Abstract
The ability of bacteria to adapt to a changing environment is essential for their survival. One mechanism bacteria have evolved to sense environmental cues and translate these signals into phenotypic changes uses the second messenger signaling molecule, cyclic diguanosine monophosphate (c-di-GMP). In addition to several classes of protein receptors, two classes of c-di-GMP-binding riboswitches (class I and class II) have been identified as downstream targets of the second messenger in this signaling pathway. The crystal structures of both riboswitch classes bound to c-di-GMP were previously reported. Here we further investigate the mechanisms that RNA has evolved for recognition and binding of this second messenger. Using a series of c-di-GMP analogs, we probed the interactions made in the RNA-ligand complex for both classes of riboswitches to identify the most critical elements of c-di-GMP for binding. We found that the structural features of c-di-GMP required for binding differ between these two effectors and that the class II riboswitch is much less discriminatory in ligand binding than the class I riboswitch. These data suggest an explanation for the predicted preferential use of the class I motif over the class II motif in the c-di-GMP signaling pathway.
INTRODUCTION
Cyclic diguanosine monophosphate (c-di-GMP) is a ubiquitous second messenger signaling molecule used by many bacterial species to translate diverse environmental cues into phenotypic changes essential for survival1-5. This signaling pathway regulates many important bacterial processes including the transition from a motile, planktonic lifestyle to a sessile, biofilm forming state6, 7 as well as playing a role in the virulence response of pathogenic organisms8-10. The concentration of c-di-GMP in the cell is tightly regulated by proteins that either synthesize (diguanylate cyclases)11-13 or degrade (phosphodiesterases)14 the second messenger primarily in response to extracellular signals.
To initiate the phenotypic changes necessary for cellular adaptation, c-di-GMP must interact with downstream macromolecular targets4-6. Several c-di-GMP-binding proteins have been identified within the bacterial domain that act as effectors in this pathway6. These include members of the PilZ domain containing family,15-21 transcription factors,22-24 and degenerate diguanylate cyclases and phosphodiesterases,25-29 which have lost catalytic function but retain their ability to bind c-di-GMP. Despite the progress made towards elucidating the downstream targets of this second messenger, the mechanism of action of many of these c-di-GMP-binding proteins is still unknown.
Additionally, two classes of c-di-GMP-binding riboswitches, termed class I (c-di-GMP-I) and class II (c-di-GMP-II), were identified as part of this signaling pathway30, 31. Riboswitches are non-coding RNA elements that bind small molecule ligands with high affinity and specificity32-35. They contain two domains, the aptamer domain and the expression platform. Ligand binding to the aptamer domain induces structural rearrangements within the RNA that cause changes in the expression levels of the downstream genes, typically by affecting either transcription or translation36-39. Over five hundred examples of class I c-di-GMP-binding riboswitches and 45 examples of class II riboswitches have been identified in diverse bacterial species, with a few instances of both RNA motifs present in a single organism30, 31. The identification of these riboswitches is of particular interest because unlike protein effectors, ligand binding is directly coupled to gene regulation, suggesting a mechanism for how c-di-GMP induces a cellular response upon binding this class of effectors. In addition, the broad distribution of these motifs across the bacterial domain indicates that c-di-GMP-binding riboswitches are likely one of the primary targets of this second messenger30, 31.
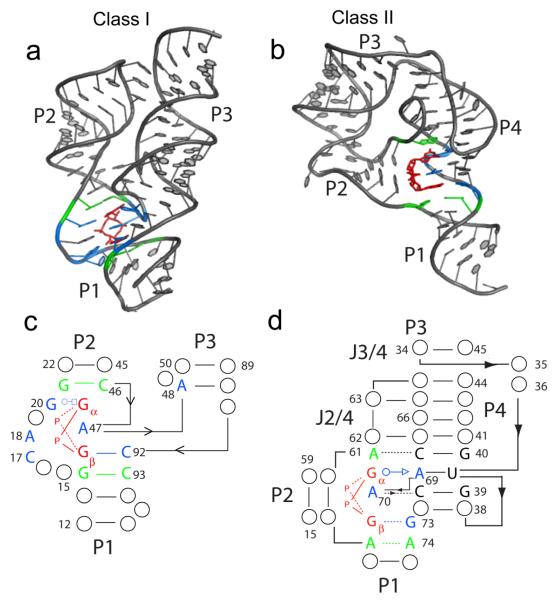
The crystal structures of c-di-GMP bound to both the class I and class II riboswitch have been determined40-43 (Figure 1). Consistent with the different predicted secondary structures for the two riboswitches, the overall three dimensional architecture of these RNAs is also distinct, with each providing unique binding pockets for c-di-GMP and therefore utilizing different modes of ligand recognition. Class I riboswitches consist of three helices that adopt a y-shape40-42, whereas class II folds into a more compact structure containing a kink-turn and pseudoknot43. The second messenger ligand is incorporated into structural RNA elements when bound to both riboswitches, a double helix in class I and a triple helix in class II, likely contributing to the extremely tight binding affinities observed for these RNA receptors (10 pM to low nanomolar)30, 31, 40 as compared to protein receptors (approximately 50 nM to low micromolar)21-23, 26.
Figure 1.
Crystal structures of the class I and class II riboswitch aptamer domains bound to c-di-GMP. c-di-GMP is colored in red, nucleotides in direct contact with the ligand are show in blue, and nucleotides that stack directly above and below the ligand are shown in green. (a) Structure of the class I Vc2 aptamer from V. cholera bound to c-di-GMP (PDB ID 3MXH). (b) Structure of the class II Cac-1-2 aptamer from C. acetobutylicum bound to c-di-GMP (PDB ID 3Q3Z). (c) Binding pocket of the class I aptamer. (d) Binding pocket of the class II aptamer. Nucleotides that form base triples with residues directly contacting c-di-GMP are shown.
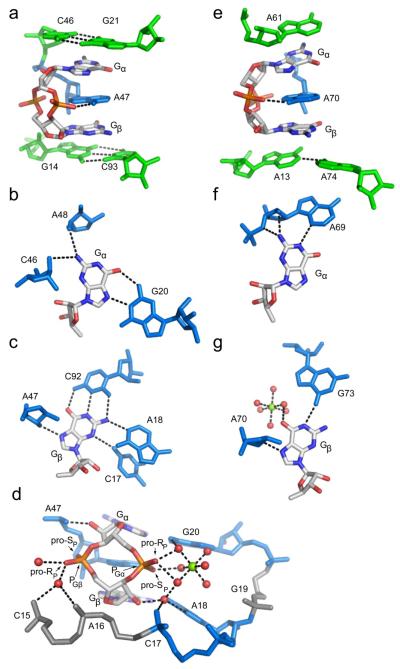
c-di-GMP binds to the class I riboswitch at the junction of three helices, P1, P2, and P3. Two critical nucleotides specifically recognize the guanine bases of the ligand in an asymmetric fashion40-42 (Figures 1a,c; 2b,c). G20 contacts the top base, designated Gα, along its Hoogsteen face, while C92 interacts with the second base, Gβ, in a canonical Watson-Crick pair. A third critical nucleotide, a highly conserved adenosine (A47), intercalates between the two bases of the ligand, resulting in extensive stacking interactions that bridge the P1 and P2 helices (Figures 1a,c; 2a). Recognition of the phosphodiester backbone also plays a role in c-di-GMP binding by the class I riboswitch. Both metal coordination and hydrogen bonding contacts to the phosphates are predicted, with the phosphate 5′ of Gα more heavily recognized than that of Gβ (Figure 2d). Furthermore, the class I riboswitch uses the ribose 2′ hydroxyls for second messenger recognition as evidenced by specific hydrogen bonds made to this functional group of c-di-GMP (Figure 2d).
Figure 2.
c-di-GMP recognition by the class I and class II riboswitches. Coloring of RNA residues is the same as in Figure 1. c-di-GMP is colored by atom with carbon shown in white, oxygen in red, nitrogen in blue, and phosphorus in orange. Hydrogen bonds are shown as blacked dashed lines. (a) Side view of c-di-GMP bound to the class I aptamer. The G21-C46 base pair is the first base pair of the P2 helix and the G14-C93 base pair is the first of the P1 helix. A47 intercalates between Gα and Gβ. (b) Gα recognition and (c) Gβ recognition by the class I riboswitch. (d) Backbone recognition of c-di-GMP by the class I riboswitch. The phosphate 5′ of Gα (PGα) is more heavily contacted than the phosphate 5′ of Gβ (PGβ) and each ribose 2′-OH is involved in a single hydrogen bonding contact. PGα is coordinated by a magnesium ion, shown as a green sphere. Coordinated water molecules are shown as red spheres. (e) c-di-GMP is bound to the class II riboswitch in a similar conformation to class I, with adenosines stacking above (A61), below (A13 and A74), and in between (A70) Gα and Gβ. The hydrogen bond between the exocyclic amine of A70 and a non-bridging phosphate oxygen of c-di-GMP is the only backbone contact. (f) Interactions made by the class II aptamer to Gα and (g) Gβ.
