, 30, 40 or 50% 500 mM ammonium acetate solution, or a 22 min flow for the last salt step at 100%. Each salt bump eluted a subset of the complex peptide mixture inline onto the reverse phase portion of the column, which was then eluted using a 0–80% acetonitrile gradient over 3 h. Xcalibur 1.2 software was used for data collection.
2.4. Peptide Sequence Identification
Tandem mass spectra were filtered using ChargeCzar (Klammer et al., 2005) and analyzed twice using a normalized implementation of the database searching program Sequest (Eng et al., 2008) creating two separate sets of output files. As described previously (Wu et al., 2004), the first database search (standard) used a sequest.params file containing only a static modification of +57 m/z on cysteine resulting from disulfide reduction and alkylation during the protein digest. The second database search (corrected for 15N incorporation) used a parameter file that contained a static modification on each amino acid, shifting the average mass to account for the enriched heavy nitrogen atoms. The average enrichment of the sample was assessed using the program Atomizer (MacCoss et al., 2005) and determined to be 93 ape (atom percent excess). All tandem mass spectra were searched against an in-house compiled mammalian database from March 2007 concatenated to a shuffled decoy database. The program DTASelect (Tabb et al., 2002) was used to filter for peptide sequences while maintaining an FDR<5% at the protein level as previously described (Blackler et al., 2006).
2.5. Ion Chromatogram Extraction
For each peptide exceeding the DTASelect filter criteria in both the standard and 15N corrected database searches, ion chromatograms were extracted from the Xcalibur data file for the unlabeled and 15N-enriched peptide isotope distributions as described previously (Wu et al., 2004).
2.6. Calculation of Ion Current Ratios and Estimation of Protein Ratios
Each pair of ion chromatograms extracted from the Xcalibur data file was analyzed using the program RelEx (MacCoss et al., 2003). An in-house matching algorithm was then applied to the identified proteins in each group to collapse peptides from orthologous proteins into a single protein entry (Epperson et al., 2004). Briefly, proteins that shared at least one peptide or the species-truncated protein name were grouped together as a single ground-squirrel protein. These within-season unique protein groups were then recursively matched by peptide and truncated name between seasons to create the list of common proteins, along with the relative expression of the 15N-enriched peptides found in each season for each protein group. Statistical differences between the ratio of unlabeled and 15N-enriched peptides of SA compared to Ent samples were assessed as previously described (Wu et al., 2004); a t-test p-value of ≤0.05 was used for reporting (Tables 1, 2 and S1). Any proteins below this threshold or with only one peptide found in either state were eliminated from further consideration, as were the single peptide false-positive entries from the scrambled database and all keratin and protease contaminants.
Table 1.
Proteins significantly increased in liver extracts from Ent hibernators
| GI | protein | Ent/SA | p-value | gene | species | function |
|---|---|---|---|---|---|---|
| 28872725 | proteasome 26S non-ATPase subunit 11 | 2.68 | 6.63E-03 | PSMD11 | H. sapiens | protein catabolism |
| 19527028 | high density lipoprotein binding protein | 2.08 | 9.62E-04 | HDLBP | M. musculus | cellular cholesterol homoeostasis |
| 52345385 | protein disulfide isomerase-related protein | 1.92 | 2.55E-06 | PDIA6 | R. norvegicus | protein folding |
| 34577110 | fructose-bisphosphate aldolase A | 1.85 | 2.57E-02 | ALDOA | H. sapiens | gluconeogenesis |
| 4507677 | tumor rejection antigen (gp96) 1 | 1.71 | 5.46E-23 | HSP90B1‡ | H. sapiens | protein folding |
| 4758950 | peptidylprolyl isomerase B, cyclophilin B | 1.68 | 9.20E-03 | PPIB | H. sapiens | protein folding |
| 16507237 | heat shock 70kDa protein 5 | 1.66 | 2.89E-18 | HSPA5‡ | H. sapiens | protein folding |
| 27807407 | ferritin, light polypeptide | 1.65 | 2.60E-03 | FTL | B. taurus | iron transport and storage |
| 27806321 | aldehyde dehydrogenase 1A1 | 1.64 | 6.57E-03 | ALDH1A1 | B. taurus | aldehyde metabolism |
| 9910128 | aldehyde dehydrogenase 9A1 | 1.57 | 9.21E-04 | ALDH9A1 | M. musculus | amino aldehyde metabolism |
| 32189392 | peroxiredoxin 2 | 1.55 | 1.31E-04 | PRDX2 | H. sapiens | oxidative stress protection |
| 11415030 | H4 histone family, member E | 1.51 | 1.52E-03 | HIST1H4J | H. sapiens | histone |
| 5453832 | oxygen regulated protein precursor | 1.49 | 1.05E-02 | HYOU1 | H. sapiens | protein folding |
| 6679687 | protein disulfide isomerase associated 3 | 1.47 | 2.53E-07 | PDIA3‡ | M. musculus | glycoprotein folding |
| 12083607 | ribosomal protein S14 | 1.46 | 2.43E-03 | RPS14 | R. norvegicus | ribosomal protein |
| 13384620 | heterogeneous nuclear ribonucleoprotein K | 1.40 | 3.78E-05 | hnRNPK‡ | M. musculus | mRNA metabolism |
| 5031751 | 3-hydroxy-3-methylglutaryl-CoA synthase 2 | 1.38 | 6.55E-07 | HMGCS2‡ | H. sapiens | ketone body formation |
| 20849516 | similar to 60S ribosomal protein L12 | 1.35 | 2.21E-02 | RPL12 | M. musculus | ribosomal protein |
| 13386034 | ribosomal protein S13 | 1.35 | 8.02E-03 | RPS13 | M. musculus | ribosomal protein |
| 6680179 | hemoglobin Z, beta-like embryonic chain | 1.34 | 8.98E-03 | HBE1 | M. musculus | oxygen transport |
| 38090038 | similar to Rpl7a protein | 1.31 | 2.03E-02 | RPL7A | M. musculus | ribosomal protein |
| 16758398 | acyl-CoA synthetase long-chain 5 | 1.30 | 4.52E-05 | ACSL5 | R. norvegicus | lipid/fatty acid metabolism |
| 13540663 | betaine-homocysteine methyltransferase | 1.24 | 9.44E-04 | BHMT | R. norvegicus | methionine regeneration |
| 18677763 | hydroxyacyl-Coenzyme A dehydrogenase | 1.24 | 3.15E-06 | HADHA | R. norvegicus | fatty acid metabolism |
| 4503497 | enoyl-CoA hydratase/3-hydroxyacyl CoA DH | 1.24 | 5.13E-02 | EHHADH | H. sapiens | lipid/fatty acid metabolism |
| 16933546 | ribosomal protein P0 | 1.20 | 4.19E-02 | RPLP0 | H. sapiens | ribosomal protein |
| 22122797 | 3-ketoacyl-CoA thiolase B | 1.15 | 2.21E-04 | ACAA1 | M. musculus | lipid/fatty acid metabolism |
Proteins are sorted by fold change, Ent>SA;
previously found by DiGE
Table 2.
