Abstract
Microarray technology has facilitated many powerful high-throughput studies in the fields of genetics and proteomics, among others. However, preparation of microarrays composed of cell-derived membranes with embedded receptors has proven difficult. Here we describe a new method for forming microarrays composed of synthetic lipid vesicles and natural cell membranes. The method is based upon assembly of vesicles and natural membranes into recessed micro- and nanowells and using a polydimethylsiloxane (PDMS) block as a “squeegee.” This method is used to assemble phospholipid vesicles into arrays with micron and nanoscale dimensions. Native myelin and neuronal lipid raft arrays are also formed in 30 minutes or less. We show the natural membrane arrays can be used for sensing lipid-protein interactions by detecting cholera toxin binding to ganglioside GM1 in neuronal lipid rafts. In multicomponent arrays myelin can be distinguished from neuronal rafts by antibody binding to cell-specific surface antigens. Finally, myelin arrays formed in gold nanowells are used for surface plasmon resonance sensing. This assembly approach is simple, broadly applicable and opens up new avenues of research not easily accomplished with standard microarray technology.
Keywords: Microarray, nanoimprint lithography, atomic layer deposition, vesicles, cell membranes, plasmonic biosensing, surface plasmon resonance
Microarray-based assays are commonplace for high-throughput interaction screening for numerous types of biomolecules like proteins, nucleotides and carbohydrates.1–6 Spotting thousands of different probes on a surface enables high-throughput experiments where a multitude of different parameters can be evaluated simultaneously. However, array-format assays for evaluating how molecules bind to their native membrane targets, such as transmembrane proteins and lipid-protein complexes in rafts, are challenging. Membrane-bound proteins can be removed from their native membranes, reconstituted into proteoliposomes and immobilized on solid substrates, but this is not ideal from a biosensing standpoint. Transmembrane proteins can denature when removed from their natural membrane environment and interact with solid substrates when embedded in solid supported lipid bilayers causing denaturation.7 Therefore they are problematic in a conventional microarray format where probe molecules are immobilized directly on a surface. Nearly half of the top selling pharmaceuticals have membrane-bound receptors as targets, therefore new techniques for evaluating these interactions, such as advanced optical biosensing, are crucial.8, 9 Also, some potential therapeutic molecules, like monoclonal IgM antibodies may bind to native lipid-protein complexes making standard array assays difficult.10 Formation of arrays of unmodified natural membranes by simple methods could enable new studies that are difficult to carry out with traditional methods.
To facilitate microarray studies with membrane-bound receptors, many strategies are used to form membrane arrays, both in the form of immobilized vesicle arrays and supported lipid bilayers arrays.11 Many of these approaches require chemical modification of the substrate and lipid membranes to selectively immobilize vesicles in defined arrays. For example, preferential adsorption of attractive and repulsive surface modifiers has been used to direct vesicles to nanometric holes.12–14 Microcontact printing was used by Vogel and coworkers to immobilize single intact vesicles separated by less than 1 μm with a total density of 106 vesicles per mm2.15 Other groups have stamped polyethylene glycol (PEG) on to the top surface of microwell arrays, leaving the interior of the wells PEG-free. The PEGylated surfaces are resistant to vesicle adsorption, but the hole interiors, after functionalization with neutravidin, have high affinity for biotinylated vesicles.16 Another strategy to form vesicle arrays uses hybridization between DNA-conjugated vesicles and immobilized complementary strands.17–19 Other groups used standard pin-based microarray printing techniques20 or patterned self-assembled monolayers21 to define membrane arrays with embedded G-protein coupled receptors. Also, various methods were demonstrated to form supported monolayer22, 23 or bilayer arrays.24–29
A facile, broadly applicable method for forming biomembrane arrays requiring no surface modification, no recognition elements or modification of the lipids, and amenable to naturally derived membranes would facilitate array-based screening for many types of membrane receptor-ligand interactions. In the present study we describe a simple method that can be used to form high-density vesicle and natural membrane arrays. An advantage of this method is that it requires no chemical modification of the substrate beyond microfabrication. Furthermore, there is no need to modify the vesicle membranes to direct assembly, unlike other methods which require incorporation of biotinylated, PEGylated or DNA-conjugated lipids. This provides a great advantage as it facilitates assembly of unmodified natural membrane arrays, as we demonstrate by forming single and multicomponent microarrays of myelin and lipid rafts extracted from cortical neurons. These arrays are used for detecting the presence of membrane-bound receptors in natural membranes. Finally, nanoscale myelin arrays in gold films are used for surface plasmon resonance (SPR) sensing of antibody binding to cell-specific surface antigens. The simplicity of this approach makes it applicable to a large variety of medical and therapeutic molecules and their membrane-bound targets of action.
RESULTS AND DISCUSSION
Assembly of phospholipid vesicle arrays
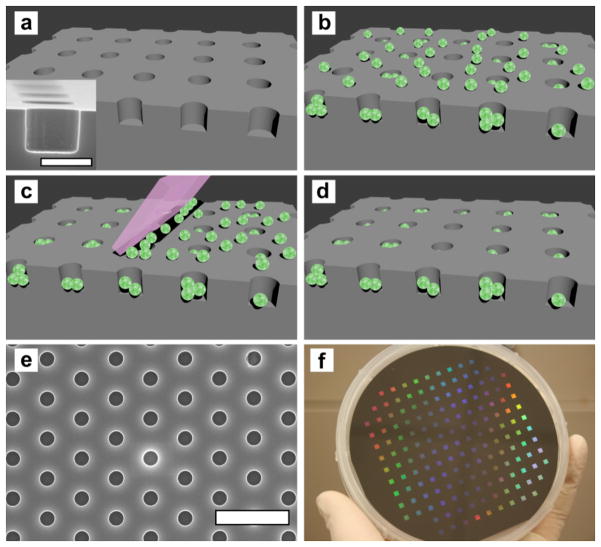
The assembly process is similar to first step of Gravure-type printing, which has been used for assembly of solid nanomaterials into defined patterns.30, 31 Figures 1a-d shows a schematic of the assembly process and a scanning electron microscope (SEM) image of a microwell cross section, while Figure 1e shows a SEM image of a hexagonal microwell array substrate with 1 μm well diameter and depth and 3 μm periodicity. Figure 1f shows a 4-inch silicon wafer with 157 microwell arrays. The silicon microwell arrays are coated with 10 nm thick Al2O3 to facilitate vesicle adhesion but prevent vesicle rupture. The Al2O3 overlayer conforms to the contours of the microwell array, similar to conformal deposition of Al2O3 on plasmonic nanopores shown in our previous work.32 The starting materials for the simplest arrays are fluorescently labeled phospholipid vesicles. Vesicle solutions were placed on the substrates and allowed to settle, then washed with buffer and immersed in a buffer bath. (Figure 1b) To clean vesicles from the top of the microwell array the PDMS squeegee was placed in firm contact with the substrate in the buffer bath and translated by hand across the substrate surface at least 10 times. (Figure 1c)
Figure 1.
Schematic illustration of the array assembly process and images of microwell substrates. (a) Illustration of an array of wells in an Al2O3-coated silicon substrate. Inset: A cross sectional SEM image of a microwell. The scale bar is 1 μm. (b) Vesicles are deposited on the substrate and fill the wells as well as populate the top surface. (c) The PDMS block “squeegee” is translated across the substrate, removing vesicles that are not immobilized in the recessed wells. (d) After using the squeegee, the top surface of the substrate is devoid of vesicles, while the recessed wells are filled. (e) SEM image of a hexagonal microwell array with 1 μm well diameter and 3 μm periodicity. The scale bar is 5 μm. (f) Photograph of a 4 inch wafer patterned with 157 microwell arrays, each with 1 μm well diameter and depth and 3 μm periodicity.
