Abstract
Objective
To semi-automatically induce semantic categories of eligibility criteria from text and to automatically classify eligibility criteria based on their semantic similarity.
Design
The UMLS semantic types and a set of previously developed semantic preference rules were utilized to create an unambiguous semantic feature representation to induce eligibility criteria categories through hierarchical clustering and to train supervised classifiers.
Measurements
We induced 27 categories and measured the prevalence of the categories in 27,278 eligibility criteria from 1,578 clinical trials and compared the classification performance (i.e., precision, recall, and F1-score) between the UMLS-based feature representation and the “bag of words” feature representation among five common classifiers in Weka, including J48, Bayesian Network, Naïve Bayesian, Nearest Neighbor, and Instance-based Learning Classifier.
Results
The UMLS semantic feature representation outperforms the “bag of words” feature representation in 89% of the criteria categories. Using the semantically induced categories, machine-learning classifiers required only 2,000 instances to stabilize classification performance. The J48 classifier yielded the best F1-score and the Bayesian Network classifier achieved the best learning efficiency.
Conclusion
The UMLS is an effective knowledge source and can enable an efficient feature representation for semi-automated semantic category induction and automatic categorization for clinical research eligibility criteria and possibly other clinical text.
Keywords: Clinical Research Eligibility Criteria, Classification, Hierarchical Clustering, Knowledge Representation, Unified Medical Language System (UMLS), Machine Learning, Feature Representation
1. Background
Eligibility criteria specify the characteristics that a human volunteer must or must not possess to participate in a clinical study or to be treated according to a standard clinical care guideline. Each criterion is an independent sentence describing a patient characteristic, often with a temporal constraint. Examples are “Age of at least 18 years and 75 years or less” or “Have a CD4 cell count of 200 copies/ml or higher within 60 days of study entry.” With more and more patient health information being available electronically, especially through the expanding adoption of electronic health records (EHR) worldwide, it is appealing to link eligibility criteria to electronic patient information to automatically match patients to clinical research opportunities or to automatically screen patients for personalized clinical care. However, the unstructured format of eligibility criteria is a big barrier to this goal1. On ClinicalTrials.gov2, the official public registry of clinical trials, clinical eligibility criteria are organized as a paragraph, a bullet list, or arbitrary user-specified topic categories. In contrast, patient information is often structured and encoded in various clinical terminologies by category. For example, disease diagnoses are often encoded by The International Classification of Diseases (ICD) version 9, while the Current Procedure Terminology (CPT) encodes laboratory test results. In order to improve the efficiency for eligibility determination in the vast patient information space, ideally eligibility criteria should be categorized and structured in the same way as the corresponding patient information. The various templates for structuring eligibility criteria of different categories also necessitate automatic criteria categorization to facilitate efficient eligibility criteria template selection.
Currently there is no standard categorization of clinical research eligibility criteria. Various task-dependent criteria categories have been defined according to certain criteria features, such as the purpose, clinical topic, disease area, or syntactic complexity of eligibility criteria1. For example, the most common categories for eligibility criteria include inclusion criteria and exclusion criteria. The Trial Bank Project defines three categories of clinical eligibility queries: age-gender-rule, ethnicity-language-rule, and clinical-rule3. ERGO classifies criteria by syntactic variations into simple statement, complex statement, and comparison statement4. The ASPIRE project categorizes eligibility criteria as either pan-disease queries or disease-specific queries5. Tu6 viewed an eligibility criterion as a dynamic property and differentiated eligibility criteria by their objectiveness, variability, and controllability of the underlying clinical conditions. Specifically, Tu classified eligibility criteria as (1) stable requisite; (2) variable routine; (3) controllable; (4) subjective; and (5) special. Metz et al.7 classified eligibility criteria into five categories: demographic, contact information, personal medical history, cancer diagnosis, and treatment to date. The existing approaches to categorizing eligibility criteria are largely task-dependent and hence may not generalize across application domains. Furthermore, most of the categorization processes are manual and hence are time consuming and expensive8–9. More efficient, generic semantic categorization of eligibility criteria is much needed.
In fact, clinical research eligibility criteria are ideally suited for automatic categorization. Ross et al. analyzed clinical eligibility criteria and discovered that about 92% of clinical eligibility criteria contained only one content topic (e.g., patient characteristic, behavior, or treatment) in each sentence, with about 71% of queries describing patient characteristics, 34% describing the treatments or procedures patients have received or will receive, and 4% specifying patient behaviors10. With this observation, we hypothesize that it is feasible to automatically categorize clinical research eligibility criteria using machine-learning classifiers.
We have previously presented a methodology for inducing semantic categories from free-text clinical research eligibility criteria on the AMIA Fall Symposium 201011. Extending that work, in this paper, we describe the design and evaluation of a novel approach to dynamic categorization of clinical research eligibility criteria based on hierarchical clustering. Our design fully integrates semi-supervised hierarchical clustering and supervised machine-learning classifiers via a shared semantic feature representation for eligibility criteria based on the UMLS semantic types. Our research question was “would the UMLS semantic knowledge be effective to facilitate machine-learning approaches to categorization of clinical research eligibility criteria?” We measured the prevalence and distribution of the semi-automatically induced criteria categories among 27,278 criteria extracted from 1,578 clinical studies. We also evaluated the classification efficiency by using five common classifiers available in the open-source Weka package12, including J4813, Bayesian Network14, Naïve Bayesian15, the nearest-neighbor (NNge)16, and the instance-based learning classifier IB117. With the use of the Unified Medical Language System (UMLS) semantic types for feature representation, classifier-learning efficiency was significantly improved over the use of the traditional “bag of words” feature representation. More design and evaluation details follow next.
2. An Integrated Framework for Categorization based on Clustering
Figure 1 illustrates our machine-learning framework for integrated category induction and criteria classification, both sharing the same feature representation by annotating each eligibility criterion with a UMLS-based semantic lexicon18. Instead of using manually defined categories, we semi-automatically generated eligibility categories using a hierarchical clustering algorithm to augment humans for category induction. Then a supervised machine-learning classifier achieves automatic classification. More design details are provided below.
Figure 1.
