Abstract
The current understanding of the mechanism of anti-tuberculosis drug resistance has been shaped by the history of development of anti-tuberculosis drugs in the past 60 years and was arrived at as part of inductive generalization. Recently, these standard beliefs have been tested in controlled hollow fiber systems experiments. Drug resistance in Mycobacterium tuberculosis was shown to be related to pharmacokinetic-pharmacodynamic (PK/PD) factors, and factors such as pharmacokinetic variability. Poor PK/PD exposures due to our current non-optimized dosing regimens initiate a chain of evolution driven events, starting with induction of multi-drug efflux pumps, followed by the development of chromosomal mutations in time, which together lead to high level resistance multi-drug resistant tuberculosis and extremely drug resistant tuberculosis.
Introduction
Global efforts to eliminate tuberculosis (TB) by 2050 have been threatened by recent worldwide emergence of multidrug-resistant TB (MDR-TB) and extensively drug resistant-TB (XDR-TB), estimated at 440 000 and 25 000, respectively, in 2008 [1]. This is despite injection of enormous resources to support the World Health Organization (WHO) recommended directly observed treatment strategy (DOTS) programs which are meant to prevent TB drug resistance. MDR-TB refers to simultaneous resistance to rifampin and isoniazid, while XDR-TB refers to MDR-TB plus additional resistance to at least one injectable drug plus a quinolone. Both forms of TB are difficult to treat, are expensive, long, more likely to fail and more likely to result in death. The need for extended therapy using combinations of drugs remains a practical obstacle to effective TB control. We review the clinical, laboratory and pharmacokinetic/pharmacodynamics (PK/PD) factors associated with development of drug resistant TB and propose a new scenario based on evolution and PK/PD science. There has been a general paucity of data in this area; however the few pivotal studies in the past two years point towards a departure from standard beliefs on how Mycobacterium tuberculosis resistance arises.
Current beliefs of how M. tuberculosis resistance emerges
The understanding of the mechanism of anti-TB drug resistance has been shaped by the history of development of anti-TB drugs in the past 60 years, and was arrived at as part of inductive generalization. Unfortunately, this approach is prone to bias. Based on observations in regimens tested between 1952 and 1980, each drug in the regimen was assigned special roles in treatment of M. tuberculosis. Pyrazinamide, isoniazid, ethambutol, rifampin and streptomycin are each thought to target certain specific populations of the M. tuberculosis such as bacilli under acidic, aerobic and/or hypoxic conditions within caseous foci, at the edge of pulmonary cavities and inside macrophages, respectively [2;3]. Resistance suppression is defined as one drug preventing resistance to another, not one drug preventing resistance to itself. The resulting belief, almost universally accepted, is that if patients take these multi-drug regimens without defaulting, then MDR-TB and XDR-TB emergence would be ameliorated. Accordingly, it is believed that missing drug doses leads to ‘effective monotherapy’ for some bacillary populations because of different drug half-life’s and differential drug penetrations into effective compartments. It has been theorized that resistance evolves independently for each drug one at any one time through ‘unlinked processes’, leading to the standard step-wise pick up of mutations that leads to sequential acquisition of resistance [2;4]. Finally, the belief has been that resistance arises from replicating bacilli, so that non-replicating persistent bacilli (NRP) do not mutate and cause drug resistance. Recently, each of these staple beliefs has been challenged in well designed experiments that applied both PK/PD and none PK/PD methodology.
Just what do you mean by “resistant”?
The term “drug resistance” is ambiguously defined in many situations. What is drug resistance, especially in the context of M. tuberculosis? The WHO defines drug resistance as “the ability of certain microorganisms to withstand attack by antimicrobials.” In the context of M. tuberculosis, this is defined as the ability of >1% proportion of a bacilli to grow in the presence of critical concentration of drug [5]. The critical concentrations themselves are defined as the concentration of antibiotic that inhibit growth in 95% of wild type strains that have hitherto not been exposed to drug. Thus, these are essentially epidermiologic cut-off values. The current critical concentrations that define resistance are shown in table 1.
Table 1.