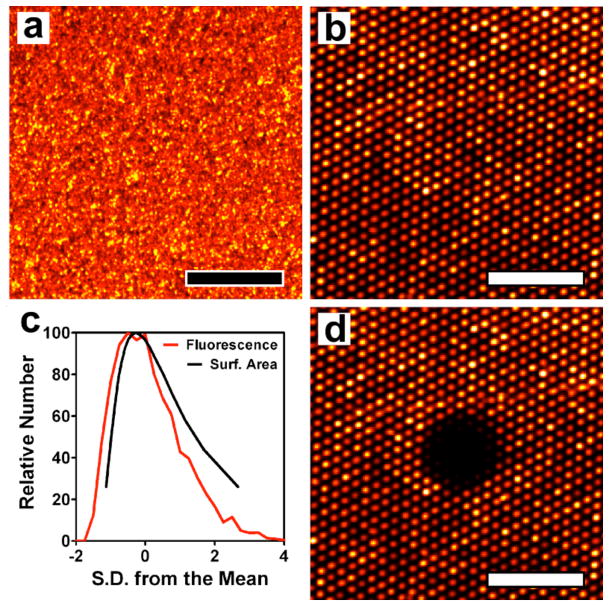
After the vesicles adsorb to the surface they are uniformly distributed over the array. (Figure 2a) After carrying out at least 10 passes with the squeegee, the top surface of the array is cleared but the vesicles remain in the microwells. A fluorescence image of a microarray with 1 μm well diameter and 3 μm periodicity resulting from assembly of fluorescent egg phosphatidylcholine (egg PC) vesicles is shown in Figure 2b. Nearly all of the holes are filled with vesicles, and the interstitial spaces between wells are largely devoid of lipids. An image from a larger area of an array is shown in Figure S1a in the Supporting Information. While it is not straightforward to experimentally determine the exact number of vesicles per well, it is possible to determine the relative amount of lipid material occupying the wells by analyzing the distribution of fluorescence intensity. To determine the distribution of amount of lipid in each well, the fluorescence intensity from 5200 wells was determined, a histogram was constructed and then plotted against the deviation from the mean fluorescence intensity of all holes. (Figure 2c) Also plotted in Figure 2c is the distribution of surface area (determined by dynamic light scattering (DLS)) for vesicles free in solution after extrusion through 200 nm pores. DLS measures the distribution of vesicle diameter, which is transformed into surface area by assuming the vesicles are spherical. Comparing these two distributions indicates that a microarray formed by this method is no less uniform, in terms of the amount of membrane material per well, than if the array were composed solely of individual vesicles. (Detailed discussion in the Supporting Information)
Figure 2.
Fluorescence images of a phospholipid vesicle microarray. (a) A microarray uniformly covered with Rho-PE-labeled egg PC vesicles before applying the PDMS squeegee. (b) A microarray after applying the squeegee showing 1 μm-diameter wells with 3 μm periodicity filled with vesicles. (c) Distribution of average fluorescence intensity (red) from 5200 microarray wells and the distribution of surface area (black) of vesicles extruded 21 passes through a 200 nm polycarbonate filter determined by dynamic light scattering. (d) Fluorescence image of the microarray in (b) 2 minutes after photobleaching a 25 μm-diameter area showing no fluorescence recovery. This indicates the material in the wells is isolated and that a supported lipid bilayer does not form spontaneously. The scale bars in a, b and d are 25 μm.
It is important that no supported lipid bilayer forms over the array and microarray wells are isolated from one another to prevent crosstalk of immobilized vesicles. To determine if membrane materials in the well arrays are isolated from one another, the immobilized vesicles in a 25 μm diameter area were photobleached. Two minutes after bleaching another image taken showed that none of the fluorescence recovered, thus there is no lipid diffusion on the surface and the holes are indeed isolated. (Figure 2d) We repeated the process by photobleaching two individual 1 μm-diameter wells and the fluorescence did not recover after two minutes. (Figure S1b-c in the Supporting Information) The absence of a supported bilayer is expected because the wells are coated with an Al2O3 overlayer deposited by atomic layer deposition (ALD). Others groups have showed that PC vesicles will not spontaneously rupture on Al2O3 surfaces.24, 33 While vesicles will adhere to Al2O3 surfaces, unlike glass34 or SiO2-coated surfaces,35 Al2O3-coated surfaces do not facilitate strong surface-membrane interactions which lead to vesicle rupture. In general, vesicles adsorbed on substrates that do not promote rupture are thought to remain intact on the surface.36 To further illustrate this, we repeated the photobleaching experiment on 1 μm well arrays coated with Al2O3 and SiO2 without using the PDMS squeegee. On Al2O3-coated microwell arrays a photobleached spot shows no recovery of fluorescence indicating that non-ruptured vesicles are adsorbed on the surface. (Figure S2a-c in the Supporting Information) However, when the microwell array is coated with SiO2, the vesicles spontaneously rupture and form a supported lipid bilayer, which is expected based on previous research.35, 37 When a spot of the membrane on SiO2 is photobleached, the fluorescence recovers in a fashion consistent with supported bilayer formation. (Figure S2d–e in the Supporting Information) Therefore to successfully form arrays of isolated vesicles it is important to coat micro- and nanowell arrays with a material that does not facilitate spontaneous vesicle rupture.
Assembly of natural membrane arrays and ligand-receptor binding
A major advantage of this method of array formation is the ability to immobilize natural membranes that cannot be functionalized with recognition elements (e.g. biotin or DNA) or PEG. To demonstrate this capability, microarrays were formed with myelin and the lipid raft membrane fraction isolated from cortical neurons. Myelin is the lipid-rich material produced by oligodendrocytes that insulates axons to facilitate fast neuronal signal transduction in the nervous system. Lipid rafts are nanoscale membrane structures that are rich in cholesterol, sphingolipids and glycosphingolipids, such as ganglioside GM1.38 Many important cellular signaling pathways are dependent on lipid rafts, as they serve to transiently compartmentalize transmembrane proteins that are crucial for many cellular processes.39 However, lipid rafts (and the proteins associated with them) are difficult to interface with traditional biosensors.
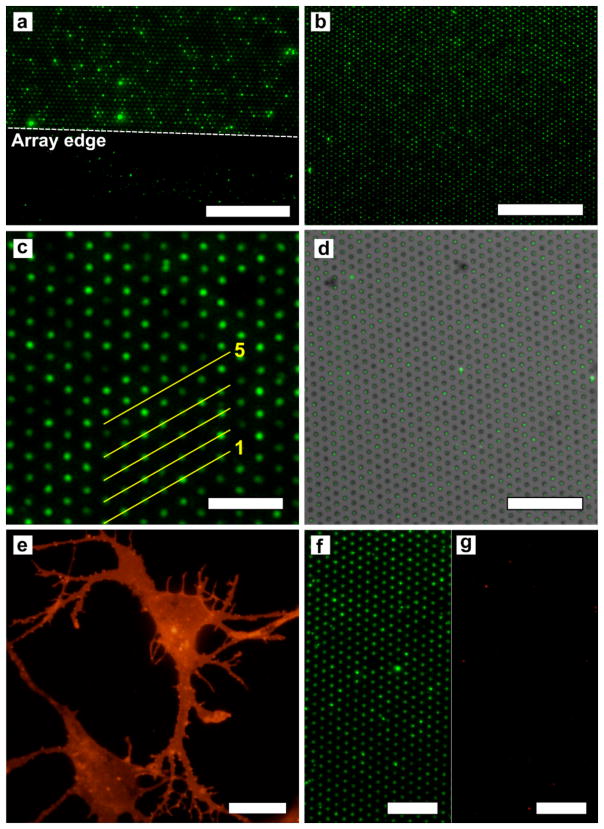
Before applying the squeegee to form the membrane microarrays, the myelin and lipid rafts were uniformly distributed over the substrate surface. (Figure S3a–b in the Supporting Information) Figure 3a shows a fluorescence micrograph of a myelin microarray stained with FM1-43. The field of view shows the edge of the microwell array, demonstrating that the non-array areas are largely devoid of myelin due to removal by the squeegee. Figure 3b shows an FM1-43-stained microarray formed from the lipid raft fraction of membranes isolated from cortical neurons. The microarray has high degree of uniformity and well occupancy approaching 100%. Line scans of the fluorescence intensity across a number of wells in Figure 3c show that there is some variability in the amount of material per well but very little lipid material between wells. (Figure S3c in the Supporting Information)
Figure 3.