Similar to the class I riboswitch, c-di-GMP recognition by the class II riboswitch is also achieved through asymmetric contacts to the bases, however, the nature of these interactions differ significantly from class I43 (Figures 1b,d; 2f,g). No canonical pairings are observed for c-di-GMP binding to class II. Instead, Gα is recognized as part of a base triple with A69 and U37, and Gβ is contacted by hydrogen bonds from RNA residues A70 and G73, as well as by a hydrated magnesium ion (Figure2f,g). Stacking interactions are also important for ligand recognition, as evidenced by three conserved adenosine nucleotides that stack between (A70), above (A61) and below (A13) the bases of c-di-GMP (Figures 1b,d; 2e). These observed stacking interactions in the binding pocket are the only direct similarities in ligand recognition between the two riboswitch classes, suggesting that this is an important mechanism for c-di-GMP binding. In contrast to the extensive backbone recognition of c-di-GMP by the class I riboswitch, only a single hydrogen bond between the intercalating adenosine (A70) and a non-bridging phosphate oxygen of the ligand is observed for class II (Figure 2e). This interaction was also observed in the class I riboswitch40, 42. Based on the molecular views of c-di-GMP bound to these riboswitch effectors, it is evident that RNA has evolved two distinct mechanisms for recognition of the same second messenger ligand.
The crystal structures of c-di-GMP bound to both the class I and class II aptamers indicate which interactions made to the ligand are important for binding42, 43. Given that these two riboswitches use different strategies for c-di-GMP recognition, we anticipated that the ligand specificity between these two RNAs would also differ. We previously reported the selective targeting of the class II riboswitch using a 2′-O-methyl analog of the second messenger, demonstrating that these two riboswitches differ at least in their use of the c-di-GMP ribose rings for ligand binding43. Furthermore, c-di-GMP-binding riboswitches are prevalent in a large number of pathogenic organisms30, 31 and the ability to manipulate these riboswitches and the biological processes they regulate is therefore an advantageous goal44-48. Here, we set out to identify which moieties of the ligand are most important for binding by each riboswitch class and to understand how the structural features of c-di-GMP required for binding differ between the two classes. To investigate these questions, we used a series of c-di-GMP analogs to systematically perturb the interactions made between the aptamer domain of each riboswitch class and the bases and ribosyl-phosphate backbone of the ligand. This work reveals which elements of c-di-GMP necessary for binding by class I differ from those required by class II and shows that the class II riboswitch is much less discriminatory in ligand recognition. These second messenger analogs could potentially be used to control RNA-mediated c-di-GMP signaling pathways.
MATERIALS AND METHODS
Materials
c-di-GMP was synthesized enzymatically as previously described43, 49. Nucleotide analogs of c-di-GMP were chemically synthesized on solid support using phosphoramidites purchased from either ChemGenes or Glen Research. The controlled pore glass (CPG) solid support, 3-(4,4′-dimethoxytrityloxy)-2,2-(dicarboxymethlyamido)propyl-1-O-succinoyl-long chain alkylamino-CPG (3′-CPR-II CPG), was purchased from Glen Research. DNA/RNA synthesis grade acetonitrile (ACN), anhydrous pyridine, triethylamine (TEA), 1-(mesitylene-2-sulfonyl)-3-nitro-1,2,4-triazole (MSNT) and triethylamine-trihydrofluoride (HF-TEA) were purchased from Sigma-Aldrich. The oxidation reagent tert-buytl hydroperoxide/toluene was prepared as previously described50. All RNA molecules were cloned and transcribed in vitro using T7 RNA polymerase as previously described40, 42, 43. Synthetic RNA oligonucleotides were purchased from Dharmacon and deprotected according to the manufacturer’s protocol. Synthetic DNA oligonucleotides were purchased from the W.M Keck Facility (Yale University). T4 RNA Ligase 2 and T4 polynucleotide kinase (PNK) were purchased from New England Biolabs.
Chemical synthesis of c-di-GMP analogs
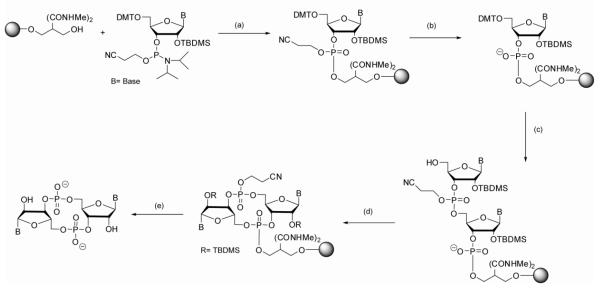
Phosphorothioate modified analogs were synthesized and characterized as previously described51. All analogs containing a standard phosphodiester backbone were synthesized using solid phase chemistry (Scheme 1) on a 2 μmol scale as previously described43, 52-55 with the following adaptations. 5′-DMTr-2′-OTBDMS cyanoethyl phosphate protected phosphoramidites (50 mM in acetonitrile (ACN)) were coupled to the solid support using 5-benzylmercaptotetrazole (125 mM in ACN) as the activator. The phosphate linkage was oxidized using a 1 M solution of tert-butyl hydroperoxide in toluene, followed by capping of unreacted sites on the support using a 1:1 mixture of 10% acetic anhydride/ACN and 10% 1-methylimidazole/ACN. To allow for cyclization of the dinucleotide on-bead, the cyanoethyl phosphate protecting group was removed in 50% TEA/ACN prior to 5′-detritylation in 3% DCA/DCM. After detritylation, coupling of the second phosphoramidite was performed, followed by oxidation, capping and detritylation. The dinucleotide was cyclized on bead under dilute conditions with 0.1 M MSNT in anhydrous pyridine for 12-24 hours. Beads were washed with pyridine and dried under argon and the cyclization procedure was repeated for another 12-24 hours until the reaction proceeded for a total of 72-96 hours. Global deprotection and cleavage from the solid support was afforded by incubation of the beads with a 1:1 mixture of ammonium hydroxide and 40% aqueous methylamine at 65°C for 10 minutes. For 2′-OH analogs, treatment with HF-TEA for 90 minutes at 65°C resulted in TBDMS deprotection. All molecules were purified by HPLC on a C18 reverse phase column using a gradient of 0% to 5% ACN in 50 mM triethylammonium acetate, pH 6.0. The identity of all compounds was confirmed by ESI-MS in negative ion mode (supplemental info., Table S1) and purity determined by analytical HPLC analysis (supplemental info., Figure S1). The chemistry employed here is compatible with 2′-tert-butyldimethylsilane (TBDMS), 5′-dimethoxytrityl (DMTr) and cyanoethyl (CNE) phosphate protected RNA phosphoramidites, which are the most common commercially supplied forms of the starting monomers. This scheme allows access to any c-di-GMP derivative for which the standard phosphoramidite is commercially available.
Scheme 1.
Solid-phase synthesis of base and ribose modified c-di-GMP analogs containing a standard phosphodiester backbone. The first phosphoramidite was coupled to the bead followed by oxidation, capping and selective removal of the cyanoethyl phosphate protecting group to provide the free 3′-hydroxyl necessary for the cyclization reaction. After coupling of the second phosphoramidite and deprotection of the 5′-OH group, the linear dinucleotide was cyclized on bead. Cleavage from the solid support and deprotection yielded the pure cyclic dinucleotide following HPLC purification. (a) i) tetrazole/ACN ii) tBuOOH iii) acetic anhydride/methylamine; (b) i) 50% TEA/ACN, 2 hours; (c) i) 3% DCA/DCM ii) tetrazole/ACN + CNE phosphoramidite iii) tBuOOH iv) acetic anhydride/methylamine (d) 0.1M MSNT, 72-96 hours; (e) i) ammonium hydroxide ii) HF-TEA (for 2′-OH analogs only).
Preparation of class I RNA with site-specific incorporation of 2-aminopurine
The wild-type class I Vc2 riboswitch from Vibrio cholerae of the sequence 5′-GGAAAAAUGUCACGCACAGGGCAAACCAUUCGAAAGAGUGGGACGCAAAGCCUCCGGCCUAAACCAGAAGACAUGGUAGGUAGCG85GGGUUACCGAUGGCA-3′ was transcribed in vitro up to and including G85, followed by the HDV ribozyme sequence for production of homogeneous 3′-ends for subsequent ligation. Cleavage by the ribozyme produced a 2′-3′-cyclic phosphate on the 3′-end of the RNA that was removed by treatment with T4 PNK (100 μM ATP, 100 mM imidazole, pH 6.0, 5 mM β-ME, 20 μg/mL BSA, 10 mM MgCl2) for 6 hours at 37°C as previously described56. Dephosphorylated RNA was ligated to a 5′-phosphorylated chemically synthesized RNA oligonucleotide containing a 2-aminopurine (2AP) fluorescent base analog (5′-GGGUUACC(2AP)AUGGCA-3′). Enzymatic ligation was performed with T4 RNA ligase 2 by splinted ligation using a 24 nucleotide DNA splint (5′-CATCGGTAACCCCGTTACCTACCA-3′). The RNA substrates and DNA splint were mixed in 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 1 mM DTT, and 400 μM ATP and heated to 70°C for 5 minutes followed by a 10 minute incubation at room temperature (22°C) to promote annealing57, 58. T4 RNA ligase 2 was added and the reaction was incubated at 37°C for 6 hours. Ligated RNA was purified by denaturing polyacrylamide gel electrophoresis (PAGE), excised from the gel and eluted in 300 mM sodium acetate, pH 5.2 for 18 hours at 4°C. RNA was concentrated and washed with water in a centrifugal filter (Amicon Ultra, 10K MWCO). The final sequence of the ligated class I aptamer, designated G94(2AP), was 5′-GGAAAAAUGUCACGCACAGGGCAAACCAUUCGAAAGAGUGGGACGCAAAGCCUCCGGCCUAAACCAGAAGACAUGGUAGGUAGCGGGGUUACC(2AP)AUGGCA-3′.