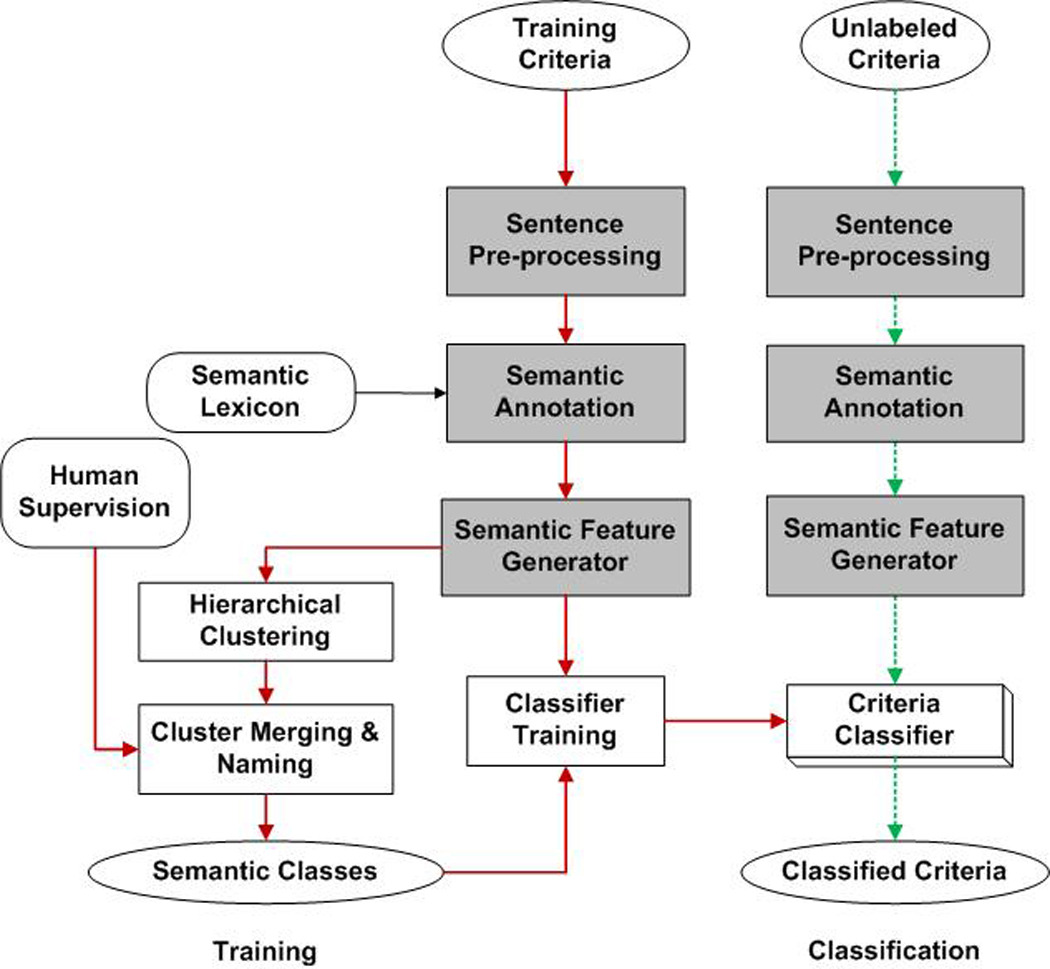
A framework for dynamic categorization of free-text clinical eligibility criteria by UMLS-based hierarchical clustering:
solid arrows show the machine learning process for classifier development;
dotted arrows show the automatic criterion classification process using the classifier;
shadowed blocks indicate the shared modules between the training and classification stage.
2.1 Semantic Feature Representation
Each criterion was first parsed by a previously published and freely available semantic annotator18 to identify the UMLS-recognizable terms, many of which were associated with multiple semantic types and hence resulted in ambiguity. We removed such ambiguities by using a set of predefined semantic preference rules19 to select specific types over general types18. For example, the UMLS concept pericardial was associated with two UMLS semantic types, Body Location or Region and Spatial Concept. In the UMLS Semantic Network, Body Location or Region was one of the sub-types of Spatial Concept. Hence, the semantic type Body Location or Region was more specific than Spatial Concept. Therefore, for the UMLS concept pericardial, the type Body Location or Region was retained as the preferred semantic type.
We also created three new types to label terms that were not covered in the UMLS but frequently occurred in clinical research eligibility criteria. We defined the Numeral type for numbers (e.g. 18, 60, two). We also defined the Symbol type for comparison connector (e.g. +, >=, @) and the Unit type for measurement units (e.g. mm3, ph, kg). The example criterion “Prior adjuvant therapy for metastatic disease allowed” would be annotated as “prior /Temporal Concept | adjuvant therapy/Therapeutic or Preventive Procedure | for/ | metastatic disease / Neoplastic Process | allowed /Social Behavior”, in which each UMLS concept (underlined) was separated from its corresponding UMLS semantic type (italic) by a slash.
Figure 2 shows the process of transforming eligibility criteria into a semantic feature representation matrix. A criterion was denoted as S and it was mapped into a semantic type vector T (Figure 2-A). The value of a semantic feature in the vector T was weighted by its frequency of occurrence, , where i = {1,2,3⋯k}, Fj being the frequency of the semantic type j and k being the number of all the different semantic types in sentence S. For example, the above sentence contained 4 different UMLS semantic types, each occurring once; therefore, the weighted frequency of each semantic type was 0.25. All the criteria instances were parsed and transformed into a feature representation matrix to support classifier learning, where each row was a criterion and each column was a UMLS semantic type, as shown in Figure 2-B.
Figure 2.
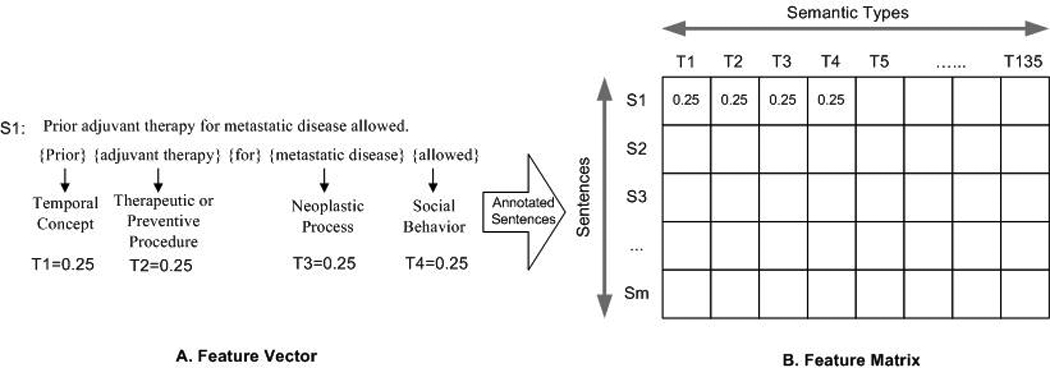
The process of transforming eligibility criteria into a UMLS-based semantic feature matrix.
