Abstract
Objective
To evaluate global gene expression patterns in the common iliac arteries of monkeys with varied extent of atherosclerosis.
Methods
The left common iliac artery was removed from ovariectomized cynomolgus monkeys (n=12) after 6.5 years of consuming a diet containing fat and cholesterol at levels comparable to that consumed in western populations. Arterial gene expression was analyzed by DNA microarray and real time RT-PCR.
Results
Significant differential expression of 986 genes was observed in iliac arteries containing moderate to large atherosclerotic plaques compared to normal/minimally affected reference group arteries. Atherosclerosis-associated genes included cytokines, chemokines, components of signal transduction pathways, and transcriptional activators and repressors, as well as other functional categories. Real time RT-PCR confirmed differential expression of genes chosen from a variety of functional categories. Specifically, expression of genes for estrogen receptor 1 (ESR1), claudin 11, and BH protocadherin 7 (PCDH7) were reduced, whereas expression of genes for apolipoprotein E (ApoE), growth differentiation factor 15 (GDF15), superoxide dismutase 2 (SOD2), SET domain, bifurcated 2 (SETDB2), phospholipase A2 group IIA (PLA2IIA), phospholipase A2 group VII (PLA2VII), and ring finger protein 149 (RNF149) were increased in atherosclerotic arteries.
Conclusions
The gene expression environment in arteries containing atherosclerotic plaques is profoundly different from that of relatively unaffected arteries and reflects the cellular and molecular complexity of atherosclerosis and associated arterial remodeling processes.
Keywords: atherogenesis, gene expression, diet, cholesterol, intervention potential, atherosclerosis
Introduction
The sequelae of atherosclerosis are leading causes of morbidity and mortality. Much is known about the pathogenesis of atherosclerosis1, and many studies of gene expression in atherosclerotic lesions have been performed, both at the level of single gene analysis2,3 and by global gene expression profiling technologies such as DNA microarray4–13. In spite of the applications of global gene expression technologies and the availability of large gene expression data sets, there is only partial agreement on gene expression patterns in atherosclerosis5, and the need exists for more complete data sets.
The cynomolgus monkey is an ideal model for the study of the effects of atherosclerosis in the cardiovascular system. Female cynomolgus monkeys develop atherosclerosis when fed an atherogenic diet, and, like women, their susceptibility varies with their reproductive stage; premenopausal monkeys are relatively protected from atherosclerosis while postmenopausal monkeys lose such protection.14 Moreover, the ovariectomized cynomolgus monkey has proven to be an important animal model for the postmenopausal human vasculature.15,16
Previous studies in the cynomolgus monkey model have demonstrated strong parallels between atherosclerosis progression and expression of mRNA for inflammatory mediators, cell adhesion molecules, and chemokines, as well as markers indicative of the presence of immune cell populations such as monocyte-macrophages and T cells.17,18 Under certain circumstances the expression of these markers may be more sensitive indicators of responses to treatment than atherosclerotic lesion size.17–19 In the current study, the iliac artery was used as a surrogate for the coronary artery because atherosclerosis extent in the right and left iliac arteries is very highly correlated with atherosclerosis in the coronary arteries.14 In this study we explored the effect of atherosclerosis on global gene expression patterns in the common iliac arteries of ovariectomized female cynomolgus monkeys with varied extent of atherosclerosis.
Methods and Materials
Animals and Study Design
Twelve adult female surgically menopausal cynomolgus macaques (Macaca fascicularis), estimated average 21 years of age (range 11–27), were used for this study. At the initiation of the study presented here, the monkeys had been housed in stable social groups of 3–4 animals each and consumed various semi-purified diets for 6.5 years. The diets were formulated to mimic a typical diet consumed by people in North America, containing ~0.20 mg cholesterol/Calorie of diet, up to 30–35% fat, and the protein source varied between various formulations of animal (casein/lactalbumin) and vegetable (soy). The monkeys had been surgically postmenopausal (ovariectomized) for 4–6 years prior to the initiation of this study.
The left common iliac artery was obtained from each monkey for evaluation as described.17 Briefly, monkeys were anesthetized and the left common iliac artery was isolated via laparotomy. One ligature was placed at the point of bifurcation of the iliac artery from the abdominal aorta, a second ligature was placed proximal to the bifurcation into internal and external iliac arteries, and the section of iliac artery between ligatures was removed. One segment was placed in RNA later® (Sigma, St. Louis, MO) for preservation of RNA, one segment was fixed in 4% paraformaldehyde for measurement of plaque intimal area as described20, and one segment was preserved in OCT for a separate study.
For data analysis, the left iliac artery biopsies were stratified into 3 groups based on the extent of atherosclerotic plaque. Arteries that were normal (no lesion) or minimally affected by atherosclerosis (plaque extent 0.04 ± 0.02 mm2 (mean ± SEM), range 0–0.113 mm2), n=4, were designated reference group arteries. The group designated medium contained lesions of 0.44 ± 0.04 mm2 (range 0.30 ± 0.542 mm2), n=5. The extent of plaque in the category designated large lesions was 0.83 ± 0.12 mm2 (range 0.61–1.003 mm2), n=3.