Kd measurements of analogs to the class I riboswitch using 2AP fluorescence
All fluorescence measurements were taken at room temperature (22°C) on a Photon Technology International (PTI) scanning spectrofluorometer with excitation and emission slits set to 5 nm. For Kd measurements, a 250 μL reaction volume with 200 nM G94(2AP) RNA in buffer containing 10 mM NaCl, 10 mM MgCl2, and 10 mM sodium cacodylate, pH 6.8 was prepared in a quartz cuvette. Fluorescence spectra were obtained using an excitation wavelength of 310 nm and emission was recorded from 325 nm to 425 nm. A concentrated solution of ligand was added directly into the cuvette and the fluorescence intensity at 360 nm was monitored after each addition to determine when the binding reaction reached equilibrium59, 60. Ligand was titrated from a concentration of 60 nM to approximately 15 μM, and longer incubation times were required to reach equilibrium for the lower ligand concentrations. For analogs with a Kd ≥ 15 μM, a complete binding curve could not be obtained and instead an estimation of the Kd is reported based on the amount of fluorescence quenching observed at the highest concentration tested (see supplemental info.). The emission spectrum for each ligand concentration was recorded and the Kd was determined by monitoring the decrease in fluorescence intensity (FI) at 360 nm. The FI at 360 nm was normalized to the fluorescence observed in the absence of ligand and plotted against ligand concentration. Data were fit to the quadratic equation as follows:
| (1) |
where FI=fluorescence intensity observed at 360 nm, FI0= fluorescence intensity at 360 nm in the absence of ligand, FI∞= difference between the fluorescence intensity at saturation and fluorescence in the absence of ligand, LT= concentration of ligand, RT= total RNA concentration (200 nM), and Kd= binding affinity of ligand.
Kdmeasurements of c-di-GMP by gel-shift and Kd measurements of analogs by competition gel-shift
Radiolabeled c-di-GMP was enzymatically synthesized using the purified PleD diguanylate cyclase protein as previously described61. The wild-type class II aptamer from Clostridium acetobutylicum of the sequence 5′-GUAUUUGUUUGGAAACAAUGAUGAAUUUCUUUAAAUUGGGCACUUGAGAAAUUUUGAGUUAGUAGUGCAACCGACCAACGAUUA-3′ was transcribed in vitro using T7 RNA polymerase as previously described43. RNA was folded in the presence of trace amounts of radiolabeled c-di-GMP by heating to 70°C and slow cooling in folding buffer (10 mM NaCl, 10 mM MgCl2, and 10 mM sodium cacodylate, pH 6.8). Binding reactions were incubated at room temperature (22°C) until equilibrium was reached, (24 hours for class I (G94(2AP) RNA) and 1 hour for class II (WT RNA)). The Kd of c-di-GMP for each riboswitch was then measured by separating free c-di-GMP from RNA-bound c-di-GMP by native PAGE (100 mM Tris/HEPES pH 7.5, 10 mM MgCl2, 0.1 mM EDTA) at 4°C and the fraction of c-di-GMP bound at each RNA concentration was determined. Gels were scanned using a STORM phosphorimager (GE Healthcare) and ImageQuant (GE Healthcare) was used to quantify bands. Data was fit to the following equation to determine Kd values:
| (2) |
with FB∞= fraction of c-di-GMP bound at RNA saturation and RT= total RNA concentration.
Competition experiments to determine the Kd of analogs were performed under similar conditions. In this case, radiolabeled c-di-GMP and unlabeled competitor analog were premixed in folding buffer before adding RNA to a final concentration of 25 nM (class I (G94(2AP) RNA)) or 50 nM (class II (WT RNA)). RNA was heated to 70°C for 3 min and slow cooled in the presence of both labeled and unlabeled ligand and incubated at room temperature for 4-24 hours (class II) or 48-72 hours (class I) before resolving free and bound c-di-GMP by native PAGE. We observed no changes in the measured binding affinities for incubation times longer than 4 hours for class II or 48 hours for class I, indicating that equilibrium had been achieved. The fraction bound (FB) of labeled c-di-GMP was quantified and the Kd of the unlabeled competitor analog was determined from the following equation as previously reported43, 62:
| (3) |
where FB∞= fraction bound of c-di-GMP at saturating concentrations of competitor analog, FB0= fraction bound in the absence of competitor, CcdiG= concentration of labeled c-di-GMP (estimated as 0.025nM based on the efficiency of the enzymatic labeling reaction), KdcdiG= affinity of c-di-GMP for the riboswitch, CT= concentration of unlabeled competitor analog, KdC= affinity of competitor analog, and RT=total concentration of riboswitch RNA.
The C92U class I mutant RNA was transcribed and purified as previously described40. The affinity of c-GMP-AMP for the C92U mutant was determined by competition with radiolabeled c-di-GMP as described above using 250 nM C92U RNA. The affinities of the c-GMP-AMP deoxy derivatives for the C92U RNA were also measured using the competition assay. Binding reactions were incubated at room temperature for 48 hours before analysis by native PAGE.
To determine the change in binding energy (ΔΔGbind), the binding energy (ΔGbind) of each analog was first obtained from the following equation:
| (4) |
where R=universal gas constant and T= temperature. ΔΔGbind was then calculated according to:
| (5) |
RESULTS
Design and synthesis of c-di-GMP analogs
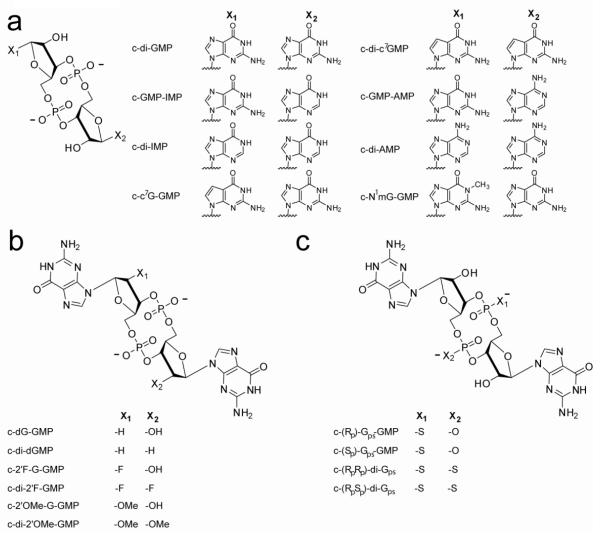
The crystal structures of c-di-GMP bound to the class I and class II riboswitch reveal the interactions made with the ligand42, 43, yet it remains unclear which of these interactions with c-di-GMP are most important for binding. To identify the interactions most critical for binding by each riboswitch class, we designed and chemically synthesized a series of c-di-GMP analogs, systematically modifying the bases (Figure 3a), ribose rings (Figure 3b), and phosphate backbone (Figure 3c) of the second messenger. All analogs were cyclic di-nucleotides, and we prepared both the symmetric (modification of both GMP units) and the asymmetric (modification of a single GMP unit) versions of each analog, except for the N1-methyl guanine derivative (c-N1mG-GMP). Base and ribose modified analogs were synthesized on solid-phase (Scheme 1) and phosphate modified analogs were synthesized in solution51. Both the symmetric and asymmetric analogs were tested on the class I riboswitch and a subset of these analogs were tested on class II.
Figure 3.
Structures of the c-di-GMP analogs used in this study. X1 and X2 indicate where modifications were made within each series. (a) Base modified, (b) ribose modified, and (c) phosphate modified analogs.
Affinity measurements of c-di-GMP and its analogs for the class I and class II riboswitch
In order to measure the binding affinities of c-di-GMP analogs, we used two techniques, a competition gel-shift experiment and a fluorescence based assay. In the case of the class I riboswitch, the high affinity (Kd 10 pM) and slow rates of ligand binding cause the approach to equilibrium to be extremely slow and essentially unattainable on an experimental timescale40. For this reason, the wild-type sequence could not be used for affinity measurements. To overcome this challenge, we used a version of the class I riboswitch that was modified for fluorescence in both assays because it has a weaker affinity for c-di-GMP such that equilibrium can be achieved at a faster rate.
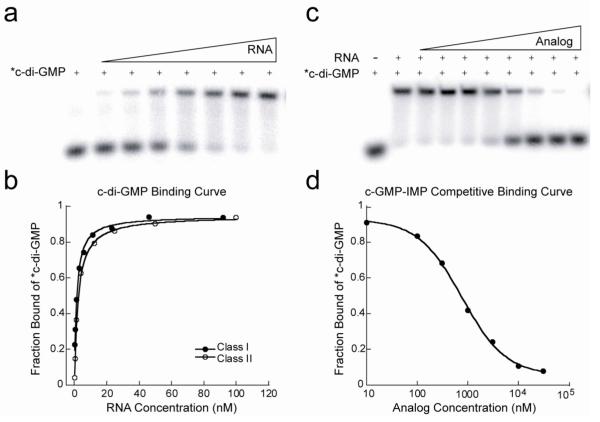
We have previously reported a gel-shift assay using radiolabeled c-di-GMP to characterize the binding of the second messenger to its riboswitch targets40, 42, 43. Using this method, the affinity of the class I riboswitch (G94(2AP) variant, see below) for c-di-GMP was 1.4 nM and that for the class II riboswitch was measured as 2.2 nM (Tables 1,2, Figure 4a,b). Radioactive versions of c-di-GMP were prepared enzymatically but it is not feasible to prepare cyclic dinucleotide analogs using enzymatic synthesis. To measure the affinities of the unlabeled analogs, we used a competition gel-shift assay with radiolabeled c-di-GMP. RNA and labeled c-di-GMP were incubated in the presence of increasing concentrations of the unlabeled competitor analog and the fraction of radiolabeled c-di-GMP displaced from the riboswitch was monitored to determine the affinity of the competitor43 (Figure 4c,d).