2.2 Semi-Supervised Hierarchical Clustering
After the semantic annotator transformed the criteria into a UMLS-based semantic feature matrix, the hierarchical agglomerative clustering algorithm (HAC) algorithm20 was applied on the semantic feature matrix to induce the semantic categories of criteria. HAC used a bottom-up approach that initially created a cluster for each criterion and then progressively agglomerated clusters based on their semantic similarity, one pair each time, until all criteria were joined into a giant cluster. To assess the similarity of eligibility criteria, the Pearson correlation coefficient21 was applied to quantify the relationship between the semantic representations of two criteria with the value ranging between −1 and 1. For example, if criterion A and B contained completely different sets of UMLS semantic type features, their correlation value would be −1.0 because they were perfectly divergent. If the two sets were not perfectly divergent, but still diverged, the correlation would remain negative, but would be greater than −1.0. In contrast, if criteria A and B were convergent, then their correlation would be 1.0. If there was no relationship, their correlation would be 0. A Pearson correlation coefficient was initially computed for every possible criterion comparison to create a table of correlation values between every possible pair of the criteria being clustered. Then the criterion pair that had the highest correlation value was first selected to merge into one cluster to form a new "pseudo-criterion". The new pseudo-criterion would contain the same number of feature representation, but each feature would now be the arithmetic mean of the two original feature sets. The two original criteria that were merged would be removed from the table of correlation values, and new correlations would be found between the new pseudo-criterion and all of the remaining criteria. The next highest remaining correlation value in the table would be identified and that pair of criteria would be joined for form a new pseudo-criterion. This process continued until all that remains was a single pseudo-criterion containing the arithmetic mean of all the original criteria at each feature. When clustering an original criterion with a previously formed pseudo-criterion, the newly formed pseudo-criterion must be an arithmetic mean of all the criteria' features that it contains, not a simple average between the pseudo-criterion's and the original criterion's features. By retracing the order in which the criteria were progressively joined into clusters and by knowing the correlation value of each step, we identified the criteria related to each other closely and the criteria related only distantly by setting a threshold from the [−1,1] value range, where any criteria paired with correlations greater than that threshold were considered a cluster, and any criteria or clusters with correlations less than that threshold were not. In this way, all criteria had a correlation greater than the threshold were considered as a cluster.
A manual review was performed to merge and label these clusters to form semantic categories based on their semantic similarity18. For example, one cluster contained the criterion “SGOT <=2 times ULN”, while the other cluster contained the criterion “Neutrophil count larger than 1000 per mm3”. Both criteria had similar scores of the semantic similarity between instances in each cluster, which were typical laboratory test results; therefore, these two clusters were semantically related. In addition, the syntax of the instances in the two clusters was similar, both including a clinical object, a comparison symbol, numerical values, and measurement units. A manual review concluded that the two clusters were semantically similar and could be merged into one category: Diagnostic or Lab Results. Meaningful category names were created manually and supplied to the classification module. We purposely reused the labels of the UMLS semantic types to name criteria categories since they are familiar to many informatics researchers.
2.3 Supervised Machine-learning for Criteria Classification
Two human raters independently labeled a set of criteria using the developed semantic categories and reached consensus on the categorization results. These instances were also parsed by the semantic annotator and transformed into semantic features. The manual categorization results and their semantic features were used to train five very commonly used supervised classifiers in the open-source Weka package12, including J4813, Bayesian Network14, Naïve Bayesian15, the nearest-neighbor (NNge)16, and the instance-based learning classifier IB117. We compared the classification performance using the UMLS-based semantic feature representation and the standard “bag of words” representation respectively among all the five classifiers.
3. Results
3.1 The 27 Semantic Categories for Eligibility Criteria
ClinicalTrials.gov is a public registry of all clinical trials and their eligibility criteria2. From this web site, we randomly extracted 5,000 sentences. Excluding 179 non-criterion sentences and using the remaining 4,821 criteria sentences, the hierarchical clustering module initially recommended 41 clusters whose pair-wise similarity was above the threshold 0.75. We manually induced 27 eligibility criteria categories, whose organizational hierarchy includes 6 topic groups: Demographics (e.g., age or gender), Health Status (e.g., disease or organ status), Treatment or Health Care (e.g., therapy or medication), Diagnostic or Lab Tests, Ethical Consideration (e.g., willing to consent), and Lifestyle Choice (e.g. diet or exercise). More category information can be found in our previous publication11. Two independent human raters manually labeled 2,718 criteria sentences. The Cohen’s Kappa22 was 0.82, which indicated an excellent inter-rater agreement. Table 1 shows the 27 criteria categories with their topic groups and frequency, and example instances. The most frequent category, Disease, Symptom or Sign, covered 29.21% of all the instances, followed by Diagnostic and Lab Tests, covering 14.63% of all the instances. The next two most frequent categories were Pharmaceutical Substance and Drug (12.84%) and Age (5.91%).
Table 1.
Criteria categories and groups and their distributions. For instance, 29.21% of eligibility criteria belong to the semantic category “Disease, Symptom and Sign.”