All procedures involving animals were conducted in compliance with State and Federal laws, standards of the U.S. Department of Health and Human Services, and guidelines established by the Wake Forest University Animal Care and Use Committee (ACUC). The facilities and laboratory animal program of Wake Forest University are fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
RNA extraction
Total RNA was extracted from iliac artery biopsy segments (2.2–18.8 mg) as described.21
Protein extraction and immunoblot
Proteins were extracted from the TRI reagent organic phase and resuspended using the SDS method as described.22 The protein content of the extracts was measured using the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Proteins (25 µg per lane) were resolved by electrophoresis on 10% polyacrylamide SDS gels and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (BioRad Laboratories, Hercules, CA). AquaBlock (East Coast Biologics, North Berwick, ME) was used to block nonspecific binding sites in the membranes overnight. The membranes were incubated overnight with goat anti-apolipoprotein E (apoE) primary antibody diluted in AquaBlock (1:1000, Millipore, Temecula, CA). The membranes were washed and incubated overnight with IRDye 800-conjugated secondary anti-goat antibody (1:1000, Rockland Immunochemicals, Gilbertsville, PA) for 3 hours. The membranes were washed again and the apoE bands were imaged on an Odyssey Infrared Imager (LI-COR, Lincoln, NE). The membranes were then incubated overnight with mouse anti-β-actin antibody (1:8000; Calbiochem, La Jolla, CA), washed, and incubated overnight with IRDye 800-conjugated secondary anti-mouse antibody (1:8000, Rockland Immunochemicals, Gilbertsville, PA). The membranes were washed and the β-actin bands were imaged on the Odyssey Infrared Imager. The Odyssey densitometry program (LI-COR, Lincoln, NE) was used to analyze the apoE and β-actin images. The density of the apoE protein band was normalized to the β-actin loading control; data are expressed as relative protein density.
DNA microarrays
The CodeLink Whole Human Genome Bioarrays (Applied Microarrays, Tempe, AZ; formerly GE CodeLink)23–25 were used for the DNA microarray analysis of gene expression of the cynomolgus monkey RNA as documented.26 These microarrays contain 53,000 single-stranded 30-mer oligonucleotide probes for human genes/transcribed sequences. Biotinylated probes were prepared using 0.275 µg iliac artery total RNA as the starting material. Hybridization and processing were carried out as described.21,23,27 The microarray images were acquired and aligned with GenePix Pro software (MDS, Inc., Toronto, Canada). Background correction was applied, and the individual gene specific spots were identified and quantitated using CodeLink software (Applied Microarrays). Acuity software (MSD, Inc.) was used to obtain fold expression values. The expression of each gene was normalized to the median gene expression and each slide to the 50th percentile of gene expression using GeneSpring 7.0 software (Agilent, Santa Clara, CA). GeneSpring software was also used for statistical analysis of the microarray data.
Real time RT-PCR
Real time reverse transcription-polymerase chain reaction (RT-PCR)23 was used to confirm the differential expression of 9 genes that had been shown to be differentially expressed by DNA microarray. Complementary DNA (cDNA) was synthesized from 0.1 µg of total RNA by reverse transcription using the High Capacity cDNA Archive Kit from Applied Biosystems (Foster City, CA). The PCR reaction used cDNA from the reverse transcription reaction and thermal stable AmpliTaq Gold DNA polymerase for DNA amplification. Predesigned primers and FAM-labelled minor groove binding probes were obtained from TaqMan Gene Expression Assays (Applied Biosystems). Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as the housekeeping gene. All samples were run in duplicate. Data from the real time RT-PCR reactions were analyzed by qBase software.28
Statistical analysis
GeneSpring GX 7.0 software (Agilent, Santa Clara, CA) was used for the statistical analysis of DNA microarray data. The p value was set at 0.05 for the 1×3 analysis of variance. The Benjamini and Hochberg False Discovery Rate was used for multiple testing correction. Approximately 5% of the identified genes would be expected to pass this restriction by chance using the Benjamini and Hochberg False Discovery Rate. Real time RT-PCR and immunoblot data were analyzed by t-test or by analysis of variance followed by Newman-Keuls post hoc test when a significant F was found (GraphPad Prism 4, GraphPad Software, San Diego, CA).
Results
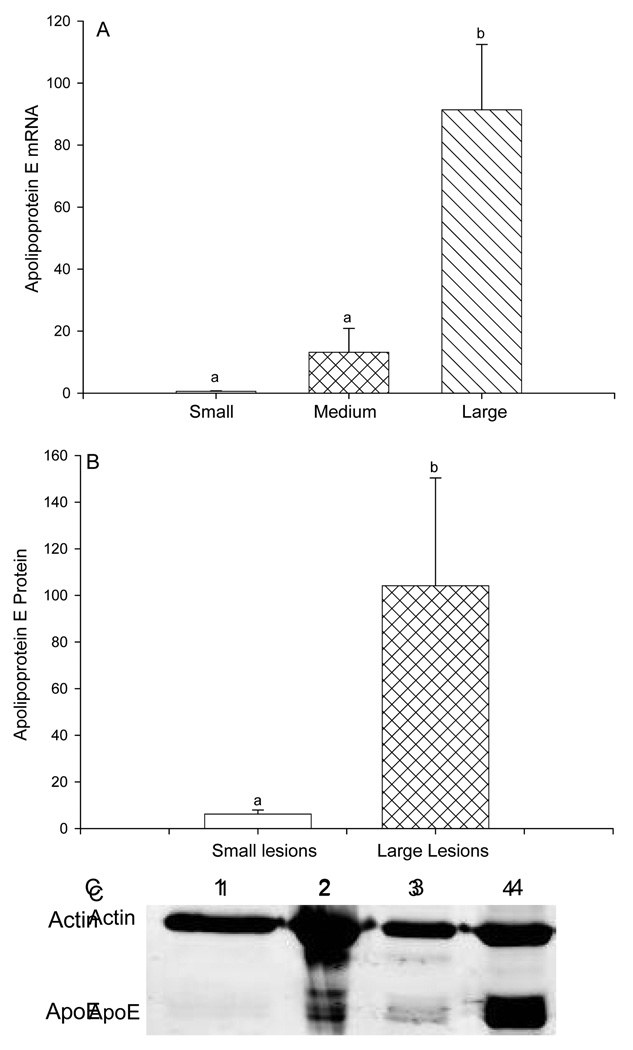
Extent of atherosclerotic plaques was used to stratify arterial gene expression data for analysis into the reference group with no or minimal lesions, and groups with medium or large atherosclerotic plaques as described above.. This analysis identified a list of probes for which expression in arteries with atherosclerotic plaques was significantly different from expression in the reference group arteries with p values less than 0.05. Of those differentially expressed genes, probes that represented transcribed sequences with no name and no known function were excluded from further analysis, as were genes for which the expression values were less than 1.0, and those for which the fold expression values were between 0.5-fold and 2-fold of the control values. The list of 986 genes that fulfilled all criteria for further analysis is shown in Supplemental Table 1 (see Table, Supplemental Digital Content 1, http://links.lww.com/MENO/A5). The differentially expressed genes were sorted into 30 gene ontologies based on function (Table 1). Relative to the reference arteries, gene expression levels within the atherosclerotic plaque-containing arteries were significantly higher in 84% of the genes and significantly lower in 16% of the genes. Genes chosen for follow-up by immunoblot or real time RT-PCR reflect the wide range of functional categories that exhibited differential expression in the presence of atherosclerotic plaque. Apolipoprotein E expression showed the greatest increase with a 152-fold higher expression in arteries with large plaque compared to the reference arteries (Figure 1A). Increased expression of Apo E gene in arteries with large atherosclerotic plaques was confirmed by immunoblot of Apo E protein (Figure 1B, C).