Fluorescence images of natural membrane microarrays stained with FM1-43 and GM1 detection with CTX. (a) Myelin microarray showing the array edge, which is indicated by the dashed line. Beyond the edge of the array there is little adherent myelin. The scale bar is 50 μm. (b) Microarray formed with neuronal lipid rafts showing > 99% occupancy. The scale bar is 50 μm. (c) Magnified image of a microarray formed with neuronal lipid rafts. The line scans for lines 1 – 5 can be seen in Supporting Information Figure S5c. The scale bar is 10 μm. (d) A bright field image of a microarray with 1 μm-diameter wells with 3 μm periodicity overlaid with a fluorescence image of FM-143-stained lipid rafts assembled into the wells. In this image there are 975 wells, all of contain fluorescent lipid rafts, therefore the occupancy for this image is 100% over the approximately 80 μm × 80 μm area. The scale bar is 30 μm. (e) Cortical neurons in culture stained with TRITC-conjugated CTX. The scale bar is 10 μm. (f) Neuronal lipid raft microarray rendered fluorescent by CTX binding to GM1. The scale bar is 15 μm. (g) The same array as in (f) showing the red fluorescence channel, which indicates that very little SAPE binds to the neuronal lipid raft microarray. The scale bar is 15 μm.
To quantify well occupancy for natural membrane microarrays, brightfield images of the microwells were compared to fluorescence images of the lipid that occupies the microwells. In many arrays occupancy approaching 100% was achieved. Occupancy is defined as the number of fluorescent spots divided by the number of microwells in a given area. Figure 3d shows a bright field and fluorescence overlay for a lipid raft microarray that has 100% well occupancy over an approximately 80 μm × 80 μm area.
To demonstrate the functionality of the membrane microarrays for sensing ligand binding to membrane bound receptors, we immobilized lipid raft membranes and detected specific binding of the B-subunit of cholera toxin (CTX). Binding of CTX to the membranes of cells in vitro is a common marker for the location of lipid rafts since they are enriched in GM1 as shown in Figure 3e. Likewise, the lipid raft membrane fraction extracted from neurons should also be enriched in GM1 and strongly bind CTX. To investigate this, a microarray was formed using the lipid raft fraction isolated from cortical neurons then exposed to 10, 50 and 200 nM CTX conjugated to Alexa-488. Figure 3f shows a fluorescence image of the resulting array when lipid rafts were exposed to 50 nM CTX. CTX clearly binds to the lipid raft membranes that are immobilized in the microwells. To evaluate the response with different concentrations of CTX, we compared the responses for individual array spots from each of the three treatment groups (10, 50, 200 nM CTX). The intensity of each individual array spot was determined and the distributions of fluorescence intensities for the three concentrations were plotted in a histogram shown in Figure S4a in the Supporting Information. The distributions clearly shift to larger values with increasing concentration of CTX. The mean intensities of array spots for the three groups are significantly different, determined by ANOVA and post-hoc mean comparisons. (Figure S4b in the Supporting Information) For each concentration of CTX between 2100 and 3200 individual array spots were analyzed, which makes for a large sample size and increased statistical power. As a negative control lipid rafts were exposed to 50 nM streptavidin-R-phycoerythrin (SAPE). Figure 3g shows that minimal SAPE binds to the array, indicating that there is little nonspecific binding.
Multicomponent arrays for immunofluorescence
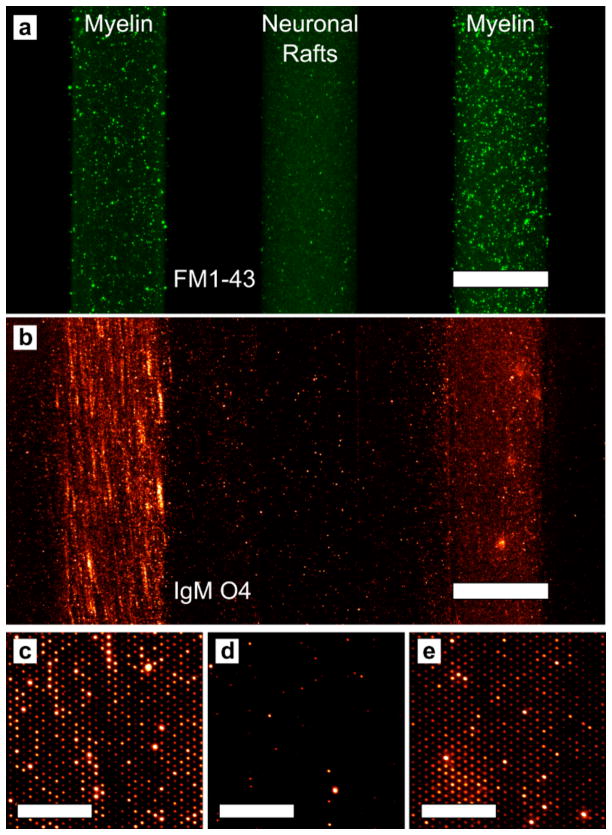
To fully exploit the advantages of a microarray, multiple components can be immobilized in defined patterns. Thus we created three microarray stripes containing myelin and neuronal rafts and used immunofluorescence to identify the myelin. The myelin and neuronal raft membranes were delivered to the substrate via a PDMS microfluidic chip. Prior to injection, the natural membranes were incubated with FM1-43 to render them fluorescent. (Figure 4a) After the myelin and rafts were adherent to the surface, the channels were flushed with PBS to remove weakly immobilized membrane particles. Then the microfluidic chip was detached from the substrate and the PDMS squeegee was employed to form stripes of arrays. The arrays were then incubated with mouse anti-oligodendrocyte marker O4 (IgMO4), an IgM antibody that binds to sulfatide, a major component of myelin but absent from neuronal lipid rafts. Figure 4b shows a low magnification image of the three stripes where the myelin arrays are labeled with fluorescently-conjugated IgMO4. The array stripes, while not as sharply defined after the squeegee are clearly separated from each other. Also, there is no crosstalk between the channels after using the squeegee, indicating that the stripes could be formed much closer to each other. This would greatly increase the number of different membrane types that could be incorporated into a single array. The individual array spots are not visible at low magnification, however, when viewed with higher magnification the individual myelin array spots are clearly visible in Figure 4c–e, due to IgMO4 binding to sulfatide in myelin. A similar experiment was carried out with fluorescently-labeled phospholipid vesicles and can be seen in Figure S5 in the Supporting Information.
Figure 4.
Multicomponent arrays formed by microfluidic delivery of myelin and neuronal raft membranes. (a) Fluorescence image after myelin and rafts were deposited via microfluidic channels on the microarray substrate. The left and right stripes contain myelin and the middle stripe contains neuronal raft membranes. The membranes are stained with FM1-43. The scale bar is 250 μm. (b) Fluorescence image of the same three stripes after applying the PDMS squeegee and incubating with IgMO4. The scale bar is 250 μm. (c–e) Magnified images from the three stripes showing that IgMO4 only binds to the myelin microarrays. (c) is from the left stripe, (d) is from the middle stripe and (e) is from the right stripe. The scale bars in (c–e) are 30 μm.