| Topic Groups | Semantic Classes | Distribution | Example |
|---|---|---|---|
| Health Status (43.72%) |
Disease, Symptom and Sign | 29.21% | No coagulation disorders. |
| Pregnancy-related Activity | 5.17% | Pregnancy ongoing or planned within 3 years. | |
| Neoplasm Status | 3.67% | Presence of rapidly progressive, life-threatening metastases. | |
| Disease Stage | 2.20% | No stage IIIB breast cancer. | |
| Allergy | 2.15% | Allergy to fluorescein. | |
| Organ or Tissue Status | 0.73% | Adequate renal function. | |
| Life Expectancy | 0.59% | Life expectancy of at least 3 months. | |
| Treatment or Health Care (20.74%) |
Pharmaceutical Substance or Drug | 12.84% | No prior Gabapentin. |
| Therapy or Surgery | 7.61% | Prior chemotherapy allowed. | |
| Device | 0.29% | Have an active implantable device. | |
| Diagnostic or Lab Test (14.85%) |
Diagnostic or Lab Results | 14.63% | Neutrophil count >= 1000 / mm3. |
| Receptor Status | 0.22% | ER and PGR negative. | |
| Demographics (8.79%) |
Age | 5.91% | Age 23–47. |
| Special Patient Characteristic | 1.18% | Accept Healthy Volunteers. | |
| Literacy | 0.65% | Able to understand and speak English. | |
| Gender | 0.41% | Sex: Female. | |
| Address | 0.35% | Resident of Toronto, Canada. | |
| Ethnicity | 0.29% | Patients must identify their ethnicity as Latino or Hispanic. | |
| Ethical Consideration (8.52%) |
Consent | 2.76% | Patient signs informed consent. |
| Enrollment in other Studies | 2.38% | Patients included in others clinical trials of imagery. | |
| Capacity | 1.50% | Able to perform 6 minute hall walk. | |
| Patient Preference | 1.38% | Agree to come to the clinic up to 2 times per week. | |
| Compliance with Protocol | 0.50% | Able to comply with all study procedures. | |
| Lifestyle Choice (3.38%) |
Addictive Behavior | 2.09% | Abuse alcohol or drugs. |
| Bedtime | 0.47% | Usual bedtime between 21:00 and 01:00. | |
| Exercise | 0.44% | Lack of access to regular meals. | |
| Diet | 0.38% | Use of grapefruit juice products. | |
| Total | -- | 100% | -- |
3.2 Distribution and Prevalence of the Criteria Categories
To assess the comprehensiveness of the semi-automatically induced eligibility criteria categories, we randomly selected 1,578 clinical trials and extracted 27,278 eligibility criteria to investigate the prevalence and frequency of each category in these criteria, as shown in Table 2. The column “average incidence in each trial” lists the average number of instances for each category in a typical clinical trial study; the column “prevalence in all trials” lists the percentage of trials containing instances of a particular category. For example, an average trial contains 4.5 criteria about Disease, Symptom and Sign, which are prevalent in 92.27% of trials. Eight classes, including Gender, Special Characteristics of Patients, Age, Disease or Symptoms or Signs, Diagnostic and Lab Results, Pharmaceutical Substance or Drug, Therapy or Surgery, and Pregnancy Related Activity, are each prevalent in from 45% to 99% of clinical trial studies. We call these classes “majority classes”. The remaining 19 categories are the “minority classes” that are prevalent in a smaller percent, ranging from 0.13% to 31.94%, of clinical trial studies.
Table 2.
Prevalence and average incidence of each class in 1,587 Clinical Trial Studies. For example, 99.11% studies contain 1.04 instances of gender criteria.
| Class | Average Incidence in each trial |
Prevalence in all trials |
|---|---|---|
| Gender | 1.04 | 99.11% |
| Special Characteristic of Patient | 1.20 | 96.51% |
| Age | 1.52 | 93.09% |
| Disease, Symptom and Sign | 4.51 | 92.27% |
| Diagnostic or Lab Results | 2.19 | 64.58% |
| Pharmaceutical Substance or Drug | 1.63 | 52.41% |
| Therapy or Surgery | 1.11 | 48.16% |
| Pregnancy-related Activity | 0.84 | 46.58% |
| Consent | 0.39 | 31.94% |
| Neoplasm Status | 0.80 | 31.50% |
| Allergy | 0.31 | 25.79% |
| Disease Stage | 0.37 | 22.94% |
| Addictive Behavior | 0.24 | 17.93% |
| Patient Preference | 0.22 | 16.10% |
| Capacity | 0.20 | 15.91% |
| Organ or Tissue Status | 0.18 | 13.81% |
| Enrollment in other studies | 0.15 | 13.37% |
| Life Expectancy | 0.11 | 10.58% |
| Literacy or Spoken Language | 0.08 | 7.03% |
| Address | 0.07 | 5.89% |
| Device | 0.06 | 5.07% |
| Compliance with Protocol | 0.02 | 2.22% |
| Exercise | 0.02 | 1.77% |
| Diet | 0.02 | 1.58% |
| Receptor Status | 0.01 | 0.89% |
| Ethnicity | 0.01 | 0.76% |
| Bedtime | 0.001 | 0.13% |
3.3 Feature Representation Comparison: UMLS vs. “bag of words”
A total of 3,403 randomly selected eligibility criteria were used to train the classifiers. We conducted a 10-fold cross-validation to compare the classification performance among 5 commonly used machine-learning classifiers, which were J48, Bayesian Network, Naïve Bayesian, the nearest-neighbor (NNge), and the instance-based learning classifier IB1, each using the “bag of words” representation (baseline) and the UMLS semantic type representation. Our comparison was from the following perspectives: (1) classification accuracy measured by precision, recall, and F1-score (Table 3); (2) classification accuracy by criteria category (Figure 3); (3) computational efficiency of classifiers (Figure 4); and (4) classifier learning efficiency (Figure 5). Recall, precision and F1-score are defined as follows:
| (1) |
| (2) |
| (3) |
Table 3.
F1-score is consistently higher when using the UMLS feature representation than using the “bag of words” feature representation among the five classifiers (P-value = 0.0001215, t-test). The scores are macro-averages for all criteria categories.
| Classifier | J48 | BayesNet | NaiveBayes | NNge | IB1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feature Representation | ST | BoW | ST | BoW | ST | BoW | ST | BoW | ST | BoW |
| Precision | 0.870 | 0.730 | 0.853 | 0.683 | 0.839 | 0.665 | 0.797 | 0.683 | 0.701 | 0.597 |
| Recall | 0.872 | 0.723 | 0.855 | 0.680 | 0.843 | 0.667 | 0.767 | 0.631 | 0.702 | 0.574 |
| F1-score | 0.869 | 0.717 | 0.852 | 0.667 | 0.836 | 0.646 | 0.772 | 0.622 | 0.699 | 0.566 |
Figure 3.
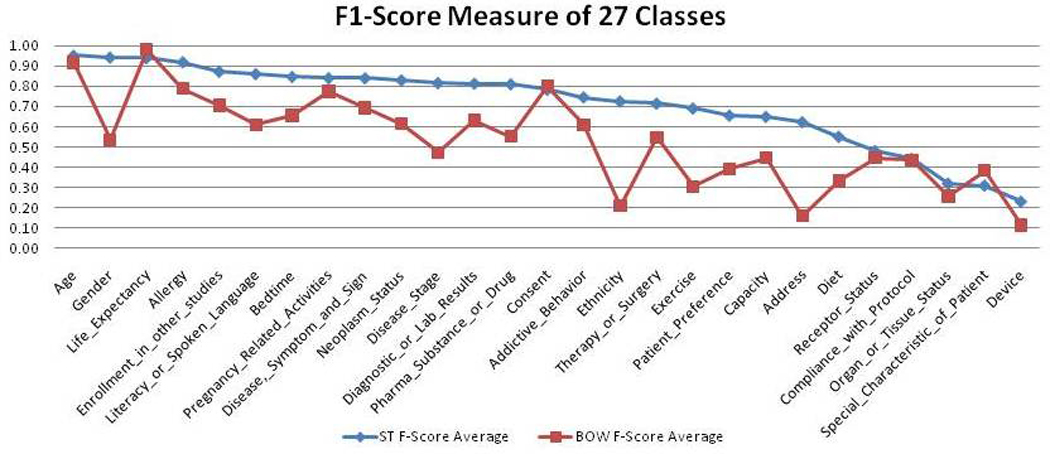
F1-scores of all categories using the UMLS and “bag of words” feature representation respectively.