Table 1.
Gene ontologies of differentially expressed genes in the monkey common iliac arteries in the presence of no or small atherosclerotic lesions (reference arteries), medium sized lesions, or large lesions.
| Gene ontology | Number of genes |
|---|---|
| Signal Transduction | 159 |
| Defense and immunity | 92 |
| Catalytic/metabolism | 86 |
| Lipid Metabolism | 62 |
| Transcriptional regulators | 60 |
| Activators (47) | |
| Corepressors (13) | |
| Nucleic acid regulation | 60 |
| Cytoskeleton | 60 |
| Receptors | 52 |
| Proteases and protease regulators | 44 |
| Channels/transporters | 44 |
| Cancer related | 29 |
| Ubiquitin system | 27 |
| Lysosomal | 25 |
| Cell fate | 25 |
| Unknown function | 22 |
| Vesicle trafficking | 21 |
| Oxidation-reduction | 18 |
| Adhesion | 18 |
| Extracellular matrix | 18 |
| Cell cycle related | 17 |
| Development | 11 |
| Adaptor proteins | 10 |
| Growth factors | 10 |
| Translation | 6 |
| Chaperonins | 6 |
| Selenium related | 5 |
| Metal metabolism | 4 |
| Angiogenesis | 4 |
Figure 1.
Expression of apolipoprotein E (apo E) gene and protein in cynomolgus monkey iliac arteries stratified by plaque size. A: Apo E gene expression from DNA microarray data of iliac arteries containing small/absent (reference group), medium, or large atherosclerotic plaque. B and C: Apo E protein expression from immunoblot of iliac arteries containing small/absent (reference group) vs. large atherosclerotic plaque. Sizes of atherosclerotic plaque in the iliac arteries used for immunoblot analysis were mean plaque size 0.010 mm2, range 0–0.041 mm2 for the small/absent (reference) group, and mean plaque size 0.757 mm2, range 0.521–0.996 mm2 for the large group. B illustrates densitometry data of immunoblot analysis of apo E normalized to β-actin expression in monkey iliac arteries, mean ± SEM, n=4 paired samples. C shows a representative immunoblot of apo E and the loading control, β-actin. Lanes 1 and 3 contain protein from arteries with small (reference group) atherosclerotic lesions. Lanes 2 and 4 contain protein from arteries with large lesions.
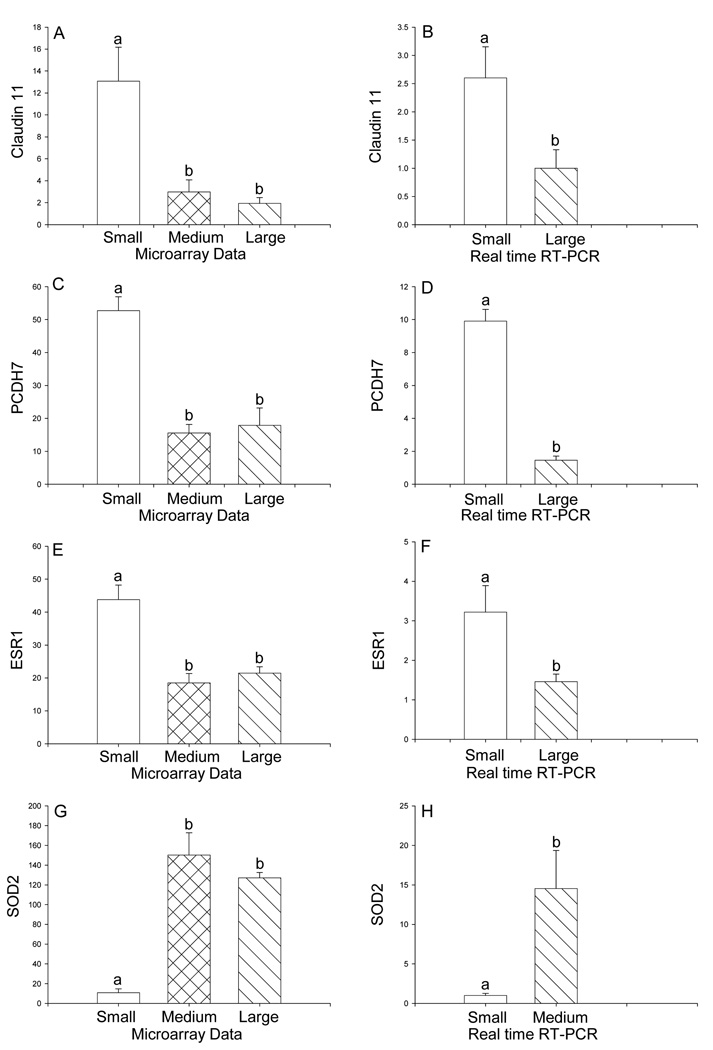
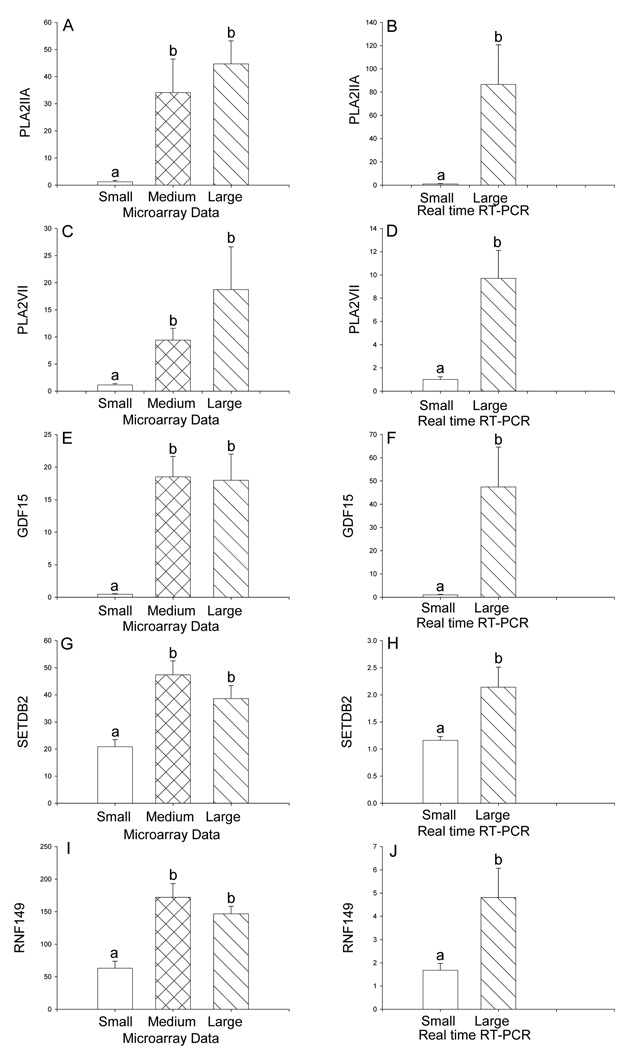
Nine differentially expressed genes in atherosclerotic arteries compared to reference arteries were confirmed by real time RT-PCR studies. All 9 of the genes tested by real time RT-PCR validated the microarray data. The gene with the greatest reduction in arteries with large plaque size was the tight junction molecule, claudin 11, with 0.1-fold of reference group expression. Reduced expression of claudin 11 was confirmed by real