Table 1.
Binding affinities of analogs measured for the class I riboswitch (G94(2AP) RNA).
| Competition | Fluorescence | ||||||
|---|---|---|---|---|---|---|---|
| Analog | Kd (nM) | Fold Loss | ΔΔGbind (kcal/mol) |
Kd (nM) | Fold Loss | ΔΔGbind (kcal/mol) |
|
| c-di-GMP | 1.4 ± 0.1a | - | - | 16b | - | - | |
|
| |||||||
| Base | c-GMP-IMP | 39 ± 2.4 | 27 | 2.0 | 75 ± 14 | 5 | 0.9 |
| c-di-IMP | 1100 ± 210 | 790 | 3.9 | ≥ 15,000 | ≥ 940 | ≥ 4.1 | |
| c-c7G-GMP | 1100 ± 80 | 790 | 3.9 | ≥ 15,000 | ≥ 940 | ≥ 4.1 | |
| c-di-c7GMP | n.b.c | - | - | n.b. | - | - | |
| c-GMP-AMP | 1600 ± 160 | 1200 | 4.0 | - | - | - | |
| c-di-AMP | n.b. | - | - | n.b. | - | - | |
| c-N1mG-GMP | 420 ± 16 | 300 | 3.4 | - | - | - | |
|
| |||||||
| Ribose | c-dG-GMP | 51 ± 5.4 | 37 | 2.1 | 140 ± 24 | 9 | 1.3 |
| c-di-dGMP | 2600 ± 88 | 1800 | 4.4 | ≥ 20,000 | ≥ 1300 | ≥ 4.2 | |
| c-2’F-G-GMP | - | - | - | 25 ± 3.7 | 2 | 0.3 | |
| c-di-2’F-GMP | 56 ± 9.0 | 40 | 2.2 | 180 ± 15 | 11 | 1.4 | |
| c-2’OMe-G-GMP | 200 ± 31 | 150 | 2.9 | - | - | - | |
| c-di-2’OMe-GMP | ≥ 30,000 | ≥ 21,000 | 5.9 | n.b. | - | - | |
|
| |||||||
| Phos. | c-(Rp)-Gps-GMP | - | - | - | 67 ± 11 | 4 | 0.8 |
| c-(Sp)-Gps-GMP | - | - | - | 280 ± 40 | 18 | 1.7 | |
| c-(RpRp)-di-Gps | 150 ± 33 | 100 | 2.8 | 320 ± 70 | 20 | 1.8 | |
| c-(RpSp)-di-Gps | 750 ± 63 | 540 | 3.7 | 1700 ± 160 | 110 | 2.8 | |
Measured by direct binding.
Value determined by Kd = koff/kon (see supplemental info, Table S2).
No detectable binding at highest ligand concentration tested (250 μM by competition, 20 μM by fluorescence).
Table 2.
Binding affinities of analogs measured for the class II riboswitch.
| Analog | Kd (nM) | Fold Loss | ΔΔGbind (kcal/mol) | |
|---|---|---|---|---|
| c-di-GMP | 2.2 ± 0.2a,b | - | - | |
|
| ||||
| Base | c-di-IMP | 4.1 ± 0.4 | 1.9 | 0.4 |
| c-c7-GMP | 33.0 ± 2.6 | 15.0 | 1.6 | |
| c-di-c7GMP | 3500 ± 630 | 1600 | 4.4 | |
| c-GMP-AMP | 271.1 ± 47.1 | 120 | 2.8 | |
| c-di-AMP | ≥ 30,000 | ≥ 13,000 | ≥ 5.6 | |
| c-N1mG-GMP | 160 ± 14 | 72 | 2.5 | |
|
| ||||
| Ribose | c-di-dGMP | 11 ± 1.2 | 5.0 | 0.9 |
| c-di-2’F-GMP | 5.8 ± 1.0 | 2.6 | 0.6 | |
| c-di-2’OMe-GMP | 4.3 ± 0.7b | 2.0 | 0.4 | |
|
| ||||
| Phosphate | c-(RpRp)-di-Gps | 3.6 ± 0.6 | 1.6 | 0.3 |
| c-(RpSp)-di-Gps | 4.0 ± 0.8 | 1.8 | 0.4 | |
Measured by direct binding.
Values previously reported43.
Figure 4.
Kd measurements of c-di-GMP by gel-shift and its various analogs using the competition gel-shift assay with radiolabeled c-di-GMP. (a) Representative gel-shift experiment for measuring the Kd of c-di-GMP for each class of RNA by direct binding. (b) c-di-GMP binding curves for the class I (G94(2AP) and class II riboswitches. (c) Representative competition gel-shift experiment. RNA, radiolabeled c-di-GMP and increasing concentrations of competitor analog are incubated until equilibrium is achieved. Free c-di-GMP is separated from RNA-bound c-di-GMP by native PAGE. (d) Sample binding curve from the competition gel-shift assay with c-GMP-IMP. Data is fit to an equation for competitive binding (equation 3) to determine the analog Kd.
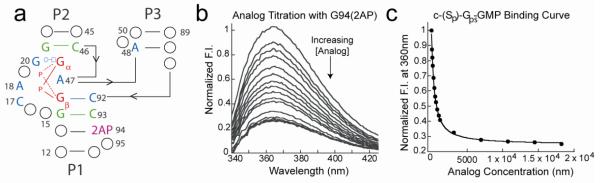
For the class I riboswitch, we site-specifically incorporated the fluorescent base analog 2-aminopurine (2AP) into the P1 helix in place of G94, yielding a fluorescently labeled class I RNA construct, G94(2AP) (Figure 5a, supplemental info). The introduction of a fluorophore within the primary sequence of the RNA was enough to weaken the Kd such that binding affinities could be easily measured using both fluorescence methods as well as the competition gel-shift assay described above. The G94(2AP) construct displays a large fluorescence signal in the absence of c-di-GMP and undergoes a 3-fold reduction in fluorescence intensity upon ligand binding (Figure 5b). To measure the binding affinities of analogs for the class I riboswitch using fluorescence, the G94(2AP) RNA was titrated with increasing concentrations of ligand and the decrease in 2AP emission at 360 nm was recorded after each ligand addition (Figures 5b,c). Using this method, the Kd of c-di-GMP for the class I riboswitch (G94(2AP) variant) was determined to be 16 nM (Table 1; supplemental info. Figure S3, Table S2). We observed an approximate 10-fold difference in the measured Kd of c-di-GMP for this RNA between the fluorescence method and the gel-shift method. To measure the affinities of analogs for the class I riboswitch, we used both the competition assay and the fluorescence assay and noted a 2-3 fold difference in the measured Kd values between these two methods, amounting to a less than 1 kcal/mol difference in binding energy (ΔΔGbind). Independent of the assay method employed, the relative ordering of ligands by affinity for the class I riboswitch remains the same. For the class II riboswitch, all binding data was obtained for the wild-type aptamer using the competition gel-shift assay.
Figure 5.
Kd measurements for the class I riboswitch by 2AP fluorescence. (a) The fluorescent G94(2AP) class I RNA construct showing the fluorescent 2AP base at position 94 in the P1 helix of the aptamer domain. (b) Binding constants (Kd) of ligands were determined by monitoring the decrease in fluorescence at 360 nm with increasing concentrations of ligand. Shown here is a sample titration with the c-(Sp)-Gps-GMP analog. (c) Kd values were determined by plotting the fluorescence intensity at 360 nm verses ligand concentration and fitting the data to a quadratic equation (Eqtn. 2). The c-(Sp)-Gps-GMP binding curve is shown here as a representative example.
Effects of base modifications on ligand affinity for the class I riboswitch
The N1 of Gα is the only atom of both guanine bases of c-di-GMP along the Hoogsteen and Watson-Crick faces that is not recognized by an RNA atom of the class I riboswitch42 (Figures 2b,c). This suggests that base recognition plays a critical role in the specificity of ligand binding. In addition, base stacking is predicted to contribute to second messenger recognition as evidenced by the universally conserved adenosine (A47) that intercalates between the guanine bases of c-di-GMP30, 42 (Figure 2a). Therefore, we expected changes to the guanine base structure that perturb the predicted hydrogen bonding contacts and base stacking interactions to result in a decreased ligand affinity. To test these hypotheses, we measured the binding affinities of base modified c-di-GMP analogs (Figure 3a).