Figure 4.
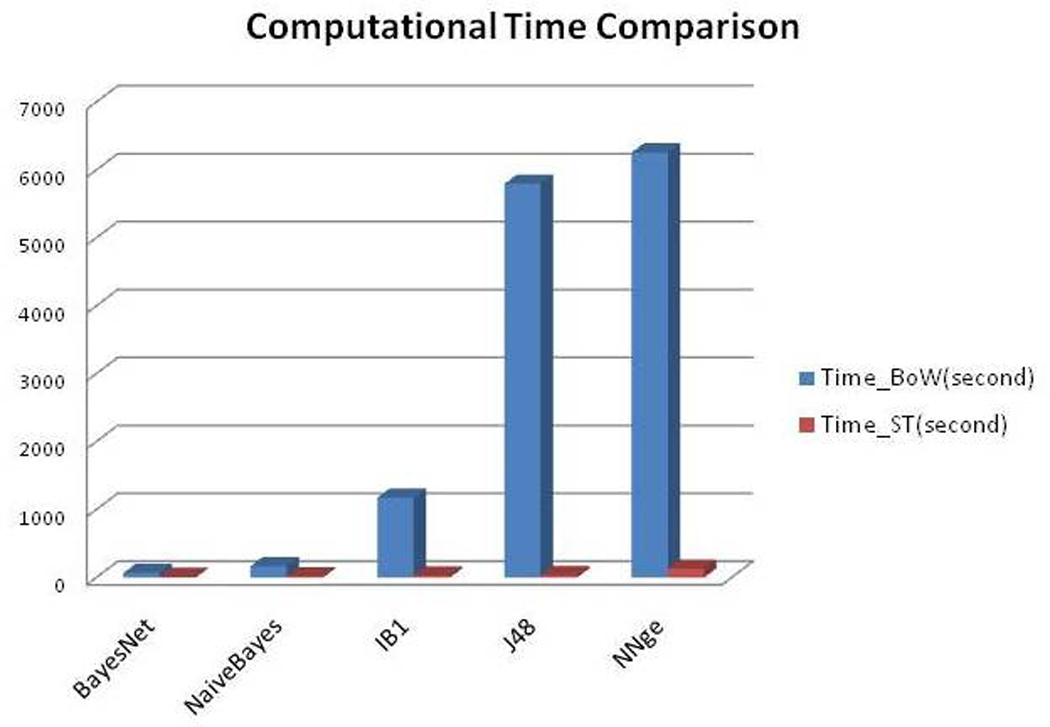
Time-efficiency between the “bag of words” and UMLS feature representation.
Figure 5.
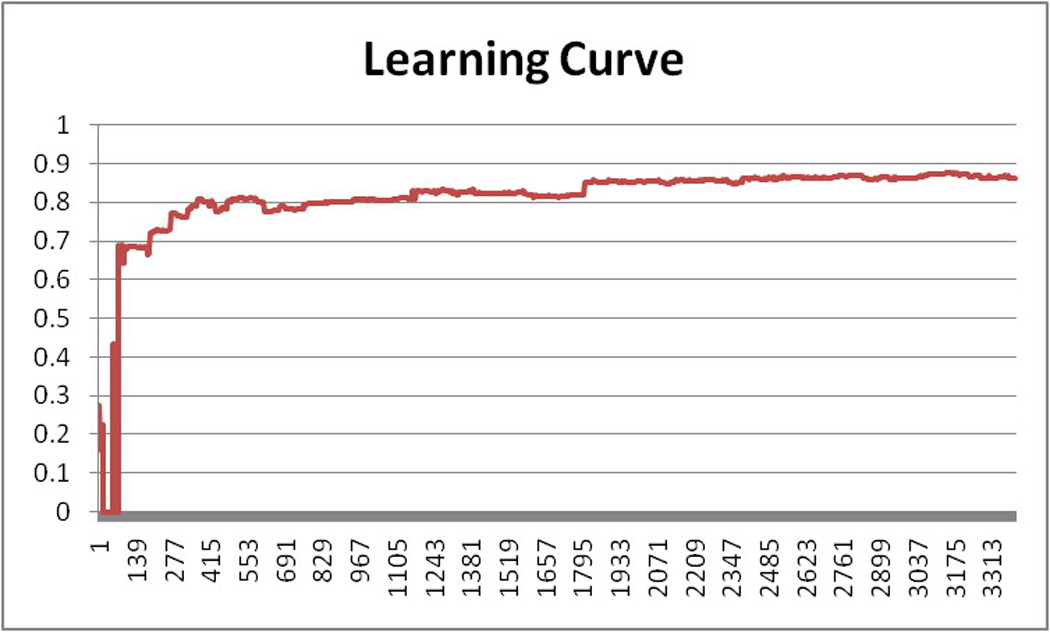
The learning efficiency of classifier J48 (X-axis: the number of training instances; Y-axis: F1-score of the J48 classifier)
As shown in Table 3, the UMLS semantic feature representation consistently outperformed the “bag of words” feature representation by achieving higher precision, recall and F1-score, with performance improvement ranging from 13.3% to 19.0% in all of the 5 classifiers. The Naïve Bayes classifier increased the F1-score by 19% by changing from the “bag of words” representation to the UMLS semantic type representation. Overall, the J48 classifier performed the best among the 5 classifiers, with a precision of 87.0%, recall at 87.2% and F1-score of 86.9% when using the UMLS semantic type representation.
Figure 3 contrasts the average F1-scores for the 27 semantic categories using the UMLS semantic type representation and the “bag of words” representation respectively. In 24 of the 27 semantic categories (89%), the UMLS semantic type representation outperformed the “bag of words” representation by using semantic knowledge that was unavailable in the “bag of words” feature representation to identify the semantic similarity between seemingly different terms. For example, criteria “Sex: Male” and “Inclusion: Female” did not share any learning feature in the “bag of words” representation because of the term variations. However, the UMLS semantic representations for both criteria shared the same UMLS semantic type Organism Attribute. Therefore, the UMLS semantic type representation was able to detect semantic similarity that was not captured in the “bag of words” representation. The P-value of the difference between the two feature representations for all 27 categories measured by a t-test23 was 0.00334 (P<0.05), indicating the statistical significance of the differences. Therefore, the performance of the UMLS semantic representation was significantly better than that of the “bag of words” representation.