Proteins significantly decreased in liver extracts from Ent ground squirrels
| GI | protein | SA/Ent | p-value | gene | species | function |
|---|---|---|---|---|---|---|
| 29293809 | ATP citrate lyase | 6.67 | 2.24E-51 | ACLY‡ | M. musculus | lipid biosynthesis |
| 41872631 | fatty acid synthase | 5.82 | 9.63E-03 | FASN | H. sapiens | fatty acid biosynthesis |
| 21614513 | formyltetrahydrofolate dehydrogenase isoform a | 2.93 | 3.12E-03 | ALDH1L1‡ | H. sapiens | biosynthetic FTHF metabolism |
| 4557585 | fatty acid binding protein 7, brain | 2.16 | 7.48E-08 | FABP7‡ | H. sapiens | fatty acid transport and metabolism |
| 29789000 | succinate-CoA ligase, GDP-forming, beta | 1.89 | 1.38E-03 | SUCLG2‡ | H. sapiens | TCA cycle |
| 6980970 | glutamate oxaloacetate transaminase 1 | 1.86 | 2.23E-02 | GOT1‡ | R. norvegicus | amino acid metabolism; urea cycle |
| 34869518 | pyruvate dehydrogenase (lipoamide) beta | 1.81 | 7.66E-03 | PDHB‡ | R. norvegicus | glycolysis |
| 16758348 | peroxiredoxin 6 | 1.79 | 2.25E-04 | PRDX6 | R. norvegicus | lipid catabolism; response to oxidative stress; redox |
| 8393215 | cystathionine gamma-lyase | 1.76 | 2.71E-03 | CTH | R. norvegicus | cysteine and glutathione biosynthesis |
| 21313536 | dihydrolipoamide S-succinyltransferase (E2) | 1.75 | 4.51E-02 | DLST‡ | M. musculus | TCA cycle |
| 16758920 | cullin-associated and neddylation-dissociated 1 | 1.69 | 2.27E-02 | CAND1 | R. norvegicus | protein ubiquitination |
| 21312260 | aldehyde dehydrogenase 1 family, member B1 | 1.68 | 4.26E-40 | ALDH1B1‡ | M. musculus | carbohydrate metabolism |
| 20070418 | aldehyde dehydrogenase family 7, member A1 | 1.67 | 2.99E-03 | ALDH7A1‡ | M. musculus | detoxification |
| 6753272 | catalase | 1.65 | 7.96E-44 | CAT‡ | M. musculus | response to oxidative stress |
| 47522786 | UDP glucose pyrophosphorylase | 1.47 | 9.34E-04 | UGP2 | S. scrofa | glycogen synthesis |
| 13699868 | methylenetetrahydrofolate dehydrogenase 1 | 1.47 | 1.78E-02 | MTHFD1‡ | H. sapiens | amino acid metabolism, one carbon pool/folate |
| 14269572 | heat-responsive protein 12 | 1.44 | 6.00E-03 | HRSP12 | R. norvegicus | regulation of translational termination |
| 51705066 | carbamoyl-phosphate synthetase 1 | 1.43 | 8.38E-24 | CPS1 | M. musculus | urea cycle (rate limiting) |
| 8659555 | aconitase 1 | 1.40 | 1.59E-02 | ACO1 | H. sapiens | TCA cycle and iron homeostasis |
| 19526790 | methionine adenosyltransferase I, alpha | 1.37 | 3.00E-02 | MAT1A | M. musculus | S-adenosylmethionine biosynthesis |
| 19743875 | fumarate hydratase precursor | 1.36 | 4.58E-05 | FH | H. sapiens | TCA cycle |
| 11095441 | aldehyde dehydrogenase 6A1 precursor | 1.36 | 3.34E-07 | ALDH6A1 | H. sapiens | valine and pyrimidine catabolism |
| 11068137 | hydroxyacid oxidase 1 | 1.34 | 9.01E-04 | HAO1 | H. sapiens | fatty acid oxidation (dietary) |
| 34858436 | prohibitin-2 | 1.33 | 6.09E-03 | PHB2 | R. norvegicus | protein chaperone;mitochondrial integrity |
| 47522692 | long-chain acyl-CoA dehydrogenase | 1.31 | 3.18E-04 | ACADL | S. scrofa | fatty acid metabolic process |
| 27806831 | nicotinamide nucleotide transhydrogenase | 1.30 | 1.82E-03 | NNT | B. taurus | electron transport/TCA cycle |
| 47522912 | arginase I | 1.30 | 3.48E-02 | ARG1‡ | S. scrofa | amino acid metabolism; urea cycle |
| 4503301 | 2,4-dienoyl CoA reductase 1 precursor | 1.29 | 5.44E-03 | DECR1 | H. sapiens | metabolism (lipid and lipoprotein, unsat'd fa) |