time RT-PCR (Figure 2 A, B). Expression of another gene involved in cell adhesion, BH protocadherin 7 (PCDH7), was also lower in the presence of atherosclerotic plaque (Figure 2 C, D). Reduced expression of estrogen receptor 1 (ESR1) was demonstrated by DNA microarray and by real time RT-PCR (Figure 2 E, F). In contrast, higher expression of superoxide dismutase 2 (SOD2) (Figure 2 G, H), phospholipase A2 group IIA (PLA2IIA) (Figure 3 A, B), and phospholipase A2 group VII (PLA2VII) (Figure 3 C, D), growth differentiation factor 15 (GDF15, also called macrophage inhibitory cytokine-1) (Figure 3 E, F), SET domain, bifurcated 2 (SETBD2) (Figure 3 G, H), and ring finger protein 149 (RNF149) (Figure 3 I, J) were all demonstrated by DNA microarray and confirmed with real time RT-PCR analysis of gene expression.
Figure 2.
Differential expression of claudin 11 (SOD2) (A, B), BH protocadherin 7 (PCDH7) (C, D), estrogen receptor 1 (ESR1) (E, F), and superoxide dismutase 2 (SOD2) (G, H) in the iliac arteries of cynomolgus monkeys on a high fat diet stratified by size of atherosclerotic lesions. Panels A, C, E, G: Differential gene expression from DNA microarray. The size of small (reference group) lesions was 0–0.113 mm2 (n=4), medium lesions were 0.30–0.542 mm2 (n=5), and large lesions were 0.61–1.003 mm2 (n=3). Panels B, D, F, H: Data from real time RT-PCR. Data are expressed as the mean ± SEM. Statistical analysis utilized ANOVA followed by Newman-Keuls post hoc test (microarray data) or Student’s t-test (RT-PCR data). Bars with different letter superscripts denote that the data for those groups are significantly different from each other, p<0.05.
Figure 3.
Differential expression of phospholipase A2, group IIA (PLA2IIA) (A, B), phospholipase A2 group VII (PLA2VII) (C, D), growth differentiation factor 15 (GDF15) (E, F), SET domain, bifurcated 2 (SETDB2) (G, H), and ring finger 149 (RNF149) (I, J) in the iliac arteries of cynomolgus monkeys on a high fat diet stratified by size of atherosclerotic lesions. Panels A, C, E, G, I: Differential gene expression from DNA microarray analysis. The size of small (reference group) lesions was 0–0.113 mm2 (n=4), medium lesions were 0.30–0.542 mm2 (n=5), and large lesions were 0.61–1.003 mm2 (n=3). Panels B, D, F, H, J: Data from real time RT-PCR analysis. Statistical analysis utilized ANOVA followed by Newman-Keuls post hoc test (microarray data) or Student’s t-test (RT-PCR data). Data are expressed as the mean ± SEM. Bars with different letter superscripts denote that the data for those groups are significantly different from each other, p<0.05.