Susceptibility breakpoints and chromosomal mutations associated with drug resistance
| Drug | Current breakpoint (mg/L) | Proposed breakpoint (mg/L) | Chromosomal mutations associated with resistance |
|---|---|---|---|
| Rifampin | 1 | 0.0625 | rpoB |
| Isoniazid | 0.2/1.0 | 0.03/0.125 | katC, inhA, oxyR, ahpC, furA |
| Streptomycin | 5 | - | rrs, rpsL |
| Pyrazinamide | 100 | 50 | pncA |
| Ethambutol | 5.0/7.5 | 4 | embB |
| Moxifloxacin | 1 | 1 | gyrB |
PK/PD dose selection and clinical application to prevent drug resistance
When a drug dose is administered to patients it becomes part of the non-deterministic process of pharmacokinetic variability. In other words, a particular dose does not lead to a specific concentration-time profile in all patients, but rather a distribution determined in part by alleles of genes encoding enzymes involved in xenobiotic metabolism, the particular physique of each patient as is the case of pyrazinamide [6], or even dietary considerations. This means that in some patients, despite patients taking all their drug doses low drug concentrations could still be encountered, which could lead to emergence of drug resistance. Thus, resistance emergence could occur in part due to non-deterministic causes that have nothing to do with DOTS or default.
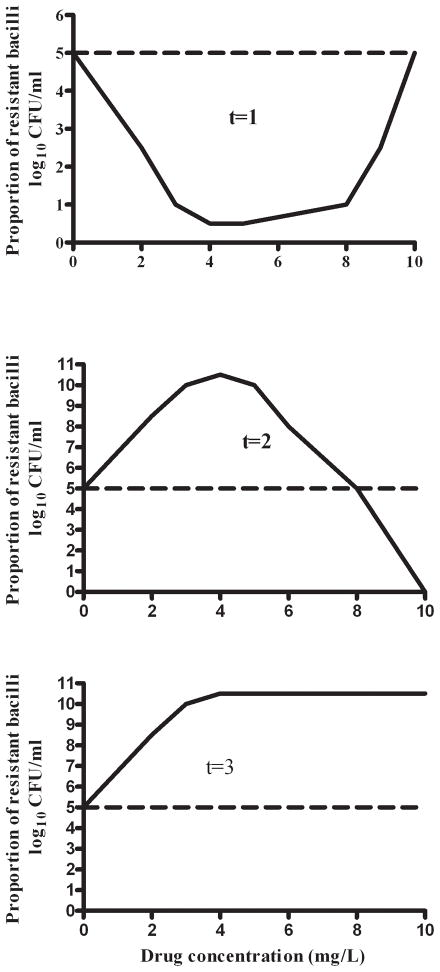
The response of the pathogen to a particular drug concentration-time profile is itself related to several PK/PD factors. For M. tuberculosis, PK/PD factors have been derived in monotherapy studies in the hollow fiber system (HFS) [7–12]. First, the shape of the concentration-time curve has been related to resistance emergence for each of the first line anti-TB drugs. Studies with isoniazid and pyrazinamide revealed that the relationship between drug exposure and population of drug-resistant M. tuberculosis was a series of curves that changed with time, starting with a “U” shaped curve, which then evolved over time to an inverted “U” curve (figure 1). In other words, the relationship is defined by a quadratic function, with time as part of the defining characteristics of the leading coefficient (see reference 11). Therefore, in interpreting indices at which resistance can be suppressed, the duration of therapy should be taken into consideration. Rifampin resistance emergence and suppression are linked to the peak concentration (Cmax) to MIC, with optimal suppression of resistance at a free drug Cmax/MIC of 175 [10]. Isoniazid resistance emergence was demonstrated to be closely linked to both Cmax/MIC and AUC/MIC [13]. On the other hand, both pyrazinamide and ethambutol resistance emergence were associated with the % time concentration persisted above MIC (%TMIC) [11;14]. The lessons are obvious, resistance emergence to a drug depends on the drug exposure achieved, and in many situations the actual shape of the concentration-time curve, which often differ from the PK/PD parameter linked to microbial kill.
Figure 1. Change in size of drug-resistant M. tuberculosis population with exposure and time.
Figure shows an upright “U” at the beginning of therapy (t=1), which evolves to an inverted “U” curve at t=2, and eventually reaches a time when no concentration of the antibiotic in question can suppress drug resistance at t=3.