Nanoarrays and surface plasmon resonance sensing
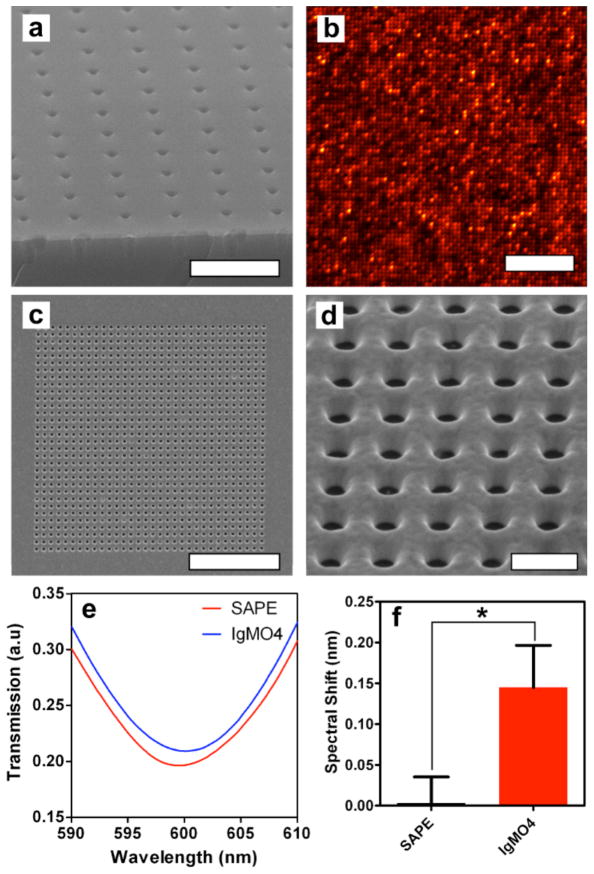
While micron-scale wells allow us to form high-density arrays (1.3×105 wells per mm2), it would be ideal to maximize the number of discrete membrane units per unit area, while making sure that enough space is left between the wells for diffraction-limited resolution with optical microscopy. Thus we fabricated wells with nanometer-scale dimensions by nanoimprint lithography followed by ALD of Al2O3. By fabricating 200 nm-diameter wells with 600 nm periodicity we are able to form an array with 2.78×106 wells per mm2, while maintaining optical resolution. The nanowell dimensions can be adjusted by depositing various thicknesses of conformal Al2O3,32 shown by shrinking the holes to 80 nm in diameter. (Figure S6 in the Supporting Information) Figure 5a shows SEM images of the 80 nm-diameter nanowells, and Figure 5b shows the resulting vesicle arrays formed from fluorescently labeled egg PC vesicles after using the PDMS squeegee. The vesicles immobilized in the nanoarrays were extruded to 100 nm in diameter, and the vesicles deform to fit into the cylindrical wells. The arrays show high occupancy with nearly all of the wells filled with vesicles. Whereas the well diameter is only 80 nm, the fluorescent spots arising from the nanoarray are approximately 400 nm across due to the optical diffraction limit, however, they are clearly resolvable due to the 600 nm periodicity. Like arrays with larger wells, photobleaching of an array with 80 nm wells shows that the individual wells are isolated from one another. (Figure S7 in the Supporting Information) Other groups have fabricated random arrays of smaller nanowells with greater surface density and immobilized vesicles and supported membranes in them.12, 17 However, the periodic nature and long-range order of wells formed by nanoimprint lithography can simplify automated image analysis and reduce sample-to-sample variability. In addition, the methods used for nanowell fabrication can be employed to form uniform well arrays with areas approaching 1 cm2 where there are more than 1.7×108 wells per array. Furthermore, the ability to tune the well diameter with ALD increases the flexibility of this platform. This could potentially allow selective capture or exclusion of vesicles or organelles based on size.
Figure 5.
Phospholipid vesicle nanoarrays and SPR sensing. (a) SEM image of a nanoarray substrate viewed in cross section. The as-fabricated nanoarrays have wells with 200 nm diameter and 600 nm periodicity, but deposition of Al2O3 by ALD (light-colored layer on surface) shrinks the well diameter to approximately 80 nm. The scale bar is 1 μm. (b) Fluorescence image of a nanoarray formed with Rho-PE labeled egg PC vesicles showing high well occupancy and that nanoarray well contents can be optically resolved. The scale bar is 10 μm. (c) A SEM image of a nanowell array (32×32) milled into a gold film. The scale bar is 5 μm. (d) Magnified SEM image of a gold nanowell array. The wells are 200 nm in diameter and have 400 nm periodicity. The scale bar is 500 nm. (e) Representative transmission spectra for SPR sensing of IgMO4 binding to myelin particles in a gold nanowell array. The red curve is the negative control spectrum where SAPE does not bind to myelin, while the blue curve is the spectrum after IgMO4 binds to myelin. IgMO4 binding results in a small red-shift of the spectrum. (f) Comparison of mean spectral shifts after incubation with SAPE and IgMO4 showing a significant difference between the two cases. The error bars are standard error of the mean and * indicates a significant difference (P = 0.03) using a one-tailed Wilcoxon matched-pairs signed rank test.
Surface plasmon resonance (SPR) is a commonly used label-free method for detecting molecular binding to surface immobilized receptors.9, 40–42 By forming vesicle arrays in nanometer wells milled into a gold film, this platform is compatible with SPR biosensing based on extraordinary optical transmission.43–51 A 32×32 array of wells with 200 nm-diameter and 400 nm periodicity were milled with a focused ion beam in a 200 nm-thick gold film on an Al2O3-coated glass slide. (Figure 5c–d) This resulted in a gold nanowell array where the well bottoms are coated with Al2O3.
Myelin particles were deposited on the gold surface and the PDMS squeegee was used to assemble nanoarrays as previously described. The transmission spectrum through the gold nanowell arrays was measured before myelin was placed on the surface, after myelin was deposited, after using the squeegee, after BSA blocking and after exposure to SAPE and IgMO4. (Figure 5e) After the vesicle arrays were blocked with BSA, they were incubated with 100 nM SAPE and the mean transmission spectrum peak position did not appreciably shift, meaning that SAPE did not bind to myelin. However, when the array was exposed to IgMO4 the spectra red-shifted 0.14 ± 0.05 nm (mean ± standard error of the mean). The mean spectral shifts after SAPE and IgMO4 exposure were determined to be significantly different (P = 0.03) using a one-tailed Wilcoxon matched-pairs signed rank test. (Figure 5f) The full spectra can be seen in Figure S6a in the Supporting Information. Such a small shift is normally difficult to measure with nanohole SPR. However, it readily and reliably measured with our experimental apparatus. The standard deviation of the noise in the spectral measurements is 1.78×10−3 nm (Figure S6b in the Supporting Information), meaning that a 0.14 nm shift has a signal to noise ratio of approximately 82. Fluorescence imaging after IgMO4 binding shows that the antibody binds primarily to the nanowell arrays where the myelin was immobilized. (Figure S6c in the Supporting Information)
GENERAL DISCUSSION AND CONCLUSIONS
We have demonstrated a unique assembly method for forming micro- to nanoscale biomembrane arrays and used the resulting arrays for sensing. This array assembly method is amenable for use with natural membranes, demonstrated by the assembly of myelin and neuronal lipid raft microarrays. Furthermore, we showed that the natural membrane microarrays can be used for detection of GM1 via CTX binding and antibody recognition of cell-specific membrane antigens. By scaling down the well size and spacing, millions of discrete membrane spots can be formed per mm2. Additionally, by forming nanowell arrays in a gold film, label-free SPR biosensing can be carried out. Because of the high-throughput fabrication methods employed, large-scale arrays were made, each with over 170 million wells per array. These fabrication methods result in perfectly uniform well arrays that have well defined long-range order and little sample-to-sample variability. This ensures that each sample has the same density of micro- and nanowells per unit area, unlike fabrication methods that result in the creation of random arrays. The uniformity of the arrays could simplify automated image analysis, compared to random arrays of nanowells. Also, increasing the density of discrete array elements increases the N for all measurements which increases statistical power.