The data set for these DNA microarrays, including all genes that were excluded from the current analysis, has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) as recommended by Minimum Information About a Microarray Experiment (MIAME) standards9 and can be accessed through GEO Series accession number GSE26326. The validity and reproducibility of these microarrays has been reported previously.23
Discussion
The most important finding of this study is the profound effect of the presence and extent of atherosclerosis on gene expression in the common iliac artery of ovariectomized female monkeys. Expression levels of a great number of genes were markedly different in the atherosclerotic arteries and the fold differences in gene expression relative to the reference group were large. The data presented here confirm and extend the findings of previous DNA microarray studies of atherosclerosis4–13, especially in regard to the elevated expression of growth factors, cytokines, chemokines and other inflammatory molecules. A variety of factors may contribute to the altered gene expression profiles in these arterial samples. Normal arteries are composed largely of vascular smooth muscle cells, with a smaller contribution from endothelial cells, fibroblasts, and relatively minor populations of monocyte-macrophages and transient immune cell subsets. During early atherogenesis, hyperlipidemia and other stimuli promote the expression of pro-inflammatory molecules which attract monocyte macrophages to bind to the endothelium and migrate into the arterial intima. During the progression of atherosclerosis these cells can accumulate, along with other cell types such as T cells and B cells as well as others. Vascular smooth muscle cell proliferation may be stimulated and a general pro-inflammatory environment within the lesion may become self-perpetuating.30,31 Thus, the arterial gene expression patterns reflect the cell populations present at the time of collection, along with any differential regulation which occurs within specific cell types in response to the cellular and physical environment of the artery. Expression patterns are also influenced by systemic factors such as hormones (e.g., steroids, insulin), plasma lipids, proteins, glucose, oxygen tension, blood pressure, and others, all of which may impart pressures on cellular physiology within the atherosclerotic plaque and surrounding artery. Our own work has demonstrated that gene expression levels of cell markers can be used to estimate cell populations within the arterial wall, and that transcript levels for a variety of inflammatory molecules are highly correlated with atherosclerosis in these arteries.17,18
These studies used an important model of the postmenopausal state, the ovariectomized cynomolgus monkey.15,16 Cardiovascular disease continues to be the leading cause of death in women, especially after the menopause. Thus, these data are relevant to the postmenopausal condition and identify mechanisms by which the presence of atherosclerosis may cause arterial damage.
The results of several previous studies with the cynomolgus monkey model gave rise to the so-called “timing hypothesis” regarding the cardiovascular risks and benefits of postmenopausal estrogen therapy. The first finding was that if estrogen replacement was begun at the time of ovariectomy and the initiation of the atherogenic diet, the monkeys were relatively protected against coronary artery atherosclerosis: whereas, in monkeys not given estrogen therapy until two years later (equivalent to 6 patient years) there was no coronary artery atherosclerosis protection.32 The second finding was that postmenopausal monkeys subjected to a baseline iliac biopsy and subsequently given estrogen replacement for 3 years varied in their therapeutic benefit depending on the extent of baseline atherosclerosis. Estrogen replacement was highly effective in inhibiting atherosclerosis progression only in those monkeys in the lowest tertile of plaque extent (p=0.0001); whereas, among those monkeys in the tertile with the largest plaques, estrogen treatment did not inhibit atherosclerosis progression (p=0.71).33 Thus, estradiol appears to have a protective effect in normal or minimally affected arteries, but is not protective, and may have adverse effects, in arteries that have already developed atherosclerotic plaques.34,35 The effect of atherosclerosis on gene expression may be responsible, at least in part, for these contrasting estrogen effects. We interpret the current data to suggest that the presence of atherosclerotic plaque in an artery results in the synthesis of factors that may interfere with the ability of estradiol, or other vascular factors, to carry out their normal protective functions. Moreover, the interaction of estradiol with atherosclerosis-associated factors may be responsible for the deleterious effects of estrogen that have been observed.34,35 In addition to the synthesis of factors that may interfere with the signaling of estradiol at its receptor, these data confirm our previous work demonstrating that there is an inverse relationship between atherosclerosis and arterial expression levels of estrogen receptor alpha (ERα, product of the ESR1 gene)17,18,36, corroborating previous findings of diminished ER immunoreactivity in atherosclerotic plaques.37 The ability of estrogen to act via its receptor may therefore be reduced in the presence of atherosclerotic plaque. Moreover, the data show that cytochrome P450 27A1 (CYP27A1) was up-regulated in vessels containing atherosclerotic lesions. CYP27A1 converts cholesterol to 27-hydroxycholesterol which may act as an endogenous selective estrogen receptor modulator in blood vessels and inhibit the protective effects of estrogen in the vasculature.38 Lastly, the pro-inflammatory environment of the atherosclerotic plaque may lead to interference of ERα transactivating capability due to transcription factor cross-talk interference from high levels of activated NFκB.18,39 Thus, the presence of atherosclerotic plaque provides multiple mechanisms that may interfere with the mechanisms of action of estradiol in the arteries. It is likely that the presence of atherosclerosis interferes with other factors that regulate gene expression in the arteries in a similar fashion to that proposed here for estrogen.
The functions of many of the proteins encoded by genes that are increased in atherosclerotic arteries appear to be tissue protective. For example, superoxide dismutase 2 and glutathione peroxidase protect against oxidation of proteins.40 In fact, all 18 genes in the ontology of oxidation/reduction were up-regulated, suggesting a compensatory response to overproduction of reactive oxygen species in the atherosclerotic lesion.41 ApoE stimulates efflux of cholesterol from macrophages, among other protective effects.42 Cytochrome P450 1A1 (CYP1A1, ↑3.4-fold) was also expressed at higher levels in atherosclerotic arteries. CYP1A1 metabolizes estradiol to 2-hydroxyestradiol, which is then methylated to 2-methoxyestradiol by catechol-O-methyltransferase. This metabolite of estradiol has been reported to exhibit cardiovascular protective effects that are independent of the estrogen receptor43 and to reduce the formation of atherosclerotic lesions in the APOE-deficient mouse.44 Thus the up-regulation of CYP1A1 may represent another potentially compensatory response to atherosclerosis. While it is likely that many of the differentially expressed genes in an atherosclerotic plaque reflect the damage occurring in the plaque tissue, these data also suggest that the tissue maintains mechanisms which may limit further damage. Using this experimental design, it is not possible to determine whether the differentially expressed genes that we observed are causally linked to atherosclerosis or whether they are a consequence of the condition. However, the tissue protective nature of many of these genes suggest that their regulation is a result rather than a cause of atherosclerosis.