These PK/PD results, as well as the exposures associated with optimal kill, can be used for several purposes. The first is to refine susceptibility breakpoints. Setting susceptibility breakpoints using the PK/PD approach does not just rely on the MIC distribution in wild type isolates, but also on the doses and the drug exposures achieved by the doses in patients, given pharmacokinetic variability. An isolate is defined as being drug-resistant if it has an MIC that precludes it from being effectively killed by antibiotic concentrations achieved in at least 90% of patients given a particular dose. Put simply, if an isolate cannot be effectively killed at site of infection in most patients by a drug after taking the maximum tolerated dose, then it is resistant to that drug. Using this approach, new critical concentrations for each of the first line anti-TB drugs, as well as moxifloxacin, were recently proposed, as shown in table 1. The most dramatic changes are proposed for isoniazid and rifampin, and thus the definition of MDR-TB itself. These are the two drugs in which PK/PD studies have been performed using at least two independent models (mice, hollow fibers and guinea pigs) [10, 13, 15–17] and population pharmacokinetic studies are available from at least 3 independent groups [18–22]; utilizing any of these studies leads to the same conclusion on breakpoints, so that it is unlikely that bias from any one PK/PD model can be invoked. Nevertheless, further work is needed to confirm these proposed susceptibility breakpoints. The breakpoints we have proposed, as well as the currently accepted breakpoints, need to be examined in large datasets of combination therapy clinical studies, and each breakpoint examined for whether it can predict microbiologic failure.
The second use of PK/PD exposures that could suppress resistance is to design doses and dosing schedules that could suppress drug resistance emergence. This is because it has been demonstrated for virtually each anti-TB agent examined; that the dose associated with maximal kill is not necessarily the one that prevents resistance emergence. Indeed, for moxifloxacin and ciprofloxacin such exposures associated with maximal bactericidal effect were also the ones associated with maximal amplification of the resistant sub-populations [7;8]. Thus, computer aided clinical trial simulations have been utilized to determine doses that best suppress resistance suppression for moxifloxacin, pyrazinamide, and rifampin. In HFS studies, a 24h AUC/MIC greater than 53 was associated with suppression of drug resistance. Monte Carlo simulations revealed that this target could be achieved by 59% of patients treated with 400 mg of drug and by 93% in patients treated with 800mg of moxifloxacin daily [7]. Similarly, Goutelle et al examined if rifampin doses of 600 mg and 1200 mg could adequately achieve the Cmax/MIC of 175 needed to suppress drug resistance [18]. The standard dose of 600 mg a day fared badly, while 1,200 mg performed better. We performed similar studies for pyrazinamide’s ability to achieve % time above MIC of ≥67% needed to suppress resistance emergence [11]. Doses ≥3G a day administered daily achieved the target in >90% of patients. All such doses advocated by this approach still need to be shown to be safe for patients. In the case of pyrazinamide, however, toxicodynamic analysis and meta-analysis recently suggested low rates of hepatotoxicity even at these high doses, provided they are administered no more than 2 months [12].
Mechanisms of resistance emergence
It is believed that during non-compliance, one of several mechanisms may lead to emergence of drug resistance. According to the pharmacokinetic mismatch hypothesis, during non-compliance the drug with the short half-life disappears quickly, leaving M. tuberculosis exposed to the drug with the longer half-life as monotherapy. In some scenarios, even without non-compliance, if the half-life of two drugs are very mismatched (e.g., rifapentine and isoniazid), then the same situation can arise especially during intermittent phases of therapy. We recently expressed this as a falsifiable hypothesis and tested it in HFS for rifampin and isoniazid with and without pre-existed resistant sub-populations (Srivastava et al. In revision). The drugs were administered as well matched regimens, or isoniazid administered 6h, 12hr, or 24 hr after rifampin. Essentially the more mismatched regimens performed better and the pharmacokinetic mismatch hypothesis was rejected. Another theory on resistance emergence has been the time in mutant suppression window hypothesis. In our work on rifampin, isoniazid, pyrazinamide, and ethambutol in the HFS this hypothesis also failed to explain emergence of resistance to these agents [10;11;13;14]. However, one mouse study confirmed this theory for moxifloxacin [23].