| 31980648 | ATP synthase, mitochondrial F1 cplx, beta | 1.28 | 2.36E-02 | ATP5B‡ | M. musculus | ATP syntheis |
| 15100179 | malate dehydrogenase 1, NAD (soluble) | 1.26 | 1.58E-02 | MDH1‡ | R. norvegicus | TCA cycle, malate-asp shuttle |
| 21313272 | phosphogluconate dehydrogenase | 1.25 | 3.37E-03 | PGD | M. musculus | pentose phosphate; CHO metabolism |
| 6678359 | transketolase | 1.22 | 3.80E-03 | TKT‡ | M. musculus | pentose phosphate; CHO metabolism |
| 5174429 | acetyl-coenzyme A acyltransferase 2 | 1.19 | 3.38E-10 | ACAA2 | H. sapiens | fatty acid metabolism; cholesterol biosynthesis |
| 27807237 | ATP synthase, mitochondrial F1 cplx, alpha | 1.15 | 1.01E-03 | ATP5A1 | B. taurus | ATP synthesis |
Proteins are sorted by fold change, SA>Ent,
abbreviations used: cplx, complex; DH, dehydrogenase;CHO, carbohydrate; unsat'd, unsaturated
previously found by DiGE
2.7. Membrane Protein Prediction
Because full-length ground squirrel sequences were generally not available, membrane spanning helices were predicted in the proteins recovered in this study in the retrieved FASTA sequences of the full-length ortholog, typically human, using TMHMM (Krogh et al., 2001, http://www.cbs.dtu.dk/services/TMHMM/).
2.8. Gene Enrichment Analysis
Functional annotation clustering was done using the DAVID webserver (http://david.abcc.ncifcrf.gov/home.jsp), version 6.7 (Dennis et al., 2003; Huang et al., 2009), to identify major biological themes underlying the phenotypic transition between SA and Ent hibernators. The proteins that increased or decreased in Ent hibernators were analyzed for overrepresented annotation features included in the default settings. Only those clusters having at least three annotations with p ≤0.05 are reported.
3. Results
This proteomics screen was designed to bypass the use of 2D gels to quantify liver protein differences between SA and Ent hibernators. Quantification was achieved by calculating the relative ratio of natural abundance peptides to their heavy-isotope counterparts in a common reference standard (Fig. 2). Liver extracts were used for several reasons: liver plays a central role in metabolic regulation; the same livers were analyzed previously using a 2D gel approach incorporating internal standards and CyDye labeling (Epperson et al., 2010) allowing a direct comparison of the protein similarities and differences revealed by the two methods; results from two previous proteomic screens of liver extracts from related hibernators, golden-mantled (Epperson et al., 2004) and arctic (Shao et al., 2010) ground squirrels, are also available and provide additional datasets for comparison. By avoiding 2D gels in this study we hoped to increase the numbers of membrane proteins evaluated between these two dramatically different physiological states.
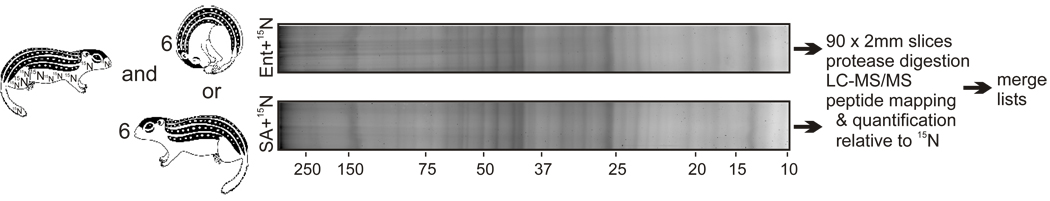
Fig. 2.
Schematic of experimental strategy. Leftmost squirrel was metabolically labeled by feeding 15N-Spirulina. Liver protein from this animal was mixed with an equal amount of liver protein from a pool of either 6 SA or 6 Ent ground squirrels, and the proteins were fractionated by size through SDS-PAGE. After electrophoresis, the proteins in each 2 mm gel piece were proteolyzed to release peptides for analysis by LC-MS/MS. Numbers below the SA lane mark the locations of protein molecular size standards in kDa.