As expected, the gene ontology of immune system/inflammation was prominently placed among the differentially expressed genes. Three genes from this ontology were chosen for follow-up by real time RT-PCR. Phospholipases A2 (PLA2) group IIA and group VII are secretory phospholipases implicated in atherogenesis.45,46 PLA2 group VII (also known as lipoprotein-associated PLA2 and as platelet activating factor acetylhydrolase) is an important risk factor and marker of atherosclerosis47,48 and its inhibition reduces the development of atherosclerosis.49 Growth differentiation factor 15 (alias macrophage inhibitory cytokine-1) is also associated with inflammation 50 and with atherosclerosis.51
Disruption of cell-cell interactions through dysregulation of cell-cell junction and adhesion molecules, as well as via proteases, has been implicated in the development of atherosclerosis.52,53 In the current study, many genes in the ontologies of cell adhesion and proteases/protease regulators were differentially expressed. Reduced expression of the tight junction molecule, claudin 11, and of the adhesion molecule, BH protocadherin 7, was shown by both microarray and real time RT-PCR. Though the mechanism is unclear, data suggest that cell-cell adhesion inhibits vascular cell smooth muscle proliferation in development of atherosclerosis.52
Another set of related gene ontologies in which substantial changes in gene expression were observed in this study were the categories of transcriptional activators, transcriptional corepressors, and nucleic acids regulation. Changes in the expression of genes in these ontologies will alter cellular responses to other upstream regulators of gene expression such as cytokines, hormones, and growth factors and the signal transduction pathways that they regulate. One gene from this category, SET domain, bifurcated 2 (SETDB2) was chosen for follow-up by real time RT-PCR. SETDB2 is a histone H3 methyltransferase and may modulate gene expression epigenetically via this mechanism.54
Differential expression of genes in the ubiquitin system gene ontology reflects the potential role of protein quality control in atherosclerosis.55 Of the 27 genes in this ontology, 25 were up-regulated. Up-regulation of ring finger protein 149, an E3 ubiquitin ligase, was confirmed by real time RT-PCR.
Summary and Conclusions
These data indicate that the presence of atherosclerotic plaque profoundly alters the gene expression environment of the artery in which it is located, reflecting both the complexity of the cellular environment present in atherosclerotic lesions as well as the array of local and systemic influences on cellular activity in atherosclerotic arteries. Some of these factors may be causative of atherosclerotic plaque, whereas others may be the result of its presence. Some of the differentially expressed genes may represent compensatory responses of the cells to the presence of plaque, whereas others are likely to represent decompensatory changes. Further work will be required to sort out the components contributing to cause and effect of gene expression in atherosclerosis.
Supplementary Material
Acknowledgments
Funding: This work was supported in part by NIH INBRE 2 P20 RR016479 (KME, CJM, AC), by unrestricted development funds of Comparative Medicine at Wake Forest University (TBC), NIH/NCRR T32 RR07009 and NIH/NHLBI PO1 HL 45666 (TBC), and AG18170 and AG28641 (TCR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Dr. Thomas B. Clarkson serves on an advisory committee to Pfizer and also has grant support from Pfizer; however, these associations do not pose a conflict of interest in connection with the Work described in this manuscript. None of the other authors have anything to disclose.
References Cited
- 1.Cullen P, Rauterberg J, Lorkowski S. The pathogenesis of atherosclerosis. Handb Exp Pharmacol. 2005;170:3–70. doi: 10.1007/3-540-27661-0_1. [DOI] [PubMed] [Google Scholar]
- 2.Boot RG, van Achterberg TA, van Aken BE, et al. Strong induction of members of the chitinase family of proteins in atherosclerosis: chitotriosidase and human cartilage gp-30 expressed in lesion macrophages. Arterioscler Thromb Vasc Biol. 1999;19:687–694. doi: 10.1161/01.atv.19.3.687. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Sukhova GK, Sun JS, Xu WH, Libby P, Shi GP. Lysosomal cysteine proteases in atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1359–1366. doi: 10.1161/01.ATV.0000134530.27208.41. [DOI] [PubMed] [Google Scholar]
- 4.Ashley EA, Spin JM, Tabibiazar R, Quertermous T. Frontiers in nephrology: genomic approaches to understanding the molecular basis of atherosclerosis. J Am Soc Nephrol. 2007;18:2853–2862. doi: 10.1681/ASN.2007040514. [DOI] [PubMed] [Google Scholar]
- 5.Bijnens AP, Lutgens E, Ayoubi T, Kuiper J, Horrevoets AJ, Daemen MJ. Genome-wide expression studies of atherosclerosis. Critical issues in methodology, analysis, interpretation of transcriptomics data. Arterioscler Thromb Vasc Biol. 2006;26:1226–1235. doi: 10.1161/01.ATV.0000219289.06529.f1. [DOI] [PubMed] [Google Scholar]