Contacts are made to the exocyclic amine and N7 on both guanine bases, suggesting that recognition of these functional groups is important for c-di-GMP binding (Figures 2b,c)42. To test this prediction, we measured the affinities of inosine (c-GMP-IMP, c-di-IMP) and 7-deaza guanine analogs (c-c7G-GMP, c-di-c7GMP). We found that removal of the exocyclic amine and N7 on one or both of the guanine bases resulted in a decreased ligand affinity, indicating that these elements of c-di-GMP are important for binding (Table 1). Because each exocyclic amine is involved in two hydrogen bonding contacts and only a single contact is observed to each of the N7’s42, we expected the inosine analogs to have a larger effect on affinity than the 7-deaza modified analogs, yet observed the opposite effects on binding. Approximately twice the amount of binding energy was lost for c-c7G-GMP (3.9 kcal/mol) as compared to c-GMP-IMP (2 kcal/mol). Riboswitch binding was completely abolished for c-di-c7GMP (Table 1). The effects of c-GMP-IMP were relatively small and this single inosine substitution was one of the most tolerated modifications by the class I riboswitch when compared across the complete series of tested analogs. c-di-IMP, which has no exocyclic amines on either guanine base, still binds the riboswitch and based on the affinity, recognition of this functional group on both bases is worth a total of 3.9 kcal/mol (Table 1). Notably, the Kd’s of c-di-IMP, which has two modifications, and c-c7G-GMP, which only has one modification, were nearly equivalent. The observed impact on the binding affinity for removal of one N7 was larger than we expected for eliminating a single hydrogen bonding interaction based on the energetic cost we measured for eliminating similar contacts to other functional elements of c-di-GMP. This suggests that additional factors important for binding are being perturbed by this modification. Taken together, these data indicate that interactions with the N7 position of the guanine bases, either direct hydrogen bonds or indirect base stacking effects, are more crucial to ligand binding than those with the exocyclic amines.
Inosine and 7-deaza guanine are structurally very similar to guanine, whereas adenine presents a different pattern of hydrogen bonding donors and acceptors along its edges and is expected to disrupt more of the specific contacts observed to the c-di-GMP bases. While it is known that the class I riboswitch can completely discriminate against c-di-AMP40, which has adenine substituted for both of the guanine bases, we wanted to investigate the effects of replacing only one of the c-di-GMP bases with adenine by testing c-GMP-AMP for binding. We found that this RNA binds c-GMP-AMP with a 1200-fold loss in affinity. Consistent with previous reports40, no binding was detected for c-di-AMP (Table 1). The relatively large loss in affinity for c-GMP-AMP suggests that specific recognition of only one guanine base is sufficient for ligand binding but recognition of both is essential for tight c-di-GMP binding. We also tested a single N1-methylG analog (c-N1mG-GMP) for binding (see below).
Effects of modifications to the ribose rings on ligand affinity for the class I riboswitch
We next looked at the effects of modifying the ribose rings of c-di-GMP on ligand affinity for the class I riboswitch. Primary recognition of this element of the ligand is mediated through hydrogen bonding contacts to the 2′-OH of both Gs42. The 2′-OH of Gα is contacted by a non-bridging phosphate oxygen of A47 and that of Gβ is recognized by a coordinated water molecule in the binding pocket42 (Figure 2d). The hydrogen bonds observed to this functional group suggest that ribose recognition contributes to high affinity ligand binding. To test this, we synthesized 2′-deoxy, 2′-fluoro and 2′-methoxy c-di-GMP variants (Figure 3b) and measured the binding affinities of these analogs for the class I riboswitch.
To determine the energetic contribution of hydrogen bonds made to the 2′-OH’s, we measured the effects of the 2′-deoxy analogs (c-dG-GMP and c-di-dGMP). Approximately 2.1 kcal/mol of binding energy was lost for c-dG-GMP and nearly twice that effect was seen for c-di-dGMP (4.4 kcal/mol). This suggests that hydrogen bonding interactions to one of the ribose rings can be sacrificed, but recognition of both is necessary to provide a large stabilizing effect to the ligand bound complex (Table 1). Based on these measurements, hydrogen bonds to both 2′-OH’s of c-di-GMP are worth a total of more than 4 kcal/mol to the binding energy (Table 1).
Analog substitutions that replace the ribose sugars with deoxyribose sugars are likely to perturb the equilibrium of the sugar pucker. The ribose rings of c-di-GMP when bound to the riboswitch are in the 3′-endo form42, which is the preferred conformation for ribose sugars. If the equilibrium of the sugar pucker shifts toward the 2′-endo conformation for the 2′-deoxy analogs, the loss in binding affinity may also reflect the consequences of altering the conformation of the ligand backbone. To differentiate hydrogen bonding effects from ribose conformational effects, we introduced 2′-fluoro substitutions into the ligand, which causes the ribose sugar to preferentially adopt the 3′-endo conformation63, 64. We found that fluorine substitutions proved to be significantly less destabilizing than the 2′-deoxy modifications. The Kd of c-2′F-G-GMP, which contains a single 2′-fluoro substitution, was within 2-fold of c-di-GMP making it the tightest binding analog identified (Table 1). Only 0.3 kcal/mol of binding energy was lost for this substitution, whereas an additional 1 kcal/mol was lost for removing the 2′-OH group from one ring (c-dG-GMP, Table 1). The c-di-2′F-GMP analog had almost the same affinity as c-dG-GMP and notably, the affinity of this analog was 50-fold tighter than that of c-di-dGMP. Approximately 2 kcal/mol in binding energy is recovered upon introducing 2′-fluoro substitutions in place of 2′-deoxy substitutions suggesting that the effects of the 2′-deoxy substitutions cannot be solely attributed to a loss of hydrogen bonding contacts (Table 1). The more favorable binding energy of c-di-2’F-GMP compared to c-di-dGMP is likely due to 2′-fluoro modified nucleotides maintaining the 3′-endo conformation, suggesting that there is no conformational penalty for binding c-di-2′F-GMP as there is for c-di-dGMP.
We expected that introduction of methyl groups on the 2′-OH’s would lead to steric clashes and therefore have large effects on binding. As anticipated, we previously found that the class I riboswitch is only able to weakly bind c-di-2′OMe-GMP with a nearly 6 kcal/mol loss in binding energy43 (Table 1). Following this observation, we expected the single 2′-OMe analog, c-2′OMe-G-GMP, to bind, but with a large energetic penalty for accommodating this additional steric bulk within the binding pocket. We found that the introduction of this single methyl group proved to be the most detrimental asymmetric ribose modification for class I riboswitch binding with a 2.9 kcal/mol loss in energy (Table 1).
Effects of modifications to the phosphate backbone on ligand affinity for the class I riboswitch
Similar to base recognition, c-di-GMP phosphate recognition by the class I riboswitch is asymmetric42. Recognition of the phosphate 5′ of Gα (PGα) is achieved through both hydrogen bonding interactions and metal coordination whereas the phosphate 5′ of Gβ (PGβ) is less heavily recognized42 (Figure 2d). Both non-bridging oxygens of PGα (pro-RP and pro-SP) are recognized while only the pro-RP oxygen of PGβ is contacted. This suggests that recognition of PGα is more important for ligand binding than recognition of PGβ.
The pro-RP oxygens of both phosphates are solvent exposed, whereas the pro-SP oxygens point into the c-di-GMP binding pocket42. Because the covalent radius of sulfur is significantly larger than oxygen, we anticipated that sulfur substitution at the pro-Sp positions of PGβ or PGα would create unfavorable electrostatic interactions with either the backbone or exocyclic amine of A47 and therefore have larger effects on binding than substitutions at the pro-RP oxgens. To test this hypothesis, we measured the affinities of the mono-thiophosphate analogs (c-(Rp)-Gps-GMP and c-(Sp)-Gps-GMP, Figure 3c) and found that binding of c-(SP)-Gps-GMP had a 18-fold effect on affinity while binding of c-(RP)-Gps-GMP had only a 4-fold effect (Table 1). Both of these mono-thiophosphate analogs can bind the riboswitch in two distinct orientations and while we do not know the preferred binding mode, the observed 4-fold preference for riboswitch binding of the Rp- substituted analog over the Sp-substituted analog indicates that sulfur substitution at the pro-RP oxygens is preferred. Interestingly, c-(Rp)-Gps-GMP was one of the tightest binding ligands for the class I riboswitch identified in this study (Table 1).
We anticipated that the di-thiophosphate analogs would have larger effects on binding than the mono-thiophosphate analogs because contacts to the more extensively recognized PGα oxygens must be perturbed with these c-di-GMP derivatives. To test this, we measured the affinities of two of the three possible diastereomers of the di-thiophosphate derivatives, c-(RpRp)-di-Gps and c-(RpSp)-di-Gps (Figure 3c). We found that c-(RpRp)-di-Gps bound with a Kd 20-fold weaker than that of c-di-GMP, while c-(RpSp)-di-Gps gave a 110-fold loss in affinity (Table 1), confirming that di-substituted thiophosphate analogs are more detrimental to binding than the mono-substituted analogs. The RpRp di-thiophosphate analog bound 5-fold tighter than the RpSp di-thiophosphate analog and had nearly the same affinity as c-(Sp)-Gps-GMP, indicating that sulfur substitutions at both pro-Rp oxygens together have nearly the same consequence on binding as a single Sp- sulfur substitution (Table 1). This further confirms that the more solvent exposed pro-RP oxygens are more tolerant to modification than the pro-SP oxygens. Although we did not test the SpSp derivative, we anticipate that this would be the weakest binding di-thiophosphate analog based upon the more unfavorable effects of Sp- substitutions as compared to Rp- substitutions. Differential binding between both the mono and di-thiophosphate analogs indicates that the class I riboswitch is able to distinguish between the phosphate oxygens.
Energetic asymmetry of ligand recognition for the class I riboswitch
While it is clear that the class I riboswitch recognizes c-di-GMP with structural asymmetry40-42, we wanted to investigate the energetic asymmetry of recognition. Comparison of the ΔΔGbind for c-dG-GMP and c-di-dGMP suggests that recognition of each hydroxyl group is worth approximately 2 kcal/mol and that energetically, binding of c-di-GMP is symmetric (Table 1). However, single modifications to c-di-GMP, such as in the c-dG-GMP analog, eliminate the symmetry of the ligand and allow the modified di-nucleotide to bind the aptamer in two different orientations. Thus, it is unclear if these asymmetric analogs adopt a single orientation in the binding pocket or if both orientations are sampled with the binding energy also reflecting a loss in symmetry. This makes the interpretation of the energetic effects from single modifications difficult. To further investigate this question, we designed an asymmetric system that would allow us to lock the orientation of the ligand in the binding pocket and make site-specific modifications that only perturb a single interaction.