Three rare categories, Life expectancy, Consent, and Special patient characteristics, did not show the advantages of the UMLS semantic type representation because the criteria belonging to those classes often contained salient keywords that were more effective than their semantic types for classification. For example, 75.53% of the criteria in the category Consent contained phrase “informed consent”. When using the “bag of words” representation, the classifier could easily tell whether a criterion belonged to the Consent category simply by looking up the keywords “informed” and “consent” in the feature set. In contrast, when using the semantic type representation, the term “informed consent” was mapped to the UMLS semantic type Regulation or Law which also included terms in criteria that were not related to consent, such as “drug regulations” or “health policy”. Using this semantic feature alone was not very sufficient to tell whether a criterion belonged to the Consent category. In this case, the use of the “bag of words” representation was more accurate than using the semantic type representation.
Figure 4 shows that the UMLS semantic type representation consistently required significantly less time than the “bag of words” representation across the 5 classifiers. The learning dimension of “bag of words” representation was much bigger than that of the semantic type representation. This might be explained by the fact that we obtained 4,413 distinct “bag of words” features but only 135 semantic-type features for the 3,403 training criteria. We also found that BayesNet classifier and NaïveBayes classifier were robust to resist the fast growth in the learning dimension and retained high efficiency, while efficiency was significantly impaired in IB1, J48 and NNge as the learning dimension expanded, as shown in Figure 4.
We also carried out an experiment to measure the learning efficiency of the best performing classifier of the five, J48, by dividing the 3,403 training criteria into groups, each containing only 3 criteria. The training process was divided into 1,134 steps, each step incrementally increasing the size of the training data set by 3. Therefore, the step-wise training sizes are 3, 6, 9,…3*K instances, where K is the sequential number of the step. The classification accuracy for each step was documented and plotted as a learning curve that grows with the size of the training data set, as shown in Figure 5. The learning speed increased fastest for the first 500 training instances. After that point, the learning performance increased relatively slowly. Finally, after about 2,000 training instances, the learning curve stabilized, indicating that a 3,403-sentence training set was sufficient to develop a stable model for classifying eligibility criteria.
3.4 A User Interface for Dynamic Categorization of Eligibility Criteria
At the point of this study, there was no standard and automatic way to categorize and organize clinical research eligibility criteria. We applied the learned classifier (J48) to organize the eligibility criteria on Clinicaltrials.gov. Figure 6 shows the eligibility criteria section of a clinical trial study on Clinicaltrials.gov, which is a bullet list. Figure 7 show the results of dynamic eligibility criteria categorization24. Using our J48 decision tree classifier, we can assign each criterion to its corresponding semantic category and organize the criteria section into a hierarchical tree, which could facilitate fast content browsing25 and faceted search26.
Figure 6.
Example of eligibility criteria narratives on Clinicaltrials.gov
Figure 7.
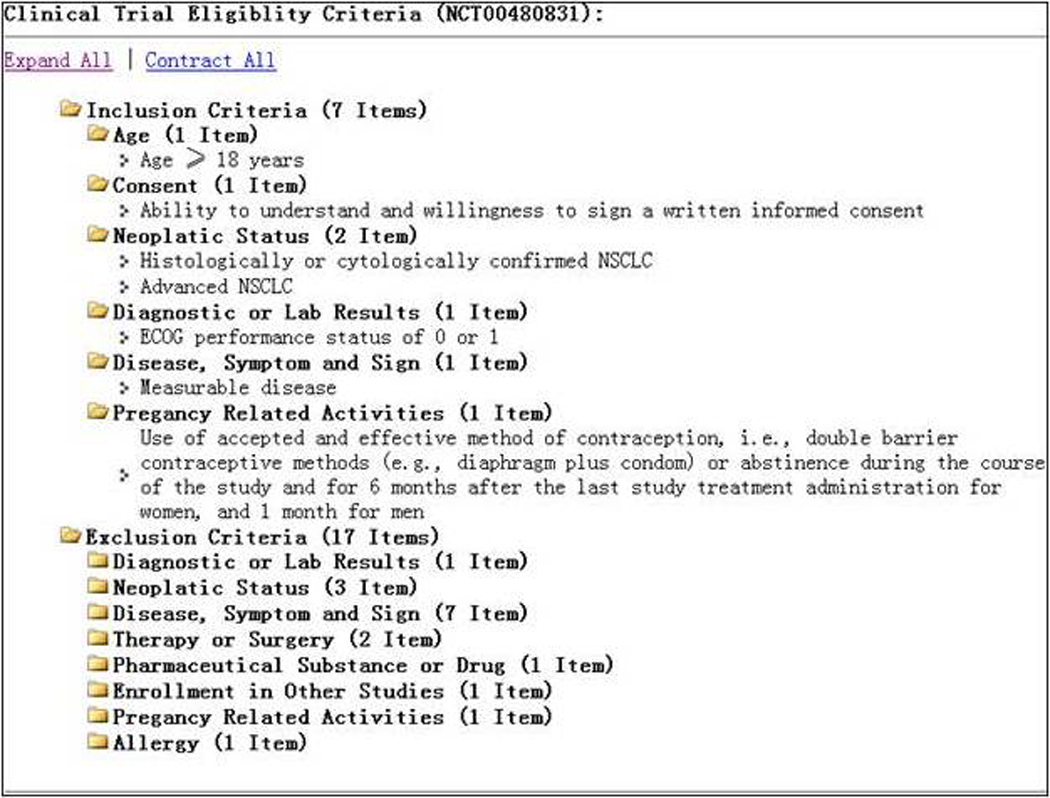
The dynamic categorization results for the example criteria in Figure 6.