New evolution based understanding
In M. tuberculosis, as in all bacteria, DNA replication allows a narrow baseline rate of chromosomal mutations, a balance between the ability to adapt to the changing environment via mutations and safeguarding genetic information from collapse if mutation rates are too high. DNA repair enzymes are central to this process. Mutations in genes that encode DNA repair enzymes would lead to hyper-mutable strains. As an example, deficiencies in M. tuberculosis MutT1 results in a 16-fold increase in spontaneous mutation frequency that leads to increased rifampin resistance in the laboratory; mutants in which two DNA repair genes ada/alkA and ogt, involved in alkylation damage repair, are inactivated increases mutation frequencies to rifampin 100 fold [24; 25]. It has been argued, convincingly that indeed non-stable mutators formed in the inflammatory lesions in patients may be even be more common [26]. Such families of hyper-mutators could lead to higher mutation rates in genes associated with anti-TB drug resistance than predicted from a stable genome approach. Epidemiologic evidence has been presented that suggests that this might be why there is a greater propensity to drug resistance in the Beijing strain [27].
One dogma has been that in latent TB, non-replicating bacilli (which have minimal DNA replication), do not generate resistant mutants. Thus, it has been assumed that most resistance arises from “luxuriantly” replicating bacilli but not in those bacteria not growing [28; 29], and that isoniazid monotherapy is appropriate for treating latent TB. Recently, Fortune and her team infected macaques with M. tuberculosis, and then examined bacteria recovered from lesions with either active TB, or latent TB, or reactivation TB [03]. Whole genome sequencing revealed that mutation rates were as high in bacteria in granulomas during latent TB as in log phase growth cultures. The mutations were a result of oxidative damage, either cytosine deamination (GC>AT) or the formation of 8-oxoguanine (GC>TA) [30]. This seminal observation suggests that drug resistant mutations arise more commonly during latency than assumed, and that treatment of latent TB may contribute to the emergence of drug resistant TB. Indeed, the highest risk factor for isoniazid monoresistance is prior treatment for latent TB [31].
Resistance has been hitherto assumed to arise from chromosomal mutations of genes encoding proteins related to either conversion of prodrug to active moiety or target proteins (table 1). The role of efflux pumps in drug resistance has been more recently examined. First, microbial kill by isoniazid in log-phase growth bacilli in the HFS was terminated by efflux pump related mechanisms [9]. This finding led to considerable controversy [28;29], especially around the meaning of “tolerance” and “resistance”. The rapid emergence of resistance to isoniazid was confirmed in a subsequent PK/PD study, and was linked to both Cmax/MIC and AUC/MIC [13]. Further insight was gained in a study in which ethambutol monotherapy led to “tolerance”, essentially low level resistance to the ethambutol that was linked to % time above the MIC [14]. Even more remarkable was the finding that isoniazid resistance could also be demonstrated after the ethambutol monotherapy, with the same “U” shaped curve encountered as if isoniazid had been administered. Administration of isoniazid after 7 days of daily ethambutol resulted in reduction in isoniazid effect in a rank order proportional to the original isoniazid dose [14]. This meant that multiple drug resistance can arise from administration of a single drug, and the resistance was proportional to drug concentrations used against the bacteria. This led us to propose a new scenario for the emergence of multi-drug resistant TB; one based on both evolution and PK/PD exposures (figure 2). In this scenario, efflux pumps are induced first, and lead to drug resistance or tolerance to more than one drug. This occurs in the face of PK/PD drug exposures that have not been optimized to suppress resistance. The bacteria continue to replicate under protection of the efflux pumps, sometimes in with a background of stable and non-stable mutator genes, and enable the bacteria to generate the canonical chromosomal mutations associated with high level resistance. We predicted that this is a common scenario, and that efflux pumps such as those encoded by Rv1258c would lead to multiple drug resistance to rifampin, quinolones and streptomycin [14]. Recently, Louw et al sequenced MDR-TB clinical isolates from Cape Town, and demonstrated these efflux pump derived clusters of resistance to rifampin and quinolones [32]. In our proposed paradigm, poor compliance and pharmacokinetic mismatching play a less significant role, while reduced exposures due to poor dosing (our current non-optimized dosing) and pharmacokinetic variability start a chain of evolution driven events rather quickly, which leads to MDR-TB and XDR-in the long run. The traditional role of combination therapy in reducing acquired drug resistance via one drug protecting another is still maintained in this scenario, provided the drugs are not expelled from bacterial cells by the same efflux pump.
Figure 2. Proposed evolution of simultaneous drug resistance to antituberculosis drugs.