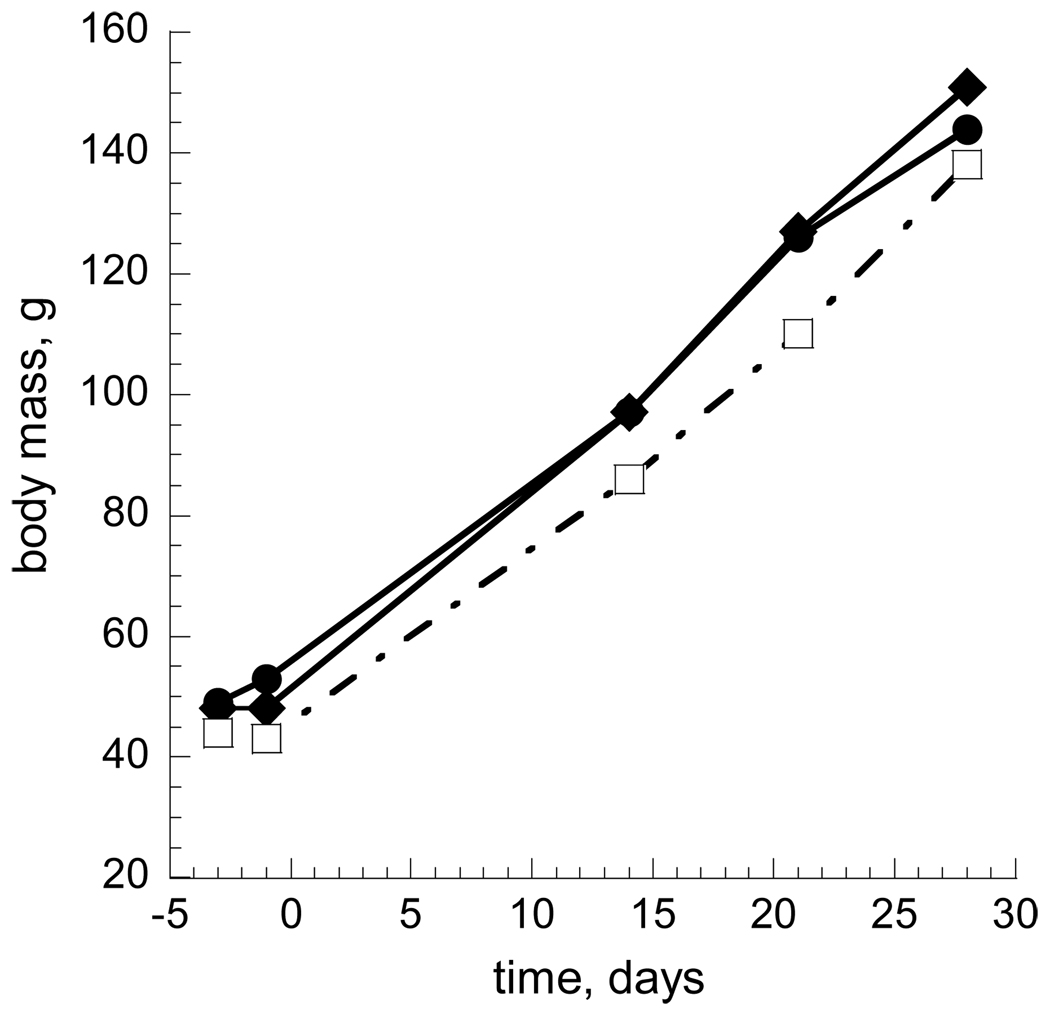
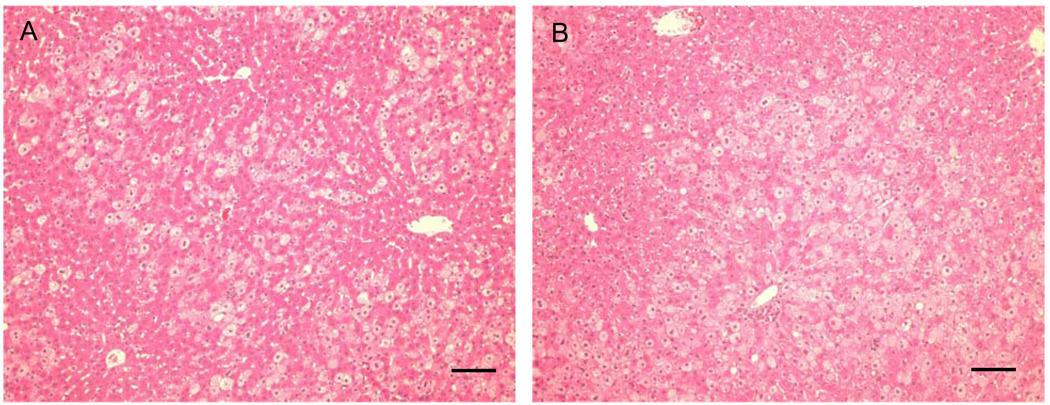
The internal reference standard labeled with 15N comprised total liver protein extracted from a young 13-lined ground squirrel fed a special diet for 4 weeks immediately after weaning in which all nitrogen came from 15N-substituted Spirulina. Ground squirrels fed the 15N diet grew to the same body weight as their littermates fed a normal diet; no differences were noted in behavior, appearance, or growth rates as assessed by body weight (Fig. 3). In the 4-week labeling period, the ground squirrels approximately doubled their body weight. Liver histology was normal (Fig. 4) in the presence of 15N, as reported previously for rats (Wu et al., 2004). Based on GC/MS analysis of hydrolyzed liver proteins in each of the two animals fed the heavy diet, the average 15N enrichment in amino acids was 92.65 and 92.98%, respectively.
Fig. 3.
Normal weight gain of ground squirrels fed 15N. Three animals were fed protein-free rat chow supplemented with Spirulina from weaning (day 0). Animal 2055 (open squares) received Spirulina containing natural abundance nitrogen isotopes, whereas the Spirulina for animals 2053 and 2054 (filled circles and diamonds, respectively) contained at least 99% 15N.
