Abstract
Ribonucleotide Reductase M1 (RRM1) is the regulatory subunit of the holoenzyme that catalyzes the conversion of ribonucleotides to 2′-deoxyribonucleotides. Its function is indispensible in cell proliferation and DNA repair. It also serves as a biomarker of therapeutic efficacy of the antimetabolite drug gemcitabine (2′,2′-difluoro-2′-deoxycytidine) in various malignancies. However, a mechanistic explanation remains to be determined. This study investigated how the alkylating agent N-ethylmaleimide (NEM) interacts with the inhibitory activity of gemcitabine on its target protein RRM1 in vivo. We found, when cells were treated with gemcitabine in the presence of NEM, a novel 110 kD band, along with the 90 kD native RRM1 band, appeared in immunoblots. This 110 kD band was identified as RRM1 by mass spectrometry (LC-MS/MS) and represented a conformational change resulting from covalent labeling by gemcitabine. It is specific to gemcitabine/NEM, among 11 other chemotherapy drugs tested. It was also detectable in human tumor xenografts in mice treated with gemcitabine. Among mutations of seven residues essential for RRM1 function, C218A, C429A, and E431A abolished the conformational change, while N427A, C787A, and C790A diminished it. C444A was unique since it was able to alter the conformation even in absence of gemcitabine treatment. We conclude that the thiol alkylator NEM can stabilize the gemcitabine-induced conformational change of RRM1, and this stabilized RRM1 conformation has the potential to serve as a specific biomarker of gemcitabine’s therapeutic efficacy
Keywords: RRM1, gemcitabine, N-ethylmaleimide
INTRODUCTION
Ribonucleotide reductase M1 (RRM1) is one of two subunits in the holoenzyme ribonucleotide reductase (RNR). It is the large subunit harboring the active site and substrate binding site. RNR synthesizes deoxynucleoside diphosphates (dNDP) from nucleoside diphosphates (NDP). It is a ubiquitous and indispensable enzyme for cell proliferation and DNA repair [1; 2]. RRM1 is also involved in other cellular functions, such as migration, invasion, and metastatic spread [3; 4; 5].
Structural investigations of RRM1 revealed that its functional form is a dimmer that contains the two active sites responsible for the reduction of ribonucleotides [1]. An alternative model suggests that the major active form may be a RRM1 hexamer [6]. Five residues in RRM1 are essential for its activity and form the active site, namely, Cys225, Asn437, Cys439, Glu441 and Cys462 in E. Coli [7; 8]. The corresponding mammalian residues are: Cys218, Asn427, Cys429, Glu431 and Cys444. In addition, two C-terminal cysteines (Cys754 and Cys759 in E. Coli, Cys787 and Cys790 in mammals) are important for the rereduction of the active site [9].
RRM1 is a predictive and prognostic biomarker in human malignancies. In advanced stage patients treated with gemcitabine-based regimens, low RRM1 levels were associated with better response or longer survival of cancer patients [10; 11; 12]. However, in early-stage patients treated with surgery alone, high RRM1 levels were positively correlated with longer survival [12; 13; 14]. These reports highlight the importance of RRM1 expression levels for therapeutic decisions in cancer patients.
Gemcitabine (F2dC) is a fluorinated deoxycytidine analog, which has been used clinically to treat a variety of malignancies [15]. Its uniquely potent cytotoxicity is attributed to multiple actions. After entering cells through nucleoside transporters (NT), it is metabolized to its monophosphate form (F2 dCMP), which is further phosphorylated to diphosphate (F2dCDP) and triphosphate (F2dCTP). F2dCDP and F2dCTP are the active cytotoxic metabolites. F2dCDP covalently binds to RRM1 and inhibits the reductase activity, resulting in reduced cellular deoxyribonucleotide (dNTP) pools. The decline in the amount of deoxycytidine triphosphate (dCTP) has a self-potentiating feedback on gemcitabine metabolism, resulting in more intracellular F2dCDP and F2dCTP. F2dCTP inhibits DNA synthesis through direct incorporation into replicating DNA, which precludes elongation, and through inhibition of DNA repair, thereby preventing the removal of incorporated F2dCTP [16; 17]. These collective effects of gemcitabine metabolites make it one of the most potent and effective drugs in the clinic.
Investigations of RNR inhibition by gemcitabine metabolites have been reported [17]. Both, a covalent modification of RRM1 by the sugar moiety of F2dCDP and the destruction of the adenosylcobalamin cofactor have been proposed as being responsible for complete inactivation [18; 19]. A recent study suggested that a tight interaction induced by F2dCDP between the two RNR subunits contributes to a complete inactivation [20].
N-ethylmaleimide (NEM) is a commonly used alkylator to covalently modify the thiol group of a cysteine residue. It is an irreversible inhibitor of cysteine proteases and has been used to investigate protein functions that involve cysteines [21]. For example, NEM can block the phosphorylation of Akt through a protein phosphatase 2A (PP2A) mediated thiol-sensitive redox regulation [22]. NEM also alkylates Cys199 of cAMP-dependent protein kinase A, resulting in its increased dephosphorylation and inactivation [23].