Formation of microarray-type chips capable of incorporating living cells or natural cell membranes is possible but usually requires chemical patterning of the substrate or specialized cell culture techniques.52 The Vogel group fractured cultured cells to form supported cell membrane sheets in vitro,53 while other groups used microcontact printing followed by the rupture of erythrocyte ghosts to form arrays of natural membranes.54 Arrays of intact bacterial cells for gene expression studies have been demonstrated,55, 56 and fibroblast coculture has allowed the formation of stem cell arrays in microwells.57 Tissue microarrays show promise, but these arrays have large spots sizes and are made from paraffin blocks, limiting their use with advanced analytical methods.58 The assembly method presented here allows facile formation of natural membrane arrays without the need to culture cells on the substrate and without post-fabrication chemical modification of the substrate surface. Because no modification steps are necessary, it is possible to form natural membrane arrays for sensing and screening in 30 minutes or less. Virtually any membrane bound compartment, ranging from reconstituted proteoliposomes to organelles and secretory vesicles could be assembled into arrays with this method. This would facilitate investigation of numerous binding interactions not currently accessible with standard microarray technology, as wells as interrogation of analytical biochemistry down to the single organelle level.
The nanofabricated well arrays used in this work have a great deal of design flexibility and could be interrogated with analytical methods beyond fluorescence imaging and SPR, such as spectroscopic imaging59 or imaging mass spectrometry.60 Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry is a powerful tool for protein analysis, but usually requires the addition of a matrix for successful sample ionization. However, matrix-free MALDI techniques, such as desorption/ionization on silicon (DIOS)61 or recently developed methods like nanostructure-initiator mass spectrometry62 could be used with these arrays for proteomic studies of membrane-bound proteins. Implementation of MALDI imaging methods could make this approach very powerful and increase throughput.63 Alternatively, secondary ion mass spectrometry (SIMS) methods could be employed for lipid profiling and chemical imaging with high spatial resolution.64–66 We envision the method to be compatible with assembly of micro- and nanoarrays composed of many different types of natural membranes and membrane-bound structures for screening molecular interactions with the potential for use as a drug discovery tool.
METHODS
Fabrication of nanowell and microwell substrates
Nanowell arrays were prepared using nanoimprint lithography. The silicon nanoimprint stamp had circular 2-dimensional posts with 195 nm diameter, 350 nm depth, and 600 nm periodicity. The surface of nanoimprint stamp was coated by (heptadecafluoro-1,1,2,2-tetrahydrodecyl)trichlorosilane, obtained from Gelest Inc. (Morrisville, PA USA), to facilitate separation from the imprinted samples. A 100 nm-thick thermal oxide layer was first grown on the silicon wafers by wet oxidation at 1100 °C for 11 minutes. The silicon wafers were spin-coated by a NXR-1025 nanoimprint thermal resist (Nanonex Corp., NJ) and cured at 150 °C for 1 minute. The resist-coated silicon wafers were imprinted by the nanoimprint stamp with a pressure of 300 psi for 2 minutes at 130 °C. After oxygen plasma ashing for 30 seconds with 2 sccm of O2 at 50 W to remove residual resist, the thermal oxide layer was etched by reactive ion etching (RIE, STS model 320) with CF4 and Ar gases at 100 W for 4 minutes with the resist as an etching mask. After removing the resist with oxygen plasma ashing, approximately 300-nm-deep circular nanoarrays were made into the silicon by a deep trench silicon etcher (Plasma Therm SLR-770) with the patterned oxide as an etching mask. After removing the oxide in buffered oxide etchant (BOE), the nanoarray samples were cleaned by 1:1 mixed solution of sulfuric acid and hydrogen peroxide for 10 minutes. The nanowell diameter was 200 nm before deposition of Al2O3. The nanowell surface was covered by a conformal Al2O3 layer deposited by atomic layer deposition (ALD) at 250 °C. The deposition rate was about 1.1 Å/cycle. The Al2O3 layer not only prevents vesicle rupture, but also reduces the nanowell diameter as desired. In the experiments, 180 and 80 nm-diameter nanowells were prepared by depositing 15 and 65 nm Al2O3, respectively.
The microwell arrays were prepared using photolithography. The same silicon wafers as for the nanowells with a 100 nm-thick thermal oxide were spin-coated with MEGAPOSIT SPR-955 photoresist (Rohm and Haas) and exposed with an i-line stepper (Canon 2500 i3) using a mask with 5 μm holes in a hexagonal array with periodicity 15 μm over a 10 mm × 10 mm area which was reduced 5× giving 1 μm holes over a 2 mm × 2 mm area and stepped across an entire wafer with 5 mm steps. The wafer was developed for 90 seconds in MF CD 26 (Rohm and Haas) using a CEE 200X (Brewer Science) spray developer. Using the resist as an etch mask, the thermal oxide layer was etched using RIE (STS model 320) with CF4 and Ar gases at 100 W for 6 minutes. The silicon was etched in a deep trench silicon etcher (Plasma Therm SLR-770) using the oxide and resist as a mask. The microwells were soaked in acetone for 5 minutes to remove the resist, cleaned in a 1:1 mixed solution of sulfuric acid and hydrogen peroxide for 5 minutes, and had the oxide mask removed in a BOE bath. Then, the microwell surface was covered with a 10 nm thick layer of Al2O3 using the same ALD procedure as for the nanowells.
Fabrication of gold nanowell arrays for surface plasmon resonance
Glass slides were sequentially cleaned by sonication in acetone, methanol, isopropyl alcohol, and deionized water for 5 minutes each. The glass surface was then coated by a 10-nm-thick layer of Al2O3 using the same ALD procedures as for the micro- and nanowells. A 200 nm-thick Au film with a 5 nm-thick Cr adhesion layer was deposited on the Al2O3 coated glass slides using an electron-beam evaporator (CHA, SEC600). Nanowell arrays with 200 nm hole size and 400 nm periodicity were patterned with focused ion beam (FIB) milling using 30 keV and 100 pA ion beam (FEI Dual Beam Quanta 200 3D). This resulted in gold nanowells where the bottom of the wells was covered with Al2O3.
Phospholipid vesicle formation
Vesicles were formed by first evaporating chloroform solvent from lipid solutions containing Egg PC and 1% (w/w) 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl, ammonium salt (Rho-PE) or 1% (w/w) 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl), ammonium salt (NBD-PE). All lipids were obtained from Avanti Polar Lipids and used as received. Evaporation of chloroform was carried out over at least 6 hours under vacuum. The dry lipid film was rehydrated with Tris buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA) to a total lipid concentration of 1 mg/mL and refrigerated overnight. The following day, the rehydrated vesicles were vortex mixed for 10 seconds then sonicated for 15 minutes in a room temperature water bath. Prior to exposure to the micro- and nanohole substrates, vesicles were extruded 21 passes through polycarbonate filters with 200 or 100 nm pore sizes using an Avanti Mini-Extruder.
Myelin isolation
Myelin was isolated from SJL/J mouse whole brain according to established procedures.67 Myelin quality was determined by Western blotting for the presence of MAG, MOG, PLP, CNP, and MBP, and via the binding of well-characterized antimyelin lipid antibodies by direct ELISA.
Neuronal lipid raft isolation
Neuron membranes were isolated using floatation ultracentrifugation in non-continuous sucrose gradient. DIV7 cortical neurons were lysed in ice-cold lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease inhibitor cocktail) for 30 minutes. The neuronal lysates were mixed with an equal volume of 100% (w/v) sucrose. The mixture was transferred to a centrifuge tube, and 8 mL of 35% sucrose and 3.5 mL of 5% sucrose were overlaid sequentially. After centrifugation at 2×105g for 20 hours at 4 °C, six fractions (2 mL of each) were collected from the top of the gradient. Each fraction and the pellet were dissolved in SDS-sample buffer and subjected to western blotting for the presence of caveolin-1, a marker of the membrane raft fraction which was fraction two of six.