Fig. 4.
Normal liver morphology in Spirulina-fed 13-lined ground squirrels. Representative micrographs of liver sections from A) 15N-enriched, or B) conventional (14N) Spirulina control as the only source of dietary nitrogen, stained with hematoxylin and eosin. Scale bar is 0.1 mm.
The 15N reference was mixed into a pooled sample containing equal amounts of protein extract prepared from six individual SA livers, and another sample containing a mixture from six Ent livers. These mixtures were fractionated by SDS-PAGE to increase the probability that multiple peptides corresponding to a single protein would be recovered. After electrophoresis, each gel lane, containing either the pooled SA or Ent sample spiked with the 15N reference standard, was excised and cut into 88 2mm sections. Each gel slice therefore contained a complex, size-restricted mixture of liver proteins. These gel sections were individually processed to release peptides, and peptide mixtures were analyzed by multi-dimensional protein identification technology (MuDPIT, Fig. 2, Washburn et al., 2002; Wu et al., 2003). We recovered 652 and 566 non-redundant orthologous protein groups from the SA and Ent gel lanes, respectively, after filtering for contaminants and redundancy as described in section 2.6. 389 of these groups were matched between seasons on the basis of shared peptides and species-truncated protein name. Of these, RelEx analysis revealed that 61 distinct liver proteins differed significantly (p<0.05) between the two states: 27 were increased in Ent hibernators (Table 1) and 34 were decreased (Table 2) relative to SA.
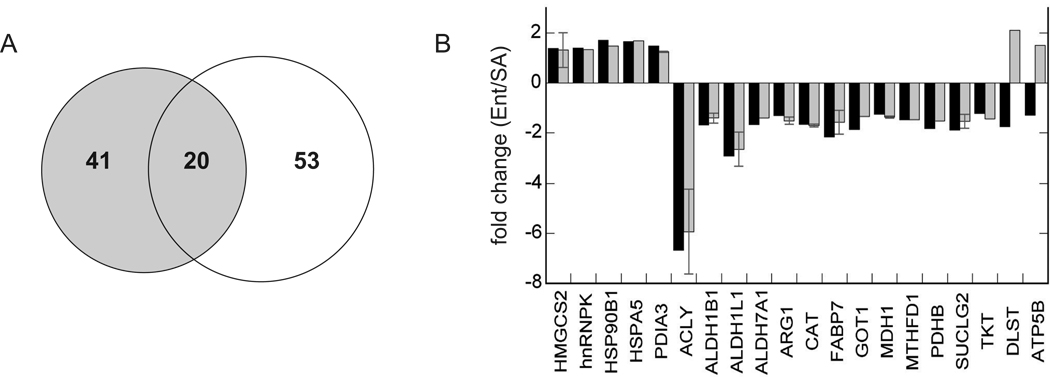
These same Ent and SA liver protein extracts were used previously to screen for protein differences using a quantitative 2D gel method (Epperson et al., 2010), providing a unique opportunity to compare the proteins recovered using the two techniques. Although the majority of protein differences between Ent and SA livers were revealed by just one of the two methods (Fig. 5A), the magnitude and direction of change of the 20 proteins recovered by both methods were highly concordant, lending validity to both methods. Specifically, 18 of the 20 were perfectly concordant for both datasets, with just two differing in the direction of change (Fig. 5B).
Fig. 5.
Comparison of liver protein differences between SA and Ent hibernators as revealed by DiGE and stable isotope quantification. A. Venn diagram depicting unique proteins that differ between Ent and SA that were revealed by each method or by both methods. B. Fold changes of the 20 proteins that were recovered by both of the two methods, quantified by 15N or DiGE (Epperson et al., 2010), black or gray bars, respectively. In the DiGE method it was not uncommon to identify the same protein in several spots, therefore, where multiple spots with the same protein were detected, the fold change is plotted as the average ± one standard deviation.
This proteomics screening approach was specifically undertaken to try to enhance recovery and assessment of membrane proteins by avoiding the use of isoelectric focusing, which is not well-suited for analysis of membrane proteins (Santoni et al., 2000). Twelve of the proteins recovered in this study were annotated to the GO cellular component category of organelle membrane and nine of these were contained in a subset annotated as mitochondrial inner membrane using the DAVID webserver (Dennis et al., 2003; Huang et al., 2009). However, database annotations regarding membrane association are often ambiguous, failing to distinguish membrane-associated proteins from true integral membrane proteins. Approximately 20–30% of cellular proteins are thought to be integral membrane proteins (Krogh et al., 2001; Stevens and Arkin, 2000), i.e., proteins that contain membrane spanning helices, which can be reliably predicted from primary sequence data (Krogh et al., 2001) using a web server tool (http://www.cbs.dtu.dk/services/TMHMM/). The proteins listed in Tables 1 and 2 were examined for membrane spanning helices using TMHMM. Of the 61 uniquely identified proteins in this study, six are predicted to contain membrane spanning helices: protein disulfide isomerase-related protein (PDIA6), cyclophilin B (PPIB), oxygen regulated protein precursor (HYOU1), acyl-CoA synthestase long-chain 5 (ACSL5), aldehyde dehydrogenase 6A1 (ALDH6A1), and nicotinamide nucleotide transhydrogenase (NNT). The latter protein contains 12 predicted transmembrane domains, whereas the other proteins each contain only one. Thus, NNT is likely the only transmembrane protein recovered in this analysis that theoretically could not have been found using 2D gels, which generally fail to resolve proteins with two or more transmembrane domains (Kline and Wu, 2009). None of the six proteins with predicted TM domains are among the list of proteins recovered by both methods (Fig. 5B), however three of the proteins recovered by DiGE: SLC27A2, CYB5A and ACMSD, are predicted to have TM domains using TMHMM. In the present study, Tables 1 and 2 combined contain 61 unique proteins; therefore, if no biases prevent membrane proteins from changing as a function of hibernation status, and they were efficiently recovered by this method that does not rely on in-gel isoelectric focusing, 12–18 transmembrane-containing proteins would be expected.