To create a constrained system for studying the effects of individual substitutions to the ligand, we exploited the fact that base recognition is a crucial determinant of ligand binding. By making a single C92U point mutation in the binding pocket, we were able to selectively bind the c-GMP-AMP analog to this mutant aptamer in a single orientation dictated by the formation of the A-U Watson Crick base pair (Figure 6a). c-GMP-AMP weakly binds the wild-type aptamer binding pocket sequence with a Kd of 1.6 μM (Table 1), whereas the introduction of the single C92U point mutation increases the affinity of the RNA for this ligand by approximately 80-fold to 19 nM (Table 3). This suggests that a productive Aβ-U92 base pair is formed, eliminating the two-fold symmetry axis in the ligand and allowing for binding in only one possible orientation.
Figure 6.
Class I riboswitch asymmetric assay with c-GMP-AMP and the C92U mutant RNA aptamer. (a) Predicted orientation of c-GMP-AMP in the binding pocket of the C92U RNA. (b) Structure of c-GMP-AMP and the 2′-deoxy derivatives synthesized. ‘A’ indicates an adenine base and ‘G’ indicates a guanine.
Table 3.
Affinites of c-GMP-AMP and the deoxy derivatives for the C92U mutant class I riboswitch.
| Analog | Kd (nM) | Fold Loss | ΔΔGbind (kcal/mol) |
|---|---|---|---|
| c-GMP-AMP | 19 ± 1.7 | - | - |
| c-G-dAMP | 200 ± 21 | 11 | 1.4 |
| c-dG-AMP | 1200 ± 110 | 64 | 2.5 |
In the background of the c-GMP-AMP molecule and C92U RNA construct, we made single site-specific 2′-deoxy modifications to assess the relative importance of each hydroxyl group. We synthesized c-dG-AMP and c-G-dAMP (Figure 6b), with the former disrupting the hydrogen bond made by A47 and the latter disrupting interactions made with the highly coordinated water molecule in the binding pocket (Figure 2d). We measured the affinities of the C92U mutant aptamer for each of these ligands and found that the two 2′-deoxy c-GMP-AMP derivatives had significantly different affinities (Table 3). Approximately 1.5 kcal/mol of binding energy was lost for eliminating contacts to the 2′-OH of adenosine (A β) and 2.5 kcal/mol were lost for removal of interactions with the 2′-OH of guanosine (Gα). This suggests that the contact made to Gα by the backbone of A47 contributes more to ligand binding than that made to Gβ and that the measured affinity for c-dG-GMP reflects a loss in energy from both breaking the symmetry of the ligand and eliminating hydrogen bonding interactions. Overall, the calculated ΔΔGbind for both ligands totaled 3.9 kcal/mol. This value is similar to the ΔΔGbind of the wild-type RNA for the doubly substituted c-di-dGMP ligand (4.4 kcal/mol, Table 1). Taken together, these data imply that recognition of the symmetrical c-di-GMP ligand by the class I riboswitch is both structurally and energetically asymmetric.
Effects of base modifications on ligand affinity for the class II riboswitch
Base recognition of c-di-GMP by the class II riboswitch is achieved through entirely different recognition motifs than those found in class I riboswitches43. No canonical Watson Crick or Hoogsteen interactions are present and in general, fewer contacts to the bases are observed. The N7, O6, and exocyclic amine are recognized on only one of the c-di-GMP bases43, whereas these functional groups are all recognized on both bases of the ligand in the class I riboswitch42 (Figures 2b,c,f,g). The N1 position of guanine is the only atom with hydrogen bonding potential along the Watson Crick and Hoogsteen faces that is contacted on both Gα and Gβ. This suggested that the class II riboswitch would tolerate a range of modifications to at least one of the guanine bases. Similar to the class I structure, a highly conserved adenosine31 (A70) intercalates between the bases of c-di-GMP indicating that base stacking also contributes to ligand binding43 (Figure 2e).
Based on structural analysis, two hydrogen bonding contacts to the exocyclic amine of Gα are predicted to be important for c-di-GMP recognition43 (Figure 2f). To test this, we measured the affinities of inosine modified analogs, expecting to see effects on binding for c-di-IMP but no change in affinity for c-GMP-IMP. Instead, we found that c-di-IMP bound with an affinity within 2-fold of c-di-GMP (Table 2) and because this analog had a near wild-type affinity, we did not test c-GMP-IMP for binding. This suggests that guanine exocyclic amine recognition does not contribute to ligand binding.
A single hydrogen bonding interaction to the N7 of Gβ is observed whereas that of Gα is not recognized43 (Figures 2f,g), leading us to hypothesize that removal of one N7 should have no effect on ligand binding. We measured the Kd of c-c7G-GMP to test this hypothesis and found that binding of this analog resulted in a 15-fold loss in affinity (Table 2). The effects on binding observed for this single base modification were unanticipated and larger than the effects for any symmetric ribose or phosphate modifications (see below) (Table 2). Following this observation, we expected to see effects on binding for c-di-c7GMP and found that the magnitude of the effect for two 7-deaza guanine substitutions (4.4 kcal/mol) was substantially greater than expected based on the magnitude of the effect for the single substitution (c-c7G-GMP, 1.6 kcal/mol). c-di-c7GMP was one of the weakest binding ligand identified for the class II riboswitch, indicating that the N7 position is crucial for high affinity binding of c-di-GMP. This is similar to what we observed for the class I riboswitch.
To test the specificity of guanine recognition by the class II riboswitch, we examined the effects of adenine substitution on ligand binding. The single adenine substitution in the c-GMP-AMP ligand only resulted in a 120-fold effect on affinity, with the Kd remaining in the nanomolar range (Table 2). Unexpectedly, we found that it is possible to replace both guanine bases of c-di-GMP with adenine and still retain some binding to the class II riboswitch. We were not able to obtain a complete binding curve because c-di-AMP binds weakly, but we estimate that the Kd is ≥ 30 μM, a nearly 6 kcal/mol loss in binding energy (Table 2). The ability of this riboswitch to bind c-di-AMP was surprising since most of the predicted contacts to the bases would be eliminated if c-di-AMP is positioned in the binding pocket in the same orientation as c-di-GMP. These results indicate that specific recognition of both guanine bases is not absolutely required for ligand binding by the class II riboswitch.
Effects of backbone modifications on ligand affinity for the class II riboswitch
The crystal structure of the class II aptamer shows that the ribosyl-phosphate backbone of c-di-GMP is minimally recognized43. The 2′-OH’s of the ribose rings are not specifically contacted, which implies that modifications could be made to this functional group with little or no effect on ligand affinity.
To test the prediction that 2′-OH recognition does not contribute to second messenger binding, we looked at the effects of the c-di-2′OMe-GMP analog on affinity. We previously reported the Kd of this ligand for class II and found that it is within 2-fold of that for c-di-GMP43 (Table 2). The ability of this riboswitch to accommodate steric bulk at these positions confirms that specific contacts are not being made to the ribose hydroxyl groups of c-di-GMP, nor are there any RNA atoms in close proximity that must undergo significant rearrangement to allow binding of this analog.
To determine if there are ribose conformational effects on binding, we introduced 2′-fluoro and 2′-deoxy modifications into c-di-GMP. We found that c-di-dGMP had a 5-fold weaker Kd than c-di-GMP and that the affinity of c-di-2′F-GMP was only 2.6-fold weaker than that of c-di-GMP (Table 2). Because the 2′-OH atoms are not specifically recognized by the riboswitch, we ascribe the observed effects for c-di-dGMP binding solely to a change in the equilibrium of the ribose sugar pucker from the 3′-endo to the 2′-endo conformation as a result of the 2′-deoxy modifications. The recovery in binding affinity for c-di-2′F-GMP to near wild-type levels is consistent with this interpretation. We did not test single 2′-fluoro, 2′-deoxy and 2′-methoxy modified analogs for binding by the class II riboswitch given the relatively small effects of the doubly modified analogs on affinity.
In addition, there is only a single hydrogen bonding interaction made to the phosphates, between the exocyclic amine of A70 and a non-bridging oxygen of the phosphate 5′ of Gα43 (Figure 2e). Therefore, we anticipated that phosphorothioate substitutions would have little effect on the stability of the ligand-bound complex. To test this hypothesis, we measured the affinities of the di-thiophosphate analogs, c-(RpRp)-di-Gps and c-(RpSp)-di-Gps and found that the Kds of both analogs were within 2-fold of that for c-di-GMP (Table 2). There was also no difference in binding between the two diastereomers. c-RpRp)-di-Gps retains symmetry and can only bind in one orientation while c-(RpSp)-di-Gps can bind in two orientations, but both analogs can bind the riboswitch such that sulfur substitution does not disrupt the interaction between A70 and the pro-SP oxygen of PGα. Thus, it was not surprising that neither of these di-thiophosphate analogs had an effect on the affinity. The SpSp substituted analog would have to be tested for binding to specifically perturb this interaction. However, the small perturbation to the binding energy for the RpRp and RpSp-substituted analogs (0.3 kcal/mol) demonstrate that the phosphates of c-di-GMP are not extensively used by this riboswitch class to sense its ligand.