4. Discussion
Clustering is an unsupervised learning technique that does not need a human labeled training set but rather identifies the similarities between instances, whereas classification is a supervised machine learning approach that needs to be trained using manually labeled examples27. In this paper, we present an effective approach to dynamic categorization of clinical research eligibility criteria by integrating hierarchical clustering and classification algorithms through the use of a shared semantic feature representation based on the UMLS semantic types. Our method demonstrates the value of using the UMLS semantic types for feature representation. To improve machine learning efficiency, various approaches have been developed to automate training data generation28–30. Our semantic annotator automatically generates features based on the UMLS semantic types and significantly reduces the learning dimension compared to the traditional “bag of words” method. Prior studies manually defined categories for clinical eligibility criteria4–5. Our method reduces the human effort required for category development and contributes a set of fine-grained semantic categories for clinical eligibility criteria. Moreover, previously proposed categories for clinical sentences were often task-dependent, such as the study that assigned Introduction, Methods, Results, or Discussion categories to sentences in MEDLINE abstracts31. To our knowledge, our research is the first of its kind to automatically categorize clinical research eligibility criteria based on the semantic similarities in the criteria. Of the 5 classification algorithms, the best performing classifier is the Decision Tree J48, which achieves an overall F1-score of 86.9%. In a different clinical domain, McKnight and Srinivasan32 reported a method incorporating sentence position and “bag of words” as learning feature and achieved results with F1-scores ranging from 52% to 79% for different categories. Compared to the existing methods, our method shows the potential to significantly improve sentence classification accuracy.
Our method for dynamic categorization of criteria sentences is inspired by and extends a notable related work for dynamic categorization of documents, which is the DynaCat system developed by Pratt25. DynaCat utilized the UMLS for knowledge-based query terms labeling and PubMed document classification. All query terms were automatically encoded with MeSH terms, but document categories and classification rules were manually specified for document categorization. We extended DynaCat by using the hierarchical clustering tools to automatically induce semantic categories for the objects to be categorized and by using a machine-learning approach to train the classifier, which was an improvement over manually defined rule-based classifiers. By using MeSH terms, DynaCat achieved standards-based query term annotation but did not reduce the feature space. As an extension, we used the UMLS semantic types to annotate eligibility criteria concepts and significantly reduced the feature dimension for machine learning-based classification. Furthermore, DynaCat performed categorization at the document level; in contrast, our method allows categorization at the sentence level.
We also compared our semi-automatically induced criteria categories to existing clinical data or clinical query categories provided by various standardization organizations, such as The Health Level Seven (HL7)33, the MURDOCK study group34, and the BRIDG group35. A significant portion of our categories overlaps with the manually defined standards. For instance, The Continuity of Care Document (CCD) defined by HL7 contains 17 clinical data types33, such as Problems, Procedures, and Medications, which are also included in our 27 categories. Those data elements that do not intersect with our categories include Header for message formatting and Payer for payment, which are not semantically interesting. The MURDOCK study group proposed 11 study variables34 for integrating clinical data. Several of them can be aligned with our categories, such as Demographics, Physical examinations, and Laboratory test results. The Biomedical Research Integrated Domain Group (BRIDG) Model was developed by a group of domain experts for clinical data modeling. The BRIDG model defined 17 eligibility criterion attributes. We were able to align 16 out of 17 BRIDG attributes with our induced semantic classes. We also identified 8 classes that were not specified by the BRIDG model. We observed that some highly prevalent criteria categories that we identified were not defined in BRIDG, such as Therapy or Surgery, which has 48% prevalence in eligibility criteria published on the ClinicalTrials.gov. These results imply that our criteria categories are comparable to those developed by clinical experts and contain categories that may be missed by clinical experts.
In this study, some classification errors were caused by the noise in the UMLS. For example, in the criterion “Alkaline Phosphatase < 2.5 times ULN,” the term Alkaline phosphatase had a UMLS semantic type Pharmacologic Substance; therefore, this criterion was classified as Pharma Substance or Drug. However, the criterion specifies the range of a lab test variable, which should be classified as Lab Test Results. Similarly, the criterion History of Cholecystectomy was mapped to a general semantic type Finding but human reviewer considered this criterion as a past surgery, whose category should be Therapy and Procedure.
We can improve our current methodology in several ways in the future. We identified two open research questions for classifying clinical sentences. One is to develop better machine learning algorithms for imbalanced training data. As we demonstrated in section 3.2, different categories achieved varying degrees of accuracy, which was partially caused by the different prevalence and incidence of these categories, as indicated in Table 2. Learning from imbalanced data sets where the number of examples of one (majority) class is much higher than others, machine-learning algorithms tend to produce better predictive accuracy over the majority classes but poorer predictive accuracy over the minority classes. This is an open research challenge for the machine learning community. Over-sampling algorithms36 can be used to improve the performance for minority classes. Another needed improvement is to develop multi-label classifier for eligibility criteria. Although the majority of eligibility criteria contain only one topic, there are still about 8% of eligibility queries containing multiple topics. For example, the criterion “pregnant women” contains two topics pregnancy and gender. Another example is “male and female with age between 18 and 65.” Multiple topics may also be present less explicitly in some examples; for instance, “positive pregnancy lab tests” could be categorized as both Lab Test Results and Pregnancy. However, our classifier only assigns one category to these eligibility criteria. The categories resulting from hierarchical clustering are not completely mutually exclusive and can contain some hidden relations (e.g., a set of lab tests for measuring pregnancy), which also could have affected the classification accuracy. These research questions are worth more studies in the future.
5. Conclusion
In this paper, we present a novel method that combines an unsupervised clustering algorithm with a supervised classification algorithm to develop a semantic classifier, which can be used to categorize clinical research eligibility criteria automatically. We also demonstrate the value of semantic knowledge such as the UMLS in improving the learning efficiency of semantic classifiers for clinical sentences such as clinical research eligibility criteria. Using the UMLS semantic types is far more effective and efficient than using words for feature representation when classifying clinical sentences, primarily because UMLS semantic knowledge matches the semantics in clinical text well.