The role of the immune system in the evolutionary scenario we have proposed for M. tuberculosis drug resistance is yet to be determined. There are some hints, especially in the case of rifamycin drug resistance, that the immune system may play an important role in suppressing drug resistance [33;34]. It seems that rifamycin monoresistance arises more commonly when there is a background of advanced immunodeficiency. However, unlike microbial kill of other bacteria by granulocytes in which the contribution of the extent to which phagocytes contribute to microbial kill have been well characterized [35; 36], no similar studies have been performed for emergence of drug resistance. Studies examining M. tuberculosis are even more scant. Studies to establish this are on-going.
Review highlights.
Mycobacterium tuberculosis resistance to pyrazinamide and ethambutol are linked to % time concentration persists above MIC while resistance to rifampin is linked to Cmax/MIC ratio and isoniazid to both Cmax/MIC and AUC/MIC.
Mutation rates are high and drug resistance emerges in Mycobacterium tuberculosis even during latency
In our current poorly optimized regimens, induction of efflux pumps might lead to multiple drug tolerance/resistance and emergence of MDR-TB
We propose a new evolutionary scenario for MDR-TB emergence that integrates PK/PD, efflux pumps, mutator genes, and standard chromosomal mutations
Acknowledgments
This work was supported by grant R01AI079497 from National Institute of Allergy and Infectious Diseases and grant 1DP2OD001886 from the national Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. WHO Report 2010. Geneva., Switzerland: Global Tuberculosis Control; 2010. [Google Scholar]
- 2.Mitchison DA. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int J Tuberc Lung Dis. 1998;2:10–15. [PubMed] [Google Scholar]
- 3.Jindani A, Aber VR, Edwards EA, Mitchison DA. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis. 1980;121:939–949. doi: 10.1164/arrd.1980.121.6.939. [DOI] [PubMed] [Google Scholar]
- 4.Morris S, Bai GH, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic Actinomycetes; Approved Standard. Wayne, PA: Clinical and Laboratory Standards Institute; 2003. [PubMed] [Google Scholar]
- 6.Wilkins JJ, Langdon G, McIlleron H, Pillai GC, Smith PJ, Simonsson US. Variability in the population pharmacokinetics of pyrazinamide in South African tuberculosis patients. Eur J Clin Pharmacol. 2006;62:727–735. doi: 10.1007/s00228-006-0141-z. [DOI] [PubMed] [Google Scholar]
- 7.Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis. 2004;190:1642–1651. doi: 10.1086/424849. [DOI] [PubMed] [Google Scholar]
- 8.Gumbo T, Louie A, Deziel MR, Drusano GL. Pharmacodynamic evidence that ciprofloxacin failure against tuberculosis is not due to poor microbial kill but to rapid emergence of resistance. Antimicrob Agents Chemother. 2005;49:3178–3181. doi: 10.1128/AAC.49.8.3178-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gumbo T, Louie A, Liu W, Ambrose PG, Bhavnani SM, Brown D, Drusano GL. Isoniazid’s bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J Infect Dis. 2007;195:194–201. doi: 10.1086/510247. [DOI] [PubMed] [Google Scholar]
- 10.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother. 2007;51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Gumbo T, Dona CS, Meek C, Leff R. Pharmacokinetics-pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob Agents Chemother. 2009;53:3197–3204. doi: 10.1128/AAC.01681-08. Studying the pharmacology of pyrazinamide has hirtherto been difficult. This documents a novel HFS model of TB that recapitulated pyrazinamide sterilizing effect and time to emergence of drug resistance in human TB. PK/PD findings led to identification of doses of 4–5G/day which would accelerate sterilzing effect and suppress resistance emergence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Pasipanodya JG, Gumbo T. Clinical and toxicodynamic evidence that high-dose pyrazinamide is not more hepatotoxic than the low doses currently used. Antimicrob Agents Chemother. 2010;54:2847–2854. doi: 10.1128/AAC.01567-09. This paper followed on reference 11, and demonstrated that high pyrazinamide doses were not associated with hepatotoxcity provided they were administered fro 2 months or less. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumbo T, Louie A, Liu W, Brown D, Ambrose PG, Bhavnani SM, Drusano GL. Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob Agents Chemother. 