Binding of an N1-methylG analog to the class I and class II riboswitches
We previously demonstrated that it is possible to preferentially bind the class II riboswitch over the class I riboswitch using the c-di-2′OMe-GMP analog43. In this work we sought to design an analog with the opposite targeting specificity. In the class II riboswitch the N1 atom of both Gα and Gβ are contacted by RNA nucleotides43 whereas only the N1 of Gβ is recognized by a binding pocket nucleotide in the class I riboswitch42 (Figure 2). Although a water molecule is coordinated to the N1 of Gα in the class I native structure42, there appears to be ample space to accommodate a methyl group at this position upon displacement of this water. However, the class II riboswitch would have to undergo binding pocket rearrangements to create space for a methyl group at this position on either guanine base. In an effort to selectively target the class I RNA over class II, we synthesized an analog containing a single N1-methyl guanine base, c-N1mG-GMP (Figure 3a).
We measured the Kd of this analog for both riboswitch classes and found that there is a larger effect on ligand binding for the class I riboswitch than for the class II riboswitch. A 300-fold loss in affinity was observed for the class I riboswitch whereas only a 75-fold loss was seen for class II (Tables 1,2). This result was unexpected based on the predicted steric clashes between the methyl group and the binding pocket nucleotides of the class II aptamer in direct contact with the N1 of Gα and Gβ. For binding by the class I riboswitch, displacement of the water molecule hydrogen bonding with the N1 position likely contributes to the observed decrease in affinity, yet the effects seem quite large for loss of this single contact. In the class II structure, the O6 of G73 hydrogen bonds with the N1 of Gβ (Figure 2f) and is not base paired with any other nucleotides of the class II RNA. Thus, this nucleotide may shift its register to accommodate this extra methyl group, explaining the observed binding by class II.
DISCUSSION
Two classes of c-di-GMP-binding riboswitches have been identified30, 31 as downstream macromolecular targets in this ubiquitous second messenger signaling pathway that regulates many diverse bacterial processes1-4, 8, 10. These two riboswitch classes have evolved different strategies for c-di-GMP recognition and consequently, differ in the structural features of c-di-GMP they require for binding and ligand specificity. Here, we have shown that the class II riboswitch utilizes fewer functional groups of both the bases and ribosyl-phosphate backbone of c-di-GMP for ligand binding and consequently is less discriminatory in second messenger recognition.
The class I riboswitch recognizes the guanine bases of c-di-GMP with greater specificity than the class II riboswitch. For example, replacing one of the guanine bases with adenine has much less of an impact on ligand binding by the class II riboswitch and adenine substitutions for both bases only abolished ligand binding by class I. In addition, we found that interactions with guanine functional groups which significantly stabilize ligand binding by the class I riboswitch are inconsequential for recognition by class II. The ability of the class II aptamer to recognize purine analogs that eliminate many of the specific contacts with the c-di-GMP bases suggests that the class II riboswitch relies largely on base stacking for ligand binding rather than specific interactions with guanine functional groups. In contrast, the class I aptamer more extensively utilizes the various structural features of guanine to sense the same ligand, resulting in increased binding specificity by this riboswitch motif.
The class I riboswitch makes specific contacts to the ribose hydroxyls of c-di-GMP whereas the class II riboswitch does not utilize this functional group for ligand binding. The specific hydrogen bonding interactions to the ribose hydroxyl groups by the class I riboswitch provide a large, stabilizing effect to the binding energy, whereas methylation of this functional group has no effect on binding by class II. However, the conformation of the ribose ring is important for ligand binding by both riboswitches. In the absence of specific recognition of the ribose hydroxyls by the class II riboswitch, 2′-deoxy substitutions still had a small effect on binding that was mitigated by 2′-fluoro substitutions. Similar effects were observed for the class I riboswitch, but the conformational penalty for 2′-deoxy sugars was much larger. Thus, both riboswitches show a preference for binding 3′-endo ribose sugars.
In accordance with these observations, contacts made to the phosphates of the ribosyl-phosphate backbone by class I are important for ligand binding whereas the class II riboswitch has not evolved a specific mechanism for c-di-GMP backbone recognition. This suggests that the c-di-GMP backbone could be modified without significant consequence to ligand affinity for the class II motif. For phosphate recognition, the class I riboswitch employs both metal coordination and hydrogen bonding interactions that allow the aptamer to differentiate between specific phosphate oxygens. These observations are again in direct contrast to what was seen for the class II riboswitch. The same phosphate backbone modifications that affected binding by the class I riboswitch had no effect on binding by class II. The greater use of the c-di-GMP ribosyl-phosphate backbone by the class I riboswitch enhances its ability relative to the class II riboswitch to discriminate between structurally similar di-nucleotide analogs.
There are a greater number of contacts made to c-di-GMP by the class I aptamer and nearly all of the interactions predicted from structural analysis42 contribute to the binding energy. While fewer specific contacts to c-di-GMP were predicted for the class II riboswitch43, some of these interactions make little, if any, contribution towards ligand binding. It is somewhat surprising that the few contacts made between c-di-GMP and the class II aptamer do not contribute more to binding since these are the only interactions predicted to stabilize the ligand-bound complex. This further supports the prediction that this riboswitch relies heavily on base stacking for ligand binding. However, the increased recognition of c-di-GMP by the class I riboswitch likely accounts for the tighter affinities of this riboswitch class for c-di-GMP compared to the class II riboswitch. Class I ribowitches have Kd’s for c-di-GMP as tight as 10 pM40, whereas the affinities for class II are weaker and vary from mid-picomolar to low nanomolar31, 43. The differential analog binding by these two c-di-GMP effectors correlates with the differences in their absolute affinities for the same second messenger ligand.
Despite the differential recognition of the c-di-GMP bases, both aptamers rely on base stacking interactions for tight ligand binding and these stacking contacts are the only conserved mechanism for guanine recognition between these two RNA effectors. The effects on binding of the 7-deaza guanine analogs were unexpectedly large for both RNAs and inconsistent with eliminating only the predicted hydrogen bonding interactions. Both of these aptamers incorporate c-di-GMP into structural elements upon binding and contain highly conserved purine nucleotides in the binding pocket that base stack with c-di-GMP42, 43. It has been shown that duplexes containing 7-deaza guanine are less stable than the corresponding duplexes with the canonical guanine base due to decreased base pairing and stacking interactions65-67. The considerable binding effects we observed for 7-deaza guanine substitutions may similarly be ascribed to the decreased ability of the di-nucleotide to stack with binding pocket residues. This interpretation is consistent with the structural prediction that stacking contacts are important for ligand binding and highlights the essential role of this mechanism for RNA recognition of c-di-GMP. Another possible explanation for the large effects on binding observed for 7-deaza guanine substitutions is that the additional steric bulk of protons at the C7 position results in steric clashes with nearby RNA atoms. While this possibility cannot be fully excluded, significant effects on binding by the class II riboswitch for the single 7-deaza guanine substituted analog are observed in the absence of both specific contacts to the N7 of Gα and nearby RNA atoms that could potentially clash with a proton at this position. This suggests that the majority of binding energy lost from the 7-deaza guanine modifications is due to decreased base stacking interactions. Taken together, this implies that the similar conformation of c-di-GMP when bound to both riboswitches is functionally relevant for maintaining these high affinity base stacking interactions.
The class II riboswitch has proven to be a more promiscuous effector of c-di-GMP than the class I riboswitch, defining the challenging goal of selectively targeting the latter motif. Because the overall recognition pattern of c-di-GMP differs between the two classes, we hypothesized that analogs could be designed to preferentially bind one riboswitch class over the other. However, the identified differences in c-di-GMP recognition strategies are more easily exploited for selectively targeting the class II aptamer. In the specific context of the two c-di-GMP-binding riboswitches studied here, the N1-methyl guanine analog could potentially target the class I riboswitch over class II. We predict that the Kd of this analog for the class I wild-type sequence (Kd c-di-GMP, 10 pM)40, is in the low nanomolar range and comparison of the absolute affinities of the class I and class II aptamers for this analog suggests that binding would be selective for class I. While this approach to selectively targeting the class I riboswitch is highly dependent on the affinities of c-di-GMP for both RNA aptamers in question, this demonstrates that it may be possible to differentiate between these two c-di-GMP effectors. Identifying an analog that is absolutely selective for class I RNA over class II remains desirable because it would provide a useful tool for manipulating RNA-mediated c-di-GMP signaling networks, particularly in those organisms that use both motifs for gene control. However, several tight binding second messenger analogs for both RNA aptamers were identified in this study and these are promising candidates for use in manipulating the diverse biological processes mediated by these c-di-GMP-binding riboswitches.
The larger effects on binding observed for the class I riboswitch over the class II riboswitch for the N1-methyl guanine analog indicate that the class II riboswitch is able to effectively accommodate steric bulk at positions of c-di-GMP that are in direct contact with RNA atoms. The effects on binding for rearrangement of the class II RNA to accommodate this added methyl group on the ligand were not as large as the corresponding effects from the predicted displacement of the water molecule coordinated to the N1 of Gα in the class I riboswitch. It is possible that the binding pocket nucleotides of the class I aptamer also in contact with this water molecule are strategically positioned by these hydrogen bonding interactions, suggesting that its displacement may disrupt a network of contacts that are necessary for maintaining the integrity of the c-di-GMP binding pocket. Taken together, this indicates that in comparison to the class I aptamer, the class II aptamer is a more flexible motif, which likely contributes to its greater promiscuity in ligand binding.
c-di-AMP was recently identified to be a bacterial second messenger signaling molecule68-70 and the ability of the class II riboswitch to bind this ligand hints at the possibility that c-di-AMP binding riboswitches may exist and participate in this emerging signaling pathway. Given that RNA has evolved two different strategies for binding of c-di-GMP, it is plausible that RNA has also evolved a strategy for specific, high affinity binding of c-di-AMP. The common theme for c-di-GMP recognition by these two distinct riboswitches is the use of base stacking and it is likely that the same approach would be employed by RNA for the specific recognition of c-di-AMP.