Assembly of phospholipid vesicle arrays
Solutions of extruded vesicles (total lipid concentration: 0.1 mg/mL) were placed on the array substrate and the vesicles were allowed to settle to the surface for 30 minutes. The substrate was thoroughly rinsed with Tris buffer to remove excess vesicles and immersed in a shallow dish containing Tris buffer. The assembly process involved sliding a PDMS “squeegee” (approximate dimensions: 25 mm × 25 mm × 5 mm) in firm contact with the submerged substrate surface at least 10 times to remove vesicles that were not adhered in the recessed wells of the substrate. After passing the squeegee at least 10 times over the array substrate, the surface was washed again with Tris buffer and then kept wet until imaging.
Assembly of myelin and lipid raft arrays
Suspensions of myelin and lipid raft membranes were diluted in PBS. After dilution, the suspensions were subjected to three 15 minute rounds of sonication at room temperature to decrease the lipid particle size. Dynamic light scattering of the myelin particles after each round of sonication showed that the mean particle size decreases from 3.37 μm before sonication to 220 nm after the third round of sonication. (Figure S7 in the Supporting Information) The sonicated suspensions were placed on the microwell array substrates for 30 minutes to allow the particles to settle to the surface. After 30 minutes the substrate with natural lipids was washed with PBS then submerged in a shallow dish containing PBS. The PDMS squeegee was passed over the substrates at least 10 times, then the substrates were again washed with PBS. The membranes in the array were stained by adding 10 μM FM1-43 (Invitrogen) to the solution.
Ganglioside GM1-cholera toxin binding assays
For CTX binding assays on natural membranes, lipid raft microarrays were prepared as described above, but without FM1-43 staining. After array formation, the surface was blocked with 1 mg/mL BSA in PBS for 30 minutes. The blocked samples were incubated with 10, 50 or 200 nM Alexa-488 conjugated CTX (Invitrogen) for 30 minutes then washed thoroughly with PBS. For the negative control, a lipid raft microarray was blocked with BSA incubated with 50 nM streptavidin-R-phycoerythrin (SAPE, Invitrogen) for 30 minutes, followed by washing and staining with FM1-43 to confirm that lipids were present in the array in the event that SAPE did not bind.
Formation of multicomponent microarrays
For multicomponent microarrays the myelin and lipid raft particles were stained with 10 μM FM1-43 then injected into PDMS microfluidic channels under the control of a syringe pump. The PDMS microfluidic was prepared by standard soft lithography methods and had channels that were 250 μm wide. After filling the channels with natural membrane particles, the solution flow was slowed to 3 μL/hour for 30 minutes to allow the particles to settle on the surface. After 30 minutes, the channels were washed with PBS and the microfluidic chip was removed from the substrate while submerged in PBS. The PDMS squeegee was translated across the substrate 10 times in a direction that was parallel to the channel direction to form microarray stripes containing natural membrane components. The microarray stripes were then blocked by incubating with 5 mg/mL BSA for 30 minutes. The stripes were exposed to IgMO4 diluted in PBS. IgMO4 conjugated to NorthernLights 557 fluorophore was supplied as a 10× solution (R & D Systems, Inc.) and diluted to 1× according to the manufacturer’s protocol for immunofluorescence assays.
Dynamic light scattering of vesicles and myelin particles
Phospholipid vesicles for DLS were prepared and extruded as described above. The vesicles were diluted to a total lipid concentration of 0.05 mg/mL in Tris buffer and then analyzed with a Brookhaven Instruments 90Plus/BI-MAS particle size analyzer. The extracted myelin (1 mg/mL in PBS) was diluted to 0.1 mg/mL in PBS and subjected to 1, 2, or 3 15-minute cycles of sonication in a bath sonicator at room temperature. A control sample was not subjected to sonication. The resulting particle solutions were analyzed with a Brookhaven Instruments 90Plus/BI-MAS particle size analyzer.
Micro- and nanoarray imaging
Arrays were imaged with one of two microscopy systems depending on desired image resolution. For routine imaging a Nikon Eclipse LV100 upright microscope with a tungsten-halogen light source and a 50× objective (0.55 NA) and a Photometics CoolSNAP HQ2 CCD camera was used. Images were captured with Photometrics Voodoo Incantation 1.2 software. For high-resolution imaging of nanoarrays and photobleaching experiments an Olympus FV1000 upright confocal system with a 60× water immersion objective (0.9 NA) was employed. Images were analyzed and colorized using ImageJ software, version 1.44j.
Surface plasmon resonance measurements
Prior to myelin addition and array assembly, the optical transmission spectra were measured for 5 arrays immersed in PBS. Myelin microarrays were formed in gold nanowell arrays in a manner similar to the formation of myelin microarrays. The arrays were blocked with 5 mg/mL BSA and then exposed to 50 nM SAPE for 30 minutes. The transmission spectra through five arrays were then recorded. The arrays were exposed to NorthernLights 557-labeled IgMO4 (diluted to 1× according to manufacturer’s protocol) for 30 minutes. The transmission spectra through the same five arrays were collected again. The transmission spectra were acquired using a Nikon LV100 upright microscope with a tungsten-halogen light source and a 50× objective with NA = 0.55. The transmitted light was collected with an optical fiber (200 μm core) and the spectra were analyzed with an Ocean Optics fiber optic spectrometer.
Data analysis and statistics
All data plotting, curve fitting and statistical analyses were carried out using GraphPad Prism version 5.04. (GraphPad Software, Inc.) The mean SPR spectra shifts for 5 separate SAPE and IgMO4 binding experiments were compared using a one-tailed Wilcoxon matched-pairs signed rank test. This nonparametric test was chosen because the normality of the data was unknown due to the small sample size. For each experiment the spectra were measured on given array after SAPE and after IgMO4 incubations, hence a matched-pairs test was employed. A one-tailed version of this test was chosen because the peak position should either remain constant or red-shift due to binding (i.e. the peak position moves in the positive direction). A negative deviation from the pre-binding peak position is not expected so a one-tailed test is appropriate.
The FRAP data points in Supporting Information Figure S2e represent the average recovery curve for three replicate photobleaching experiments. The error bars associated with the individual data points were the standard deviations of each data point for the three replicate experiments. The FRAP recovery curves were fit to a single exponential curve to determine the time at which 50% recovery had occurred (t50). The diffusion coefficient (D) was calculated by the equation D = R2/4t50, where R is the radius of the photobleached spot.68 Myelin particle distributions determined by DLS (Figure S7 in the Supporting Information) were fit to log Gaussian curves to determine the center of the distribution and the standard error of the center.