- 6.Clerin V, Shih HH, Deng N, et al. Expression of the cysteine protease legumain in vascular lesions and functional implications in atherogenesis. Atherosclerosis. 2008;201:53–66. doi: 10.1016/j.atherosclerosis.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Hagg DA, Olson FJ, Kjelldahl J, et al. Expression of chemokine (C-C motif) ligand 18 in human macrophages and atherosclerotic plaques. Atherosclerosis. 2009;204:e15–e20. doi: 10.1016/j.atherosclerosis.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Hiltunen MO, Tuomisto TT, Niemi M, et al. Changes in gene expression in atherosclerotic plaques analyzed using DNA array. Atherosclerosis. 2002;165:23–32. doi: 10.1016/s0021-9150(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 9.King JK, Ferrara R, Tabibiazar R, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 10.Lutgens E, Faber B, Schapira K, et al. Gene profiling in atherosclerosis reveals a key role for small inducible cytokines. Validation using a novel monocyte chemoattractant protein monoclonal antibody. Circulation. 2005;111:3443–3452. doi: 10.1161/CIRCULATIONAHA.104.510073. [DOI] [PubMed] [Google Scholar]
- 11.Murillo CA, Woodside KJ, Guo Q, Zhang S, O’Connor KL, Hunter GC. Integrin and matrix metalloproteinase expression in human carotid plaque. J Surg Res. 2009;155:157–164. doi: 10.1016/j.jss.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Papaspyridonos M, Smith A, Burnand KG, et al. Novel candidate genes in unstable areas of human atherosclerotic plaques. Atheroscler Thromb Vasc Biol. 2006;26:1837–1844. doi: 10.1161/01.ATV.0000229695.68416.76. [DOI] [PubMed] [Google Scholar]
- 13.Tuomisto TT, Yla-Herttuala S. What have we learnt about microarray analyses of atherogenesis? Curr Opin Lipidol. 2005;16:201–205. doi: 10.1097/01.mol.0000162325.85290.e5. [DOI] [PubMed] [Google Scholar]
- 14.Clarkson TB, Anthony MS, Morgan TM. Inhibition of postmenopausal atherosclerosis progression: a comparison of the effects of conjugated equine estrogens and soy phytoestrogens. J Clin Endocrinol Metab. 2001;86:41–47. doi: 10.1210/jcem.86.1.7151. [DOI] [PubMed] [Google Scholar]
- 15.Clarkson TB, Hughes CL, Klein KP. The nonhuman primate model of the relationship between gonadal steroids and coronary heart disease. Prog Cardiovasc Dis. 1995;38:189–198. doi: 10.1016/s0033-0620(95)80011-5. [DOI] [PubMed] [Google Scholar]
- 16.Clarkson TB, Appt SE. Controversies about HRT—lessons from monkey models. Maturitas. 2005;51:64–74. doi: 10.1016/j.maturitas.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Walker SE, Register TC, Appt SE, et al. Plasma lipid-dependent and -independent effects of dietary soy protein and social status on atherogenesis in premenopausal monkeys: implications for postmenopausal atherosclerosis burden. Menopause. 2008;15(5):950–957. doi: 10.1097/gme.0b013e3181612cef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Register TC. Primate models in women’s health: inflammation and atherogenesis in female cynomolgus macaques (Macaca fascicularis) Am J Primatol. 2009;71(9):766–775. doi: 10.1002/ajp.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Register TC, Sophonsritsuk A, Appt SA. Estrogen inhibits carotid artery inflammation in early but not late menopause in cynomolgus monkeys. Menopause. 2010;17(6):1216. S-2. [Google Scholar]
- 20.Clarkson TB. Progression and regression of nonhuman primate coronary artery atherosclerosis: considerations of experimental design. In: Malinow MR, Blaton VH, editors. Regression of atherosclerotic lesions. Vol. 79. New York: Plenum Press; 1984. pp. 43–60. NATO ASI Series A. [Google Scholar]
- 21.Eyster KM, Brannian JD. Gene expression profiling in the aging ovary. In: Park-Sarge O-K, Curry T, editors. Molecular Endocrinology: Methods in Molecular Biology. Vol. 590. Totowa, NJ: Humana Press; pp. 71–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigo MC, Martin DS, Redetzke RA, Eyster KM. A method for the extraction of high quality RNA and protein from single small samples of arteries and veins preserved in RNAlater. J Pharmacol Toxicol Methods. 2002;47:87–92. doi: 10.1016/s1056-8719(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 23.Eyster KM, Klinkova O, Kennedy V, Hansen KA. Whole genome deoxyribonucleic acid microarray analysis of gene expression in ectopic versus eutopic endometrium. Fertil Steril. 2007;88:1505–1533. doi: 10.1016/j.fertnstert.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 24.Yauk CL, Berndt ML, Williams A, Douglas GR. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shippy R, Sendera TJ, Lockner R, et al. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics. 2004;5:61. doi: 10.1186/1471-2164-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redmond DE, Jr, Zhao JL, Randall JD, et al. Spatiotemporal patterns of gene expression during fetal monkey brain development. Brain Res Dev Brain Res. 2003;146:99–106. doi: 10.1016/j.devbrainres.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Eyster KM, Mark CJ, Gayle R, Martin DS. The effects of estrogen and testosterone on gene expression in the rat mesenteric arteries. Vasc Pharmacol. 2007;47:238–247. doi: 10.1016/j.vph.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 30.Gouwy M, Struyf S, Proost P, Van Camme J. Synergy in cytokine and chemokine networks amplifies the inflammatory response. Cytokine & Growth Factor Rev. 2005;16:561–580. doi: 10.1016/j.cytogfr.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Triggiani M, Granata F, Frattini A, Marone G. Activation of human inflammatory cells by secreted phospholipases A2. Biochim Biophys Acta. 2006;1761:1289–1300. doi: 10.1016/j.bbalip.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Clarkson TB. The new conundrum: do estrogens have any cardiovascular benefits? Int J Fertil Womens Med. 2002;47:61–68. [PubMed] [Google Scholar]