To assess whether the limited number of membrane proteins recovered in this screen for protein differences between Ent and SA livers was simply a reflection of an inefficiency of the method in recovering membrane proteins, or if it instead represents a bias against membrane protein changes depending upon hibernation status, the complete lists of proteins identified in the present study were also analyzed by TMHMM. TM domains were predicted in 72 (of 652, 11.0%) and 43 (of 566, 7.6%) of the proteins in the SA and Ent lists, respectively. Both lists again had substantially fewer than the expected 20–30% of TM domain-containing proteins, indicating that a bias against recovery of proteins with TM domains simply propagated to the list of proteins that differed between the two hibernation states. Thus, from the available data it is most reasonable to conclude that proteins with TM domains are as likely to differ between SA and Ent animals as those without TM domains and that membrane proteins are underrepresented in the proteins that can be evaluated by this method.
A gene enrichment analysis was performed on the lists of proteins that were increased or decreased in Ent livers using DAVID. With the list of all liver proteins that were recovered in this experiment as background, the proteins that increased during Ent were enriched in processes of fatty acid metabolism (mainly catabolism), regulation of apoptosis (anti-apoptosis) and cellular redox homeostasis. Over-represented cellular components were endoplasmic reticulum and ribosome (Table 3). In contrast, the proteins increased in SA were involved with catabolism of nitrogen, ATP and nucleotides, and energy derived from the oxidation of organic compounds. They were also enriched in mitochondrial and lysosomal proteins (Table 4). When the human database was used as the background for the DAVID analysis instead, similar results were obtained, except that glucose catabolism became significantly enriched in SA (not shown). These findings strongly support previous results demonstrating a substantial change in metabolic and biosynthetic priorities between livers from Ent and SA 13-lined ground squirrels (Epperson et al., 2010).
Table 3.
Enriched DAVID annotation categories in the list of proteins that increased in Ent
| cluster | cluster enrichment |
number of annotations |
top enrichment |
number of proteins |
|---|---|---|---|---|
| organelle lumen | 6.1 | 3 | 5.3 | 13 |
| fatty acid metabolism | 3.4 | 4 | 66.3 | 5 |
| endoplasmic reticulum | 3.1 | 13 | 25.4 | 6 |
| regulation of apoptosis | 2.1 | 3 | 9.2 | 4 |
| anti-apoptosis | 1.8 | 4 | 25.6 | 3 |
| cell redox homeostasis | 1.5 | 4 | 12.2 | 3 |
| fatty acid beta-oxidation | 1.4 | 9 | 12.8 | 3 |
| ribosome | 1.1 | 10 | 38.7 | 4 |
Table 4.
Enriched DAVID annotation categories in the list of proteins that decreased in Ent
| cluster | cluster enrichment |
number of annotations |
top enrichment |
number of proteins |
|---|---|---|---|---|
| mitochondrion | 5.2 | 3 | 7.1 | 13 |
| nitrogen cmpd catabolic process | 3.9 | 3 | 29.0 | 5 |
| mitochondrial matrix | 3.1 | 5 | 5.3 | 9 |
| aldehyde dehydrogenase | 2.7 | 3 | 16.4 | 4 |
| lysosome | 2.1 | 3 | 27.1 | 3 |
| ATP catabolic process | 1.7 | 16 | 43.0 | 3 |
| mitochondrial envelope | 1.5 | 7 | 3.0 | 7 |
| energy from oxidation of organic cmpds | 1.3 | 8 | 2.6 | 10 |
cmpd, compound
4. Discussion
In this study we employed a novel approach to examine the liver proteome of 13-lined ground squirrel, quantifying protein differences between SA and Ent hibernators by comparison to a heavy-isotope labeled internal reference standard. This approach identified 61 unique proteins that differed significantly between the livers of hibernating and summer animals; 27 increased and 34 decreased, between Ent and SA, respectively. Previously, we reported the identification of 73 unique proteins that differed significantly between these two sets of liver protein extracts using a quantitative 2D gel method (Epperson et al., 2010), 20 of which overlapped with the present dataset. In spite of the dramatically different methods of protein quantification used in the two studies, almost all (18) of the proteins that were recovered by both methods were perfectly concordant with respect to both their direction and magnitude of change (Fig 5B), thereby strengthening the credibility of the findings from both studies. The two that differed likely reflect the differences in the methods used; i.e., with DiGE, specific isoforms were recovered, whereas these were not distinguished by the 15N method. Another important methodological difference is that the extracts underwent a chloroform:methanol extraction before analysis by DiGE which alters the protein composition of the sample.
The proteins in the present dataset increase our quantitative knowledge of the hibernating liver proteome by 56%, providing 41 new protein identifications associated with the phenotypic differences between SA and Ent animals. Thus we establish here not only the viability of this alternative method for quantitative proteomics, but also its effectiveness in providing an orthogonal method for both validation and data discovery. The availability of newer instrumentation with greater mass accuracy and sensitivity will enhance the utility of this approach in future experiments.