2007;51:2329–2336. doi: 10.1128/AAC.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14****.Srivastava S, Musuka S, Sherman C, Meek C, Leff R, Gumbo T. Efflux-pump-derived multiple drug resistance to ethambutol monotherapy in Mycobacterium tuberculosis and the pharmacokinetics and pharmacodynamics of ethambutol. J Infect Dis. 2010;201:1225–1231. doi: 10.1086/651377. HFS PK/PD study that established that efflux pump induction leads to low level multi-drug resistance, also called tolerance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayaram R, Shandil RK, Gaonkar S, Kaur P, Suresh BL, Mahesh BN, Jayashree R, Nandi V, Bharath S, Kantharaj E, Balasubramanian V. Isoniazid pharmacokinetics-pharmacodynamics in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother. 2004;48:2951–2957. doi: 10.1128/AAC.48.8.2951-2957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaram R, Gaonkar S, Kaur P, Suresh BL, Mahesh BN, Jayashree R, Nandi V, Bharat S, Shandil RK, Kantharaj E, Balasubramanian V. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother. 2003;47:2118–2124. doi: 10.1128/AAC.47.7.2118-2124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Pasipanodya J, Gumbo T. An oracle: antituberculosis pharmacokinetics-pharmacodynamics, clinical correlation, and clinical trial simulations to predict the future. Antimicrob Agents Chemother. 2011;55:24–34. doi: 10.1128/AAC.00749-10. This mini-review is the first to correlate PK/PD findings from the hollow fiber, mice, and guinea pigs with clinical pharmacological events in TB. It demonstrates that the relationships encountered in the pre-clinical models of monotherapy hold true in multiple-drug therapy in the clinic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Goutelle S, Bourguignon L, Maire PH, Van Guilder M, Conte JEJ, Jelliffe RW. Pharmacokinetics and antituberculosis pharmacodynamics of rifampin in lungs: a population modeling and Monte Carlo simulation study. Antimicrob Agents Chemother. 2009;53:2974–81. doi: 10.1128/AAC.01520-08. This is a population pharmacokinetic study with computer aided clinical trial simulations that demonstrated that standard doses of rifampin would not be able to achieve exposures known to suppress resistance based on reference 10, but instead doses of 1,200 mg a day were better. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peloquin CA, Jaresko GS, Yong CL, Keung AC, Bulpitt AE, Jelliffe RW. Population pharmacokinetic modeling of isoniazid, rifampin, and pyrazinamide. Antimicrob Agents Chemother. 1997;41:2670–2679. doi: 10.1128/aac.41.12.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Wilkins JJ, Langdon G, McIlleron H, Pillai G, Smith PJ, Simonsson US. Variability in the population pharmacokinetics of isoniazid in South African tuberculosis patients. Br J Clin Pharmacol. 2011;72:51–62. doi: 10.1111/j.1365-2125.2011.03940.x. This is a population pharmacokinetic study that demonstrates several important factors. First, the population pharmacokinetic parameter estimates in this South African population differed significantly from prior studies in Europe and the USA. Second, this study included 225 patients with TB, so that this should be considered the most definitive study on isoniazid pharmacokinetics to date. Third, there was a very high inter-occasion variability in absorption rate of the isoniazid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkins JJ, Savic RM, Karlsson MO, Langdon G, McIlleron H, Pillai G, Smith PJ, Simonsson US. Population pharmacokinetics of rifampin in pulmonary tuberculosis patients, including a semimechanistic model to describe variable absorption. Antimicrob Agents Chemother. 2008;52:2138–2148. doi: 10.1128/AAC.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, Scheidel B, Jakob V, Rodamer M, Cascorbi I, Doroshyenko O, Sörgel F, Fuhr U. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother. 2005;49:1733–1738. doi: 10.1128/AAC.49.5.1733-1738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida D, Nuermberger E, Tyagi S, Bishai WR, Grosset J. In vivo validation of the mutant selection window hypothesis with moxifloxacin in a murine model of tuberculosis. Antimicrob Agents Chemother. 2007;51:4261–4266. doi: 10.1128/AAC.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dos VT, Blazquez J, Rauzier J, Matic I, Gicquel B. Identification of Nudix hydrolase family members with an antimutator role in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2006;188:3159–3161. doi: 10.1128/JB.188.8.3159-3161.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbach SI, Springer B, Machowski EE, North RJ, Papavinasasundaram KG, Colston MJ, Bottger EC, Mizrahi V. DNA alkylation damage as a sensor of nitrosative stress in. Mycobacterium tuberculosis Infect Immun. 2003;71:997–1000. doi: 10.1128/IAI.71.2.997-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taddei F, Matic I, Godelle B, Radman M. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 1997;5:427–428. doi: 10.1016/S0966-842X(97)01157-8. [DOI] [PubMed] [Google Scholar]