Acknowledgments
The researchers were sponsored under NLM grant R01LM009886 and R01LM010815, CTSA award UL1 RR024156, AHRQ grant R01 HS019853. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. We thank Lorena Carlo and Meir Florenz for serving as reference standards for the automated semantic classifier. We also thank Yalini Senathirajah for her help with the Hierarchical Clustering Explorer software.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weng C, Tu SW, Sim I, Richesson R. Formal representation of Eligibility Criteria: A Literature Review. Journal of Biomedical Informatics. 2010;43(3):451–467. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCray AT. Better Access to Information about Clinical Trials. Annals of Internal Medicine. 2000;133(8):609–614. doi: 10.7326/0003-4819-133-8-200010170-00013. [DOI] [PubMed] [Google Scholar]
- 3.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. Journal of Biomedical Informatics. 2004;37(2):108–119. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, Sim I. A practical method for transforming free-text eligibility criteria into computable criteria. Journal of Biomedical Informatics. 2011;239(2):239–250. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niland J, Cohen E. ASPIRE: agreement on standardized protocol inclusion requirements for eligibility. 2007 [Google Scholar]
- 6.Tu SW. A Methodology for Determining Patients' Eligibility for Clinical Trials. Methods of Information in Medicine. 1993;32(4):317–325. [PubMed] [Google Scholar]
- 7.Metz JM, Coyle C, Hudson C, Hampshire M. An Internet-Based Cancer Clinical Trials Matching Resource. Journal of Medical Internet Research. 2005;7(3):e24. doi: 10.2196/jmir.7.3.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin RR. The Diabetes Prevention Program: recruitment methods and results. Controlled Clinical Trials. 2002;23(2):157–171. doi: 10.1016/s0197-2456(01)00184-2. [DOI] [PubMed] [Google Scholar]
- 9.Tassignon J-P, Sinackevich N. Speeding the Critial Path. Applied Clinical Trials. 2004 [Google Scholar]
- 10.Ross J, Tu S, Carini S, Sim I. AMIA Summit on Clinical Research Informatics. San Francisco, California: 2010. Analysis of Eligibility Criteria Complexity in Clinical Trials; pp. 46–50. [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z, Johnson SB, Weng C. American Medical Informatics Association Annual Symposium. Washington, DC: 2010. Semi-Automatic Induction of Semantic Classes from Free-Text Clinical Research Eligibility Criteria Using UMLS; pp. 487–491. [PMC free article] [PubMed] [Google Scholar]
- 12.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann, Witten I. The WEKA data mining software: an update. SIGKDD Explor. Newsl. 2009;11(1):10–18. [Google Scholar]
- 13.Quinlan R. C4.5: Programs for Machine Learning (Morgan Kaufmann Series in Machine Learning) Morgan Kaufmann; 1993. [Google Scholar]
- 14.Cooper G, Herskovits E. A Bayesian method for the induction of probabilistic networks from data. Machine Learning. 1992;9(4):309–347. [Google Scholar]
- 15.John G, Langley P. Estimating Continuous Distributions in Bayesian Classifiers; Proceedings of the Eleventh Conference on Uncertainty in Artificial Intelligence; 1995. pp. 338–345. [Google Scholar]
- 16.Martin B. Instance-Based learning : Nearest Neighbor With Generalization. Hamilton, New Zealand: Department of Computer Science, University of Waikato; 1995. [Google Scholar]
- 17.Aha DW, Kibler D, Albert MK. Instance-Based Learning Algorithms. Machine Learning. 1991;06(1):37–66. [Google Scholar]
- 18.Luo Z, Duffy R, Johnson S, Weng C. AMIA Summit on Clinical Research Informatics. San Francisco, California: 2010. Corpus-based Approach to Creating a Semantic Lexicon for Clinical Research Eligibility Criteria from UMLS; pp. 26–31. [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson SB. A Semantic Lexicon for Medical Language Processing. Journal of the American Medical Informatics Association. 1999;6(3):205–218. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SC. Hierarchical Clustering Schemes. Psychometrika. 1967;2:241–254. doi: 10.1007/BF02289588. [DOI] [PubMed] [Google Scholar]
- 21.Pearson K. Mathematical Contributions to the Theory of Evolution. XI. On the Influence of Natural Selection on the Variability and Correlation of Organs. Philosophical Transactions of the Royal Society of London. Series A, Containing Papers of a Mathematical or Physical Character. 1903;200:1–66. (ArticleType: research-article / Full publication date: 1903 / Copyright © 1903 The Royal Society) [Google Scholar]
- 22.Carletta J. Assessing Agreement on Classification Tasks: The Kappa Statistic. Computational Linguistics. 1996;22(2):249–254. [Google Scholar]
- 23.Zimmerman DW. A Note on Interpretation of the Paired-Samples t Test. Journal of Educational and Behavioral Statistics. 1997;22(3):349–360. [Google Scholar]
- 24.Weng C, Luo Z. Dynamic Categorization of Clinical Research Eligibility Criteria. Proc of AMIA Fall Symp. 2010;306 [Google Scholar]
- 25.Pratt W, Fagan L. The Usefulness of Dynamically Categorizing Search Results. Journal of the American Medical Informatics Association. 2000;7(6):605–617. doi: 10.1136/jamia.2000.0070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broughton V. Faceted classification as a basis for knowledge organization in a digital environment; the Bliss Bibliographic Classification as a model for vocabulary management and the creation of multidimensional knowledge structures. New Review of Hypermedia and Multimedia. 2001;7(1):67–102. [Google Scholar]
- 27.Mitchell TM. Machine Learning. McGraw-Hill Science/Engineering/Math; 1997. p. 432. [Google Scholar]
- 28.Nigam K, McCallum A, Thrun S, Mitchell T. Text Classification from Labeled and Unlabeled Documents using EM. Machine Learning. 2000;39(2):103–134. [Google Scholar]
- 29.Liu B, Li X, Lee WS, Yu PS. Text classification by labeling words; Proceedings of the 19th national conference on Artifical intelligence; California: San Jose; 2004. pp. 425–430. [Google Scholar]
- 30.Yetisgen-Yildiz M, Pratt W. The effect of feature representation on MEDLINE document classification; AMIA Annual Symposium Proceedings; 2005. pp. 849–853. [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal S, Yu H. Automatically classifying sentences in full-text biomedical articles into Introduction, Methods, Results and Discussion. Bioinformatics. 2009;25(23):3174–3180. doi: 10.1093/bioinformatics/btp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKnight L, Srinivasan P. Categorization of Sentence Types in Medical Abstracts; AMIA Annual Symposium; 2003. pp. 440–444. [PMC free article] [PubMed] [Google Scholar]
- 33.Dolin RH, Alschuler L, Beebe C, Biron PV, Boyer SL, Essin D, Kimber E, Lincoln T, Mattison JE. The HL7 Clinical Document Architecture. J Am Med Inform Assoc. 2001;8(6):552–569. doi: 10.1136/jamia.2001.0080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty S, Blach C, Gardner M, McCourt B, Nahm M, Tenenbaum JD. Proc of AMIA Summit for Clinical Research Informatics. San Francisco, CA: 2010. The MURDOCK Integrated Data Repository (MIDR): An integrative platform for biomarker research. [Google Scholar]
- 35.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG Project: A Technical Report. Journal of the American Medical Informatics Association. 2008;15(2):130–137. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han H, Wang W-Y, Mao B-H. Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning. In: Huang D-S, Zhang X-P, Huang G-B, editors. Advances in Intelligent Computing. Vol 3644. Heidelberg: Springer Berlin; 2005. pp. 878–887. [Google Scholar]