Supplementary Material
Acknowledgments
This work was supported by grants to S.H.O. from the National Institutes of Health (R01 GM092993), the National Science Foundation (NSF CAREER Award and DBI 0964216), and the Office of Naval Research (ONR) Young Investigator Program. S.H.O. and M.R. acknowledge the Minnesota Partnership Award for Biotechnology and Medical Genomics. This work was also supported by grants to M.R. from the National Institutes of Health (NS024180, NS032129, NS048357, R21 NS073684), the National Multiple Sclerosis Society (CA1060A11), the Applebaum Foundation, and the Hilton Foundation. The authors thank Y.S. Lin and C.L. Haynes for use of the dynamic light scattering instrument. Device fabrication was performed at the University of Minnesota Nanofabrication Center, which receives support from the NSF through the National Nanotechnology Infrastructure Network.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Discussion of array fluorescence distribution compared to vesicle surface areas, high-resolution fluorescence array images and additional images of array photobleaching, additional images of myelin particles on a microwell array and fluorescence intensity line scans, multicomponent arrays formed with fluorescent phospholipid vesicles, SEM images of nanowell dimension tuning with ALD, complete SPR transmission spectra of light through gold nanowell arrays, plots of SPR noise level and fluorescence images of IgM O4 binding to myelin in gold nanowell arrays. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.MacBeath G, Schreiber SL. Printing Proteins as Microarrays for High-throughput Function Determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 2.Kodadek T. Protein Microarrays: Prospects and Problems. Chem Biol. 2001;8:105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 3.Feizi T, Fazio F, Chai W, Wong C. Carbohydrate Microarrays - A New Set of Technologies at the Frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling Protein Microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 5.Jones RB, Gordus A, Krall JA, MacBeath G. A Quantitative Protein Interaction Network for the ErbB Receptors Using Protein Microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 6.Sassolas A, Leca-Bouvier BD, Blum LJ. DNA Biosensors and Microarrays. Chem Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 7.Salafsky J, Groves JT, Boxer SG. Architecture and Function of Membrane Proteins in Planar Supported Bilayers: A Study with Photosynthetic Reaction Centers. Biochemistry. 1996;35:14773–14781. doi: 10.1021/bi961432i. [DOI] [PubMed] [Google Scholar]
- 8.Cooper MA. Optical Biosensors in Drug Discovery. Nat Rev Drug Discov. 2002:515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 9.Cooper MA. Advances in Membrane Receptor Screening and Analysis. J Mol Recognit. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- 10.Warrington AE, Asakura K, Bieber AJ, Ciric B, Van Keulen V, Kaveri SV, Kyle RA, Pease LR, Rodriguez M. Human Monoclonal Antibodies Reactive to Oligodendrocytes Promote Remyelination in a Model of Multiple Sclerosis. Proc Natl Acad Sci USA. 2000;97:6820–6825. doi: 10.1073/pnas.97.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bally M, Bailey K, Sugihara K, Grieshaber D, Voros J, Stadler B. Liposome and Lipid Bilayer Arrays Towards Biosensing Applications. Small. 2010;6:2481–2497. doi: 10.1002/smll.201000644. [DOI] [PubMed] [Google Scholar]
- 12.Dahlin A, Zäch M, Rindzevicius T, Käll M, Sutherland DS, Höök F. Localized Surface Plasmon Resonance Sensing of Lipid-membrane-mediated Biorecognition Events. J Am Chem Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- 13.Marie R, Dahlin A, Tegenfeldt J, Höök F. Generic Surface Modification Strategy for Sensing Applications Based on Au/SiO2 Nanostructures. Biointerphases. 2007;2:49–55. doi: 10.1116/1.2717926. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer I, Petronis S, Koper I, Kasemo B, Zach M. Vesicle Adsorption and Phospholipid Bilayer Formation on Topographically and Chemically Nanostructured Surfaces. J Phys Chem B. 2010;114:4623–4631. doi: 10.1021/jp908283g. [DOI] [PubMed] [Google Scholar]
- 15.Stamou SD, Duschl C, Delamarche E, Vogel H. Self-assembled Microarrays of Attoliter Molecular Vessels. Angew Chem Int Edit. 2003;42:5580–5583. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- 16.Kalyankar ND, Sharma MK, Vaidya SV, Calhoun D, Maldarelli C, Couzis A, Gilchrist L. Arraying of Intact Liposomes into Chemically Functionalized Microwells. Langmuir. 2006;22:5403–5411. doi: 10.1021/la0602719. [DOI] [PubMed] [Google Scholar]
- 17.Dahlin A, Jonsson M, Höök F. Specific Self-assembly of Single Lipid Vesicles in Nanoplasmonic Apertures in Gold. Adv Mater. 2008;20:1436–1442. [Google Scholar]
- 18.Chaize B, Nguyen M, Ruysschaert T, le Berre V, Trevisiol E, Caminade AM, Majoral JP, Pratviel G, Meunier B, Winterhalter M, Fournier D. Microstructured Liposome Array. Bioconjugate Chem. 2006;17:245–247. doi: 10.1021/bc050273p. [DOI] [PubMed] [Google Scholar]
- 19.Stadler B, Bally M, Grieshaber D, Voros J, Brisson A, Grandin HM. Creation of a Functional Heterogeneous Vesicle Array via DNA Controlled Surface Sorting onto a Spotted Microarray. Biointerphases. 2006;1:142–145. doi: 10.1116/1.2434178. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Frutos A, Lahiri J. Membrane Protein Microarrays. J Am Chem Soc. 2002;124:2394–2395. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 21.Bieri C, Ernst OP, Heyse S, Hofmann KP, Vogel H. Micropatterned Immobilization of a G Protein-Coupled Receptor and Direct Detection of G Protein Activation. Nat Biotechnol. 1999;17:1105–1108. doi: 10.1038/15090. [DOI] [PubMed] [Google Scholar]
- 22.Czolkos I, Erkan Y, Dommersnes P, Jesorka A, Orwar O. Controlled Formation and Mixing of Two-Dimensional Fluids. Nano Lett. 2007;7:1980–1984. doi: 10.1021/nl070726u. [DOI] [PubMed] [Google Scholar]
- 23.Czolkos I, Hannestad JK, Jesorka A, Kumar R, Brown T, Albinsson B, Orwar O. Platform for Controlled Supramolecular Nanoassembly. Nano Lett. 2009;9:2482–2486. doi: 10.1021/nl901254f. [DOI] [PubMed] [Google Scholar]
- 24.Groves JT, Ulman N, Boxer SG. Micropatterning Fluid Lipid Bilayers on Solid Supports. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 25.Hovis JS, Boxer SG. Patterning and Composition Arrays of Supported Lipid Bilayers by Microcontact Printing. Langmuir. 2001;17:3400–3405. [Google Scholar]
- 26.Groves JT, Boxer SG. Micropattern Formation in Supported Lipid Membranes. Acc Chem Res. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- 27.Yee CK, Amweg ML, Parikh AN. Direct Photochemical Patterning and Refunctionalization of Supported Phospholipid Bilayers. J Am Chem Soc. 2004;126:13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Yang T, Cremer PS. Multiplexing Ligand-Receptor Binding Measurements by Chemically Patterning Microfluidic Channels. Anal Chem. 2008;80:6078–6084. doi: 10.1021/ac800912f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J, Chen J, Cremer PS. Sub-100 nm Patterning of Supported Bilayers by Nanoshaving Lithography. J Am Chem Soc. 2008;130:2718–2719. doi: 10.1021/ja077730s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus T, Malaquin L, Schmid H, Riess W, Spencer ND, Wolf H. Nanoparticle Printing with Single-Particle Resolution. Nat Nanotechnol. 2007;2:570–576. doi: 10.1038/nnano.2007.262. [DOI] [PubMed] [Google Scholar]
- 31.Sweeney CM, Hasan W, Nehl CL, Odom TW. Optical Properties of Anisotropic Core-Shell Pyramidal Particles. J Phys Chem A. 2009;113:4265–4268. doi: 10.1021/jp810837u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Im H, Lindquist NC, Lesuffleur A, Oh SH. Atomic Layer Deposition of Dielectric Overlayers for Enhancing the Optical Properties and Chemical Stability of Plasmonic Nanoholes. ACS Nano. 2010;4:947–954. doi: 10.1021/nn901842r. [DOI] [PubMed] [Google Scholar]