Overall, these data offer an explanation for why the class I motif is more frequently used for gene regulation by bacteria compared to the class II motif30, 31. The ability of the class II riboswitch to recognize the biologically relevant molecule c-di-AMP, although much weaker than its cognate ligand, could have physiological and biological consequences for the cell. Diadenylate cyclase (DAC) domains, analogous to the diguanylate cyclase (DGC) domains, are broadly distributed among the bacterial kingdom suggesting that c-di-AMP may be present in many bacterial species68-70. Initial inspection of the distribution of DAC domains (Pfam 02457) and both class I and class II c-di-GMP-binding riboswitches30, 31 across bacteria indicates that many organisms that use riboswitches for c-di-GMP signaling also have predicted DAC domains. For these organisms that potentially use both c-di-GMP and c-di-AMP signaling, the class I riboswitch would likely provide tighter genetic control than the class II riboswitch. While it has been shown that the class II riboswitch can regulate splicing in response to c-di-GMP demonstrating that this RNA can participate in complex forms of gene regulation31, the increased binding specificity of the class I riboswitch suggests that this motif is more finely tuned to explicitly respond to c-di-GMP.
c-di-GMP analogs may be capable of discriminating between RNA and protein receptors, which would be particularly useful for differentiating between the effects of RNA-mediated and protein-mediated second messenger signaling. Recently, the crystal structures of c-di-GMP bound to several protein effectors have been reported, including the degenerate EAL domain proteins LapD29 and FimX28 as well as several PilZ domain proteins17, 21, 71. When bound to both LapD and FimX, the bases of c-di-GMP are splayed apart rather than directly aligned over top of one another as they are when bound to RNA receptors, decreasing the strength of any stacking contacts formed with proteins as compared to those networks formed with RNA. This suggests that the 7-deaza guanine modified analogs would affect ligand binding by these proteins to a lesser degree than seen with riboswitch targets. In contrast, PilZ domain proteins can bind c-di-GMP as either a monomer where the guanine bases are slightly staggered overtop one another, or as an intercalated dimer. For proteins that bind the c-di-GMP dimer, interactions are not only formed between each molecule of c-di-GMP and the protein, but between the two c-di-GMP molecules as well. This suggests that ligand modifications that could potentially weaken intermolecular interactions between c-di-GMP may also affect the ability of the dinucleotide to dimerize and therefore select against PilZ domain proteins that bind the second messenger as a dimer.
These second messenger analogs may also affect the activity of the metabolic enzymes responsible for the synthesis and degradation of c-di-GMP in the cell. In particular, several of the c-di-GMP derivatives studied here may display an increased resistance to the phosphodiesterase proteins that specifically degrade this second messenger in the cell. Analogs with such properties would be especially useful for in vivo applications of riboswitch targeting, as well as for tools to further elucidate the molecular mechanisms of c-di-GMP action within the complex cellular environment.
Supplementary Material
Acknowledgements
We thank K. Smith, N. Carrasco, and other members of the Strobel Lab for advice and helpful discussions; A. Doerner for help with data collection; K. Smith and M. Griffin for critical comments on the manuscript; A. Hare for help with computational modeling; U. Jenal (University of Basel) for the gift of the PleD* expression plasmid; Z. Liang for the gift of the tDGC expression plasmid. This work was supported by National Institutes of Health Grant GM022778 to S.A.S and National Institutes of Health Grant GM79760 to R.A.J.
Footnotes
Supporting Information Available. Two figures and two tables as referred to in the text are included. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Romling U, Gomelsky M, Galperin MY. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. Annu.Rev. Gen. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Romling U, Amikam D. Curr. Opin. Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Hengge R. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 5.Pesavento C, Hengge R. Curr Opin Microbiol. 2009;12:170–6. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Schirmer T, Jenal U. Nat. Rev. Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 7.Tischler AD, Camilli A. Mol. Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamayo R, Pratt JT, Camilli A. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischler AD, Camilli A. Infect. Immun. 2005;73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter PA, Stibitz S. Curr. Opin. Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Ryan RP, Fouhy Y, Lucey JF, Dow JM. J. Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. J. Bacteriol. 2005;187:1792–8. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Proc. Natl. Acad. Sci. USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AJ, Ryjenkov DA, Gomelsky M. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amikam D, Galperin MY. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 16.Ryjenkov DA, Simm R, Romling U, Gomelsky M. J. Biol. Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 17.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pratt JT, Tamayo R, Tischler AD, Camilli A. J. Biol. Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. Mol. Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. J. Mol. Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Hickman JW, Harwood CS. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao F, He YW, Wu DH, Swarup S, Zhang LH. J. of Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. Mol. Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newell PD, Monds RD, O’Toole GA. Proc. Natl. Acad. Sci. USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro MVAS, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O’Toole GA, Sondermann H. PLOS Biol. 2011;9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandal M, Breaker RR. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 33.Winkler WC, Breaker RR. Ann. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 34.Roth A, Breaker RR. Annu. Rev. Biochem. 2009;78:305–34. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serganov A, Patel DJ. Nat. Rev. Gen. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montange RK, Batey RT. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Lau MW, Ferre-D’Amare AR. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breaker RR. Cold Spring Harb Perspect. Biol. doi: 10.1101/cshperspect.a003566. [Google Scholar]
- 39.Winkler WC. Curr. Opin. Chem. Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Smith KD, Lipchock SV, Ames TD, Wang JM, Breaker RR, Strobel SA. Nat. Struct. Mol. Biol. 2009;16:1218–U27. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulshina N, Baird NJ, Ferre-D’Amare AR. Nat. Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith KD, Lipchock SV, Livinston AL, Shanahan CA, Strobel SA. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Proc. Natl. Acad. Sci. USA. 2011;108:7757–7762. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulhbacher J, St-Pierre P, Lafontaine DA. Curr. Opin. Pharmacol. 2010;10:551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Blount KF, Breaker RR. Nat. Biotech. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 46.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Structure. 2009;17:857–68. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Nat. Chem.l Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 48.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Chem. Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Rao F, Pasunooti S, Ng YL, Zhuo WC, Lim L, Liu AWX, Liang ZX. Anal. Biochem. 2009;389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 50.Scaringe SA. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 51.Gaffney BL, Veliath E, Zhao JW, Jones RA. Org. Lett. 2010;12:3269–3271. doi: 10.1021/ol101236b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alazzouzi E, Escaja N, Grandas A, Pedroso E. Angew. Chem. Int. Ed. 1997;36:1506–1508. [Google Scholar]
- 53.Frieden M, Grandas A, Pedroso E. Chem. Comm. 1999;16:1593–1594. [Google Scholar]
- 54.Micura R. Chem. Euro. J. 1999;5:2077–2082. [Google Scholar]
- 55.Kiburu I, Shurer A, Yan L, Sintim HO. Mol. Biosys. 2008;4:518–520. doi: 10.1039/b719423d. [DOI] [PubMed] [Google Scholar]
- 56.Schurer H, Lang K, Schuster J, Morl M. Nucleic Acids Res. 2002;30:e56. doi: 10.1093/nar/gnf055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nandakumar J, Shuman S. Molec. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 58.Pljevaljcic G, Millar DP. Fluor. Spec. 2008;450:233–252. doi: 10.1016/S0076-6879(08)03411-3. [DOI] [PubMed] [Google Scholar]
- 59.Heppell B, Mulhbacher J, Penedo JC, Lafontaine DA. Methods Mol. Biol. 2009;540:25–37. doi: 10.1007/978-1-59745-558-9_3. [DOI] [PubMed] [Google Scholar]
- 60.Bradrick TD, Marino JP. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Genes. Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strobel SA, Cech TR. Biochemistry. 1993;32:13593–13604. doi: 10.1021/bi00212a027. [DOI] [PubMed] [Google Scholar]
- 63.Micklefield J. Curr. Med. Chem. 2001;8:1157–1179. doi: 10.2174/0929867013372391. [DOI] [PubMed] [Google Scholar]
- 64.Rozners E. Curr. Org. Chem. 2006;10:675–692. [Google Scholar]
- 65.Grein T, Lampe S, Mersmann K, Rosemeyer H, Thomas H, Seela F. Bioorg. Med. Chem. Lett. 1994;4:971–976. [Google Scholar]
- 66.Seela F, Ramzaeva N, Chen YM. Bioorg. Med. Chem. Lett. 1995;5:3049–3052. [Google Scholar]
- 67.Ramzaeva N, Seela F. Helv. Chim. Acta. 1996;79:1549–1558. [Google Scholar]
- 68.Romling U. Sci. Signal. 2008;1:e39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 69.Witte G, Hartung S, Buttner K, Hopfner KP. Mol. Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 70.Rao F, See RY, Zhang DW, Toh DC, Ji Q, Liang ZX. J. Biol. Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Habazettl J, Allan MG, Jenal U, Grzesiek S. J. Biol. Chem. 2011;286:14304–14. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.