The present experimental strategy was implemented with a goal of enhancing the recovery of liver membrane proteins that differ between SA and Ent hibernating ground squirrels by avoiding the use of 2D gels. Six proteins (of 61, 9%) with transmembrane domains were recovered using the 15N internal reference standard, compared to three (of 73, 4.1%) recovered by DiGE in Epperson et al. (Epperson et al., 2010). These numbers are not significantly different (p=0.17, Fisher’s exact test), although there is a trend towards an increased recovery of membrane proteins using the present method. However, both methods fall substantially short of recovering the expected 20–30% of cellular proteins that are membrane proteins.
Alterations of liver proteins during the annual hibernation cycle have been reported for two other species of ground squirrels; comparison of these datasets reveals surprising discordance. Epperson et al., (2004) used a 2D gel-based method for quantification, followed by LC-MS/MS to identify proteins that differ between SA and Ent golden-mantled ground squirrels (Spermophilus lateralis, recently reclassified as Callospermophilus lateralis, Helgen et al., 2009). Shao et al., (2010) used spectral counts from a shotgun LC-MS/MS analysis of tryptic peptides to simultaneously identify and quantify protein differences in arctic ground squirrels (Urocitellus parryii) from three stages of the circannual cycle. Discordance, both quantitatively and qualitatively, of protein changes between the golden-mantled and the 13-lined ground squirrel studies was noted previously (Epperson et al., 2010), as were differences between the arctic and the golden-mantled ground squirrel data sets (Shao et al., 2010). There are also substantial differences between the present results for 13-lined ground squirrels and those for arctic ground squirrels. Specifically, of the 27 proteins increased in Ent 13-lined ground squirrels (Table 1), 17 did not differ significantly among the three groups of arctic ground squirrels studied, 4 changed in the same direction, and 5 changed in the opposite direction. Table 2 proteins were similarly discordant: 23 of the 34 proteins that decreased in Ent compared to SA were unchanged in the arctic ground squirrel livers, 7 changed similarly, and 1 changed in the opposite direction. One protein from Table 1 and three from Table 2 were not found in the arctic ground squirrel study.
Although some of the differences among the 13-lined, golden-mantled and arctic ground squirrel data sets could be due to species or methodological differences, it is likely that the physiological status of animals in different stages of the circannual rhythm of hibernation is also a major contributing factor. In contrast to the two groups, Ent and SA, used for the 13-lined and golden-mantled proteomic screens, three animal groups were compared in arctic ground squirrels: post-reproductive (PR in May or June), representing the non-hibernating phase, plus two winter hibernation groups, late torpor (LT, near the end of a torpor bout, Fig. 1) and early arousal (EA, 1–2 hr after Tb reached 30°C in a spontaneous arousal). Considering the PR group for arctic ground squirrels to be equivalent to the SA group for 13-lined ground squirrels may be misleading. For example, liver metabolism in May-June is likely directing carbohydrate and amino acids to energy for growth rather than to fatty acid synthesis, in sharp contrast to August when the animals are beginning to fatten for hibernation. The SA group in the golden-mantled study comprised wild-trapped May-July animals, adding further complexity to their comparison with the arctic and 13-lined ground squirrel studies, which both involved animals housed for longer times in the laboratory. Hence a number of proteins may differ among the samples from non-hibernating squirrels in the three datasets because of the timing and hence physiological status of the animals. Likewise, aroused hibernators (EA or IBA) likely replenish proteins that are altered or degraded during torpor (Epperson et al., 2004; van Breukelen and Martin, 2001, 2002), providing a bona fide physiological basis for differences between LT, EA/IBA and Ent among the various studies. Perhaps the greatest lesson from these comparisons is that such screens absolutely require careful monitoring of the physiological status of the animals, and will ultimately need to be done on animals representing multiple timepoints throughout the circannual rhythm of hibernation to truly understand the phenotype.
In summary, the proteins identified as quantitatively different between SA and Ent livers using this approach substantiate and expand on the findings of our earlier proteomics screen that analyzed the same samples using a 2D gel based methodology. In the subset of proteins recovered by both approaches, there was perfect concordance in the direction and magnitude of the changes found by these different methodologies for 18 of the 20 shared proteins. Combining the two datasets reveals an informative 114 liver proteins that differ significantly between SA and Ent hibernators. The majority of these protein differences (62%) are due to decreased abundance in Ent hibernators reflecting several aspects of the metabolic differences that distinguish these two groups. Specifically, the SA animals are engaged in conversion of dietary fuel into stored fat and detoxification of dietary compounds; neither process is needed during the seasonal fast associated with hibernation. This dataset reinforces previous findings (Epperson et al., 2010) showing decreased abundance of enzymes involved in amino acid catabolism, and increased protein biosynthetic capability during hibernation. These data strengthen our understanding that hibernating ground squirrels enact a novel strategy to exploit proteins as a storage depot for amino acids, thereby preserving essential amino acids and preventing nitrogen toxicity during the long winter fast that accompanies hibernation.
Supplementary Material
Acknowledgments
We thank Drs. M. J. MacCoss and C. Wu for help with 15N enrichment, Sequest and RelEx analyses. This work was supported by Defense Advanced Research Projects Agency W81XWH-05-2-0016 to SLM and HVC and National Institutes of Health HL089049 to SLM.