In this report, we show that gemcitabine induces a conformational change of RRM1 in vivo, resulting in a 20 kD molecular shift. NEM is able to stabilize this conformational change. We were able to reveal that residues C218, C429, and E431 are essential for the gemcitabine inhibition of RRM1, while residues N427, C787, and C790 contribute to the inhibition. A C444A mutation is able to mimic the conformational change in absence of gemcitabine treatment. These mechanistic findings provide insight for the development of new drugs that might overcome gemcitabine resistance and for the development of biomarkers specific for gemcitabine efficacy.
MATERIALS AND METHODS
Cell lines and culture conditions
Human non-small-cell lung cancer (H23 and A549), breast cancer (MCF7), and embryonic kidney (HEK293) cell lines were obtained from the American Type Culture Collection (Manassas, VA). They were maintained in RPMI 1640 or DMEM medium supplemented with 10% fetal bovine serum (FBS) (Hyclone) and antibiotics at 37°C and 5% CO2. Authenticity of cell lines was confirmed by DNA finger printing.
Antibodies and reagents
Goat-anti RRM1 and RRM2 were from Santa Cruz Biotechnology. Mouse-anti actin and Flag, and rabbit-anti Flag were from Sigma-Aldrich. Gemcitabine (Eli Lilly) was prepared as 100 mM stock in PBS. NEM (Thermo Fisher) was freshly prepared in ethanol as 1 M stock. The following chemotherapy drugs were used: cytosine arabinoside, hydroxyurea, 5-fluorouracil, doxorubicin, methotrexate, and carboplatin (Sigma-Aldrich); etoposide (Bedford Laboratory); docetaxel (Sanofi-Aventis); cisplatin (Ben Venue Laboratory); pemetrexed (Eli Lilly); vinorelbine (Sicor).
Constructs and transfection
The full-length human RRM1 expression vector was described previously [4]. Seven RRM1 point mutations were created in the same vector (pCMV-Tag2) using the QuickChange® Site-Directed Mutagenesis Kit (Stratagene) or two-step PCR. The wild-type and mutation plasmids were introduced into HEK293 cells using LipofectAMINE-2000 (Invitrogen).
Westernblotting and immunoprecipitation
Cells were lysed with buffer (25 mM Tris-HCl pH 7.5, 0.5% NP-40, 150 mM NaCl) with a complete protease inhibitor cocktail (Roche), with or without 10 mM of NEM on ice for 10 min. The lysis buffer contained 1 mM of orthovanadate and 1 mM DTT as indicated. Cell lysates were cleared by centrifugation at 14,000 g, 10 min, 4°C. The resulting supernatants were used for immunoprecipitation or Western blotting. For immunoprecipitation, 1 ml of lysate was incubated with 4 μg of anti-RRM1 and 30 μl 50% slurry of immortalized protein-G (PIERCE) for 2 h, with rocking at 4°C. The immunocomplexes were then precipitated at 5,000 g, washed three times with 1 ml lysis buffer, and resuspended in 1 × SDS sample buffer (63 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 15 mM EDTA). For Western blotting, protein concentrations were determined with a protein assay kit (Bio-Rad), cell lysates were mixed with 4 × SDS sample buffer, and boiled (unless indicated) at 95°C for 5 min. Forty micrograms of total cell lysates were subject to SDS-PAGE gel separation and transferred to a nitrocellulose membrane (0.45 μm, Bio-Rad). The membrane was blocked with 25 mM Tris-HCl pH 7.5, 0.05% Tween-20, 100 mM NaCl, 5% (w/v) skim milk and incubated with primary antibody followed by secondary antibody. Target protein bands were visualized on X–ray film using SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific).
Gel staining and LC-MS/MS for protein identification
Immunoprecipitated RRM1 from treated or untreated cells was separated with SDS-PAGE gels. Bands were visualized with Bio-safe Coomassie G-250 staining (Bio-rad). Target proteins were excised, washed, and digested overnight with modified sequencing grade trypsin (Promega, Madison, WI). Following in-gel tryptic digestion, peptides were extracted and concentrated. A nanoflow liquid chromatograph (Easy-nLC, Proxeon, Odense, Denmark) coupled to an electrospray ion trap mass spectrometer (LTQ, Thermo, San Jose, CA) was used for tandem mass spectrometry peptide sequencing. Five tandem mass spectra were collected in a data-dependent manner following each survey scan. Sequences were assigned using Mascot (www.matrixscience.com) searches against human Swiss Prot entries. Results from Mascot were compiled in Scaffold, which was used for manual inspection of peptide assignments and protein identifications.
Xenografts and mice
Four week-old, nude, athymic CD1 nu/nu mice (Charles River) were subcutaneously injected with 5 × 106 H23 cells. They were injected intraperitonealy with 200 μg of gemcitabine after xenografts were established. Sixteen hours later, mice were euthanized and tumors collected. Tumor lysates were prepared by homogenization in the specified lysis buffer.