- 27.Rad ME, Bifani P, Martin C, Kremer K, Samper S, Rauzier J, Kreiswirth B, Blazquez J, Jouan M, van SD, Gicquel B. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–845. doi: 10.3201/eid0907.020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28****.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, Flynn JL, Fortune SM. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–486. doi: 10.1038/ng.811. Using macaques infected with M. tuberculosis, the authors demonstrated that bacilli recovered from lesions with either active TB, or latent TB or reactivation TB had similar mutation rates; possibly due to oxidative DNA damage. This means that resistance is easily acquired even during latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchison DA, Jindani A, Davies GR, Sirgel F. Isoniazid activity is terminated by bacterial persistence. J Infect Dis. 2007;195:1871–1872. doi: 10.1086/518046. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad Z, Klinkenberg LG, Pinn ML, Fraig MM, Peloquin CA, Bishai WR, Nuermberger EL, Grosset JH, Karakousis PC. Biphasic kill curve of isoniazid reveals the presence of drug-tolerant, not drug-resistant, Mycobacterium tuberculosis in the guinea pig. J Infect Dis. 2009;200:1136–1143. doi: 10.1086/605605. [DOI] [PubMed] [Google Scholar]
- 31**.Cattamanchi A, Dantes RB, Metcalfe JZ, Jarlsberg LG, Grinsdale J, Kawamura LM, Osmond D, Hopewell PC, Nahid P. Clinical characteristics and treatment outcomes of patients with isoniazid-monoresistant tuberculosis. Clin Infect Dis. 2009;48:179–85. doi: 10.1086/595689. This clinical study demonstrated that the risk factors for acquisition of isoniazid resistance was prior treatment for latent TB, which conferred higher risk than prior treatment for active TB. Taken together with the Fortune study (ref 28), this means that drug resistance arises in latent TB, contrary to dogma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32****.Louw GE, Warren RM, Gey van Pittius NC, Leon R, Jimenez A, Pando RH, McEvoy CR, Grobbelaar M, Murray M, van Helden PD, Victor TC. Rifampicin reduces susceptibility to ofloxacin in rifampicin resistant Mycobacterium tuberculosis through efflux. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201011-1924OC. This study demonstrated that efflux pump induction led to resistance to both rifampin and ofloxacin in MDR-TB clinical isolates from the Western Cape of South Africa, where there is a high prevalnce of the Beijing strain. This means multiple mecahnisms of resistance emergence are encountered in patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benator D, Bhattacharya M, Bozeman L, Burman W, Cantazaro A, Chaisson R, Gordin F, Horsburgh CR, Horton J, Khan A, Lahart C, Metchock B, Pachucki C, Stanton L, Vernon A, Villarino ME, Wang YC, Weiner M, Weis S. Rifapentine and isoniazid once a week versus rifampicin and isoniazid twice a week for treatment of drug-susceptible pulmonary tuberculosis in HIV-negative patients: a randomised clinical trial. Lancet. 2002;360:528–534. doi: 10.1016/s0140-6736(02)09742-8. [DOI] [PubMed] [Google Scholar]
- 34.Burman W, Benator D, Vernon A, Khan A, Jones B, Silva C, Lahart C, Weis S, King B, Mangura B, Weiner M, El-Sadr W. Acquired rifamycin resistance with twice-weekly treatment of HIV-related tuberculosis. Am J Respir Crit Care Med. 2006;173:350–356. doi: 10.1164/rccm.200503-417OC. [DOI] [PubMed] [Google Scholar]
- 35.Drusano GL, Fregeau C, Liu W, Brown DL, Louie A. Impact of burden on granulocyte clearance of bacteria in a mouse thigh infection model. Antimicrob Agents Chemother. 2010;54 :4368–4372. doi: 10.1128/AAC.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drusano GL, Vanscoy B, Liu W, Fikes S, Brown D, Louie A. Saturability of Granulocyte Kill of Pseudomonas aeruginosa in a Murine Model of Pneumonia. Antimicrob. Agents Chemother. 2011;55:2693–2695. doi: 10.1128/AAC.01687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]