Abbreviations
- ape
atom percent excess
- Ent
entrance into torpor
- IBA
interbout aroused
- MuDPIT
multi-dimensional protein identification technology
- SA
summer active
- Tb
core body temperature
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blackler AR, Klammer AA, MacCoss MJ, Wu CC. Quantitative comparison of proteomic data quality between a 2D and 3D quadrupole ion trap. Anal Chem. 2006;78:1337–1344. doi: 10.1021/ac051486a. [DOI] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Dave KR, Prado R, Raval AP, Drew KL, Perez-Pinzon MA. The arctic ground squirrel brain is resistant to injury from cardiac arrest during euthermia. Stroke. 2006;37:1261–1265. doi: 10.1161/01.STR.0000217409.60731.38. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Eng JK, Fischer B, Grossmann J, MacCoss MJ. A Fast SEQUEST Cross Correlation Algorithm. Journal of Proteome Research. 2008;7:4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Dahl T, Martin SL. Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol Cell Proteomics. 2004;3:920–933. doi: 10.1074/mcp.M400042-MCP200. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Martin SL. Quantitative assessment of ground squirrel RNA levels in multiple stages of hibernation. Physiol. Genomics. 2002;10:93–102. doi: 10.1152/physiolgenomics.00004.2002. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Rose JC, Carey HV, Martin SL. Seasonal proteomic changes reveal molecular adaptations to preserve and replenish liver proteins during ground squirrel hibernation. Am. J. Physiol. Regul Integr Comp Physiol. 2010;298:R329–R340. doi: 10.1152/ajpregu.00416.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerichs KU, Hallenbeck JM. Hibernation in ground squirrels induces state and species-specific tolerance to hypoxia and aglycemia: an in vitro study in hippocampal slices. J Cereb Blood Flow Metab. 1998;18:168–175. doi: 10.1097/00004647-199802000-00007. [DOI] [PubMed] [Google Scholar]
- Helgen KM, Cole FR, Helgen LE, Wilson DE. Generic revision in the holarctic ground squirrel genus Spermophilus. J Mammal. 2009;90:270–305. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Klammer AA, Wu CC, MacCoss MJ, Noble WS. Peptide charge state determination for low-resolution tandem mass spectra; Proc IEEE Comput Syst Bioinform Conf; 2005. pp. 175–185. [DOI] [PubMed] [Google Scholar]
- Kline KG, Wu CC. MudPIT Analysis: Application to Human Heart Tissue. Membrane Proteomics. 2009:281–293. doi: 10.1007/978-1-60327-310-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kurtz CC, Lindell SL, Mangino MJ, Carey HV. Hibernation confers resistance to intestinal ischemia-reperfusion injury. Am J Physiol Gastroint Liver Physiol. 2006;291:G895–G901. doi: 10.1152/ajpgi.00155.2006. [DOI] [PubMed] [Google Scholar]
- Lindell SL, Klahn SL, Piazza TM, Mangino MJ, Torrealba JR, Southard JH, Carey HV. Natural resistance to liver cold ischemia-reperfusion injury associated with the hibernation phenotype. Am J Physiol Gastrointest Liver Physiol. 2005;288:G473–G480. doi: 10.1152/ajpgi.00223.2004. [DOI] [PubMed] [Google Scholar]
- Ma YL, Zhu X, Rivera PM, Toien O, Barnes BM, LaManna JC, Smith MA, Drew KL. Absence of cellular stress in brain after hypoxia induced by arousal from hibernation in Arctic ground squirrels. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1297–R1306. doi: 10.1152/ajpregu.00260.2005. [DOI] [PubMed] [Google Scholar]
- MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR., 3rd A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal Chem. 2003;75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- MacCoss MJ, Wu CC, Matthews DE, Yates JR., 3rd Measurement of the isotope enrichment of stable isotope-labeled proteins using high-resolution mass spectra of peptides. Anal Chem. 2005;77:7646–7653. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Serkova NJ, Rose JC, Epperson LE, Carey HV, Martin SL. Quantitative analysis of liver metabolites in three stages of the circannual hibernation cycle in 13-lined ground squirrels by NMR. Physiol Genomics. 2007;31:15–24. doi: 10.1152/physiolgenomics.00028.2007. [DOI] [PubMed] [Google Scholar]
- Shao C, Liu Y, Ruan H, Li Y, Wang H, Kohl F, Goropashnaya AV, Fedorov VB, Zeng R, Barnes BM, Yan J. Shotgun proteomic analysis of hibernating arctic ground squirrels. Mol Cell Proteomics. 2010;9:313–326. doi: 10.1074/mcp.M900260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JT, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breukelen F, Martin SL. Translational initiation is uncoupled from elongation at 18°C during mammalian hibernation. Am. J. Physiol. 2001;281:R1374–R1379. doi: 10.1152/ajpregu.2001.281.5.R1374. [DOI] [PubMed] [Google Scholar]
- van Breukelen F, Martin SL. Molecular adaptations in mammalian hibernators: unique adaptations or generalized responses? J. App. Physiol. 2002;92:2640–2647. doi: 10.1152/japplphysiol.01007.2001. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- Williams DR, Epperson LE, Li W, Hughes MA, Taylor R, Rogers J, Martin SL, Cossins AR, Gracey AY. Seasonally hibernating phenotype assessed through transcript screening. Physiol Genomics. 2005;24:13–22. doi: 10.1152/physiolgenomics.00301.2004. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JRr. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics. 2008;32:170–181. doi: 10.1152/physiolgenomics.00075.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.