- 33.Mager MD, Almquist B, Melosh NA. Formation and Characterization of Fluid Lipid Bilayers on Alumina. Langmuir. 2008;24:12734–12737. doi: 10.1021/la802726u. [DOI] [PubMed] [Google Scholar]
- 34.Cremer PS, Boxer SG. Formation and Spreading of Lipid Bilayers on Planar Glass Supports. J Phys Chem B. 1999;103:2554–2559. [Google Scholar]
- 35.Keller CA, Kasemo B. Surface Specific Kinetics of Lipid Vesicle Adsorption Measured with a Quartz Crystal Microbalance. Biophys J. 1998;75:1397–1402. doi: 10.1016/S0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reimhult E, Höök F, Kasemo B. Intact Vesicle Adsorption and Supported Biomembrane Formation from Vesicles in Solution: Influence of Surface Chemistry, Vesicle Size, Temperature, and Osmotic Pressure. Langmuir. 2003;19:1681–1691. [Google Scholar]
- 37.Anderson TH, Min Y, Weirich KL, Zeng H, Fygenson D, Israelachvili JN. Formation of Supported Bilayers on Silica Substrates. Langmuir. 2009;25:6997–7005. doi: 10.1021/la900181c. [DOI] [PubMed] [Google Scholar]
- 38.Lingwood D, Simons K. Lipid Rafts As a Membrane Organizing Principle. Science. 2010;32:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 39.Simons K, Toomre D. Lipid Rafts and Signal Transduction. Nat Rev Mol Cell Bio. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 40.Smith EA, Corn RM. Surface Plasmon Resonance Imaging as a Tool to Monitor Biomolecular Interactions in an Array Based Format. Appl Spectrosc. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- 41.Shumaker-Parry JS, Aebersold R, Campbell CT. Parallel, Quantitative Measurement of Protein Binding to a 120-element Double-Stranded DNA Array in Real Time using Surface Plasmon Resonance Microscopy. Anal Chem. 2004;76:2071–2082. doi: 10.1021/ac035159j. [DOI] [PubMed] [Google Scholar]
- 42.Gao HW, Yang JC, Lin JY, Stuparu AD, Lee MH, Mrksich M, Odom TW. Using the Angle-dependent Resonances of Molded Plasmonic Crystals To Improve the Sensitivities of Biosensors. Nano Lett. 2010;10:2549–2554. doi: 10.1021/nl101165r. [DOI] [PubMed] [Google Scholar]
- 43.Ebbesen TW, Lezec HJ, Ghaemi HF, Thio T, Wolff PA. Extraordinary Optical Transmission Through Sub-wavelength Hole Arrays. Nature. 1998;391:667–669. [Google Scholar]
- 44.Brolo AG, Gordon R, Leathem B, Kavanagh KL. Surface Plasmon Sensor Based on the Enhanced Light Transmission Through Arrays of Nanoholes in Gold Films. Langmuir. 2004;20:4813–4815. doi: 10.1021/la0493621. [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran N, Larson DN, Stark PRH, Hainsworth E, LaBaer J. Emerging Tools for Real-Time Label-free Detection of Interactions on Functional Protein Microarrays. FEBS J. 2005;272:5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 46.Stewart ME, Mack NH, Malyarchuk V, Soares J, Lee TW, Gray SK, Nuzzo RG, Rogers JA. Quantitative Multispectral Biosensing and 1D imaging using Quasi-3D Plasmonic Crystals. Proc Natl Acad Sci USA. 2006;103:17143–17148. doi: 10.1073/pnas.0606216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang JC, Ji J, Hogle JM, Larson DN. Metallic Nanohole Arrays on Fluoropolymer Substrates as Small Label-free Real-Time Bioprobes. Nano Letters. 2008;8:2718–2724. doi: 10.1021/nl801043t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Im H, Lesuffleur A, Lindquist NC, Oh SH. Plasmonic Nanoholes in a Multichannel Microarray Format for Parallel Kinetic Assays and Differential Sensing. Anal Chem. 2009:2854–2859. doi: 10.1021/ac802276x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eftekhari F, Escobedo C, Ferreira J, Duan X, Girotto EM, Brolo AG, Gordon R, Sinton D. Nanoholes As Nanochannels: Flow-through Plasmonic Sensing. Anal Chem. 2009;81:4308–4311. doi: 10.1021/ac900221y. [DOI] [PubMed] [Google Scholar]
- 50.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh SH. Membrane Protein Biosensing with Plasmonic Nanopore Arrays and Pore-spanning Lipid Membranes. Chem Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JC, Gao HW, Suh JY, Zhou W, Lee MH, Odom TW. Enhanced Optical Transmission Mediated by Localized Plasmons in Anisotropic, Three-Dimensional Nanohole Arrays. Nano Lett. 2010;10:3173–3178. doi: 10.1021/nl102078j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandes TG, Diogo MM, Clark DS, Dordick JS, Cabral JMS. High-throughput Cellular Microarray Platforms: Applications in Drug Discovery, Toxicology and Stem Cell Research. Trends Biotechnol. 2009;27:342–349. doi: 10.1016/j.tibtech.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danelon C, Perez JB, Santschi C, Brugger J, Vogel H. Cell Membranes Suspended Across Nanoaperture Arrays. Langmuir. 2006;22:22–25. doi: 10.1021/la052387v. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka M, Wong AP, Rehfeldt F, Tutus M, Kaufmann S. Selective Deposition of Native Cell Membranes on Biocompatible Micropatterns. J Am Chem Soc. 2004;126:3257–3260. doi: 10.1021/ja038981d. [DOI] [PubMed] [Google Scholar]
- 55.Kuang Y, Biran I, Walt DR. Living Bacterial Cell Array for Genotoxin Monitoring. Anal Chem. 2004;76:2902–2909. doi: 10.1021/ac0354589. [DOI] [PubMed] [Google Scholar]
- 56.Kuang Y, Biran I, Walt DR. Simultaneously Monitoring Gene Expression Kinetics and Genetic Noise in Single Cells by Optical Well Arrays. Anal Chem. 2004;76:6282–6286. doi: 10.1021/ac049053f. [DOI] [PubMed] [Google Scholar]
- 57.Khademhosseini A, Ferreira L, Blumling J, Yeh J, Karp J, Fukuda J, Langer R. Co-culture of Human Embryonic Stem Cells with Murine Embryonic Fibroblasts on Microwell-patterned Substrates. Biomaterials. 2006;27:5968–5977. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 58.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue Microarrays for High-throughput Molecular Profiling of Tumor Specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 59.Bake KD, Walt DR. Multiplexed Spectroscopic Detections. Annu Rev Anal Chem. 2008;1:515–547. doi: 10.1146/annurev.anchem.1.031207.112826. [DOI] [PubMed] [Google Scholar]
- 60.Vickerman JC. Molecular Imaging and Depth Profiling by Mass Spectrometry—SIMS, MALDI or DESI? Analyst. 2011;136:2199–2217. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- 61.Wei J, Buriak JM, Siuzdak G. Desorption-Ionization Mass Spectrometry on Porous Silicon. Nature. 1999;399:243–246. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]
- 62.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate Nanostructures for Mass Spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 63.Schwamborn K, Caprioli RM. Molecular Imaging by Mass Spectrometry - Looking Beyond Classical Histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 64.Ostrowski SG, Szakal C, Kozole J, Roddy TP, Xu J, Ewing AG, Winograd N. Secondary Ion MS Imaging of Lipids in Picoliter Vials with a Buckminsterfullerene Ion Source. Anal Chem. 2005;77:6190–6196. doi: 10.1021/ac0508189. [DOI] [PubMed] [Google Scholar]
- 65.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Phase Separation of Lipid Membranes Analyzed with High-resolution Secondary Ion Mass Spectrometry. Science. 2006;313:1948–1951. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 66.Sjövall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Imaging of Membrane Lipids in Single Cells by Imprint-Imaging Time-of-Flight Secondary Ion Mass Spectrometry. Anal Chem. 2003;75:3429–3434. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 67.Norton WT, Poduslo SE. Myelination in rat brain: method of myelin isolation. J Neurochem. 1973;21(4):749–57. doi: 10.1111/j.1471-4159.1973.tb07519.x. [DOI] [PubMed] [Google Scholar]
- 68.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility Measurement by Analysis of Fluorescence Photobleaching Recovery Kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.