RESULTS AND DISCUSSION
Gemcitabine induces a 20 kD shift of RRM1
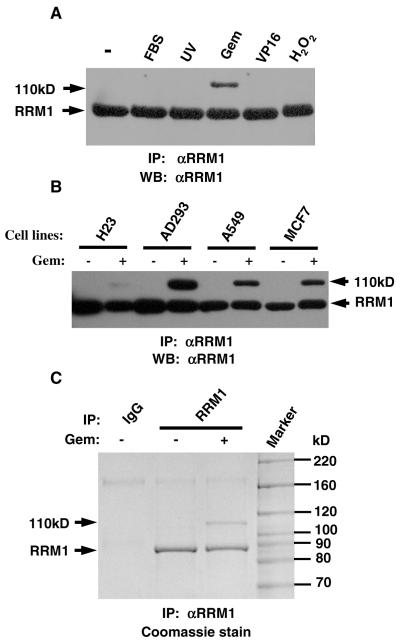
Endogenous RRM1 was immunoprecipitated with anti-RRM1 from HEK293 cells treated with different stimuli; namely, FBS, UV light, gemcitabine, etoposide, and hydrogen peroxide. Western blots showed, in addition to the expected 90 kD RRM1, an extra 110 kD band that was only present in gemcitabine treated samples (Fig. 1A). The same 110 kD band was detected in cell lines H23, A549, and MCF7 (Fig. 1B). Coomassie staining of SDS-PAGE gels also showed this band (Fig. 1C). They were identified as RRM1 by LC-MS/MS indicating that gemcitabine treatment results in a 20 kD molecular shift of RRM1.
Fig. 1.
Gemcitabine induces a 110 kD RRM1 band. (A) HEK293 cells were treated with 10% FBS, 50 J/m2 UV, 1 μM gemcitabine (Gem); 100 μM etoposide (VP16), or 300 μM H2O2. RRM1 was immunoprecipitated (IP) and Western blotted (WB) with the same antibody. (B) Four cell lines were treated with 1 μM gemcitabine. (C) HEK293 cells were treated with 1 μM of gemcitabine. Immunoprecipitated RRM1 was separated in a SDS-PAGE gel and stained with Coomassie blue. Normal goat IgG was used as a control.
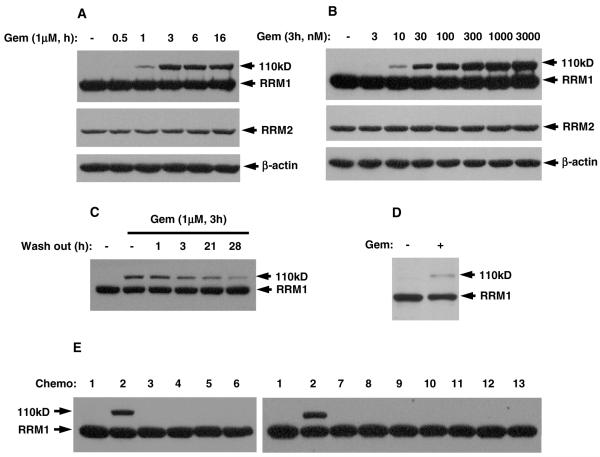
We studied the time course and dose effect of gemcitabine treatment using cell lysates. The 110 kD band could be detected as early as 1 h after 1 μM gemcitabine treatment (Fig. 2A), peaking at 3 h. It appeared at 10 nM concentrations and peaked at 3 μM of gemcitabine (Fig. 2B). Removal of gemcitabine from the growth medium led to progressive attenuation of the 110 kD band (Fig. 2C) after 3 h. H23 xenografts in mice treated with gemcitabine overnight also displayed the 110 kD RRM1 band (Fig. 2D), confirming that the molecular shift occurs in vivo. RRM2 did not show any molecular shift under the same conditions, indicating that the gemcitabine-induced shift is specific for RRM1.
Fig. 2.
Gemcitabine specifically induces 110 kD RRM1. RRM1, RRM2, and β-actin were detected in the cell lysates by Western blotting. (A) HEK293 cells were treated with 1 μM gemcitabine for the indicated time. (B) HEK293 cells were treated with the indicated doses of gemcitabine. (C) HEK293 cells were treated with 1 μM gemcitabine for 3 h, washed, and grown without gemcitabine for the indicated times. (D) Nude mice bearing H23 tumor xenograft treated with or without gemcitabine. (E) HEK293 cells were treated with drugs for 3 h: 1 – DMSO control; 2 – gemcitabine, 1 μM; 3 – cytosine arabinoside (Ara-C), 100 μM; 4 – hydroxyurea, 10 mM; 5 – 5-fluorouracil , 100 μM; 6 – doxorubicin, 10 μM; 7 – etoposide (VP16), 1 μM; 8 – docetaxel, 100 nM; 9 – carboplatin, 100 μM; 10 – cisplatin, 10 μM; 