- 33.Anthony MS, Clarkson TB. Does extent of pretreatment atherosclerosis influence the effects of conjugated equine estrogens on atherosclerosis progression? J Am Coll Cardiol. 2002;39:248A. [Google Scholar]
- 34.Clarkson TB. Estrogen effects on arteries vary with stage of reproductive life and extent of subclinical atherosclerosis progression. Menopause. 2007;14:373–384. doi: 10.1097/GME.0b013e31803c764d. [DOI] [PubMed] [Google Scholar]
- 35.Dubey RK, Imthurn B, Barton M, Jackson EK. Vascular consequences of menopause and hormone therapy: Importance of timing of treatment and type of estrogen. Cardiovasc Res. 2005;66:295–306. doi: 10.1016/j.cardiores.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Walker SA, Adams MR, Franke AA, Register TC. Effects of dietary soy protein on iliac and carotid artery atherosclerosis and gene expression in male monkeys. Atherosclerosis. 2008;196:106–113. doi: 10.1016/j.atherosclerosis.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losordo DW, Kearney M, Kim EA, Jekanowski J, Isner JM. Variable expression of the estrogen receptor in normal and atherosclerotic coronary arteries of premenopausal women. Circulation. 1994;89:1501–1510. doi: 10.1161/01.cir.89.4.1501. [DOI] [PubMed] [Google Scholar]
- 38.Umetani M, Domoto H, Gormley AK, et al. 27-hydroxycholesterol is an endogenous SERM that inhibits the cardiovascular effects of estrogen. Nature Medicine. 2007;13:1185–1192. doi: 10.1038/nm1641. [DOI] [PubMed] [Google Scholar]
- 39.Evans MJ, Eckert A, Lai K, Adelman SJ, Harnish DC. Reciprocal antagonism between estrogen receptor and NF-kappaB activity in vivo. Circ Res. 2001;89:823–830. doi: 10.1161/hh2101.098543. [DOI] [PubMed] [Google Scholar]
- 40.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Rad Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 41.Bonomini F, Tengattini S, Fabiano A, Bianchi R, Rezzani R. Atherosclerosis and oxidative stress. Histol Histopathol. 2008;23:381–390. doi: 10.14670/HH-23.381. [DOI] [PubMed] [Google Scholar]
- 42.Greenow K, Pearce NJ, Ramji DP. The key role of apolipoprotein E in atherosclerosis. J Mol Med. 2005;83:329–342. doi: 10.1007/s00109-004-0631-3. [DOI] [PubMed] [Google Scholar]
- 43.Dubey RK, Tofovic SP, Jackson EK. Cardiovascular pharmacology of estradiol metabolites. J Pharmacol Exp Therapeut. 2004;308:403–409. doi: 10.1124/jpet.103.058057. [DOI] [PubMed] [Google Scholar]
- 44.Bourghardt J, Bergstrom G, Krettek A, Sjoberg S, Boren J, Tivesten A. The endogenous estradiol metabolite 2-methoxyestradiol reduces atherosclerotic lesion formation in female apolipoprotein E-deficient mice. Endocrinology. 2007;148:4128–4132. doi: 10.1210/en.2007-0259. [DOI] [PubMed] [Google Scholar]
- 45.Rosenson RS, Gelb MH. Secretory phospholipase A2: a multifaceted family of proatherogenic enzymes. Curr Cardiol Rep. 1009;11:445–451. doi: 10.1007/s11886-009-0064-2. [DOI] [PubMed] [Google Scholar]
- 46.Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50:S237–S242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lavi S, Herrmann J, Lavi R, McConnell JP, Lerman LO, Lerman A. Role of lipoprotein-associated phospholipase A2 in atherosclerosis. Curr Atheroscler Rep. 2008;10:230–235. doi: 10.1007/s11883-008-0036-9. [DOI] [PubMed] [Google Scholar]
- 48.Carlquist JF, Muhlestein JB, Anderson JL. Lipoprotein-associated phospholipase A2: a new biomarker for cardiovascular risk assessment and potential therapeutic target. Exper Rev Mol Diagn. 2007;7:511–517. doi: 10.1586/14737159.7.5.511. [DOI] [PubMed] [Google Scholar]
- 49.Wilensky RL, Shi Y, Mohler ER, III, et al. Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nature Med. 2008:1059–1066. doi: 10.1038/nm.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karan D, Holzbeierlein J, Thrasher JB. Macrophage inhibitory cytokine-1: possible bridge molecule of inflammation and prostate cancer. Cancer Res. 2009;69:2–5. doi: 10.1158/0008-5472.CAN-08-1230. [DOI] [PubMed] [Google Scholar]
- 51.Bermudez B, Lopez S, Pacheco YM, et al. Influence of postprandial triglyceride-rich lipoproteins on lipid-mediated gene expression in smooth muscle cells of the human coronary artery. Cardiovasc Res. 2008;79:294–303. doi: 10.1093/cvr/cvn082. [DOI] [PubMed] [Google Scholar]
- 52.George SJ, Dwivedi A. MMPs, cadherins, and cell proliferation. Trends Cardiovasc Med. 2004;14:100–105. doi: 10.1016/j.tcm.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Yan S, Chai H, Wang H, Yang H, Nan B, Yao Q, Chen C. Effects of lysophosphatidylcholine on monolayer cell permeability of human coronary artery endothelial cells. Surgery. 2005;138:464–473. doi: 10.1016/j.surg.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 54.Xu P-F, Zhu K-Y, Jin Y, et al. Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc Natl Acad Sci USA. 2010;107:2521–2526. doi: 10.1073/pnas.0914396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrmann J, Soares SM, Lerman LO, Lerman A. Potential role of the ubiquitin-proteasome system in atherosclerosis. Aspects of a protein quality disease. J Am Coll Cardiol. 2008;51:2003–2010. doi: 10.1016/j.jacc.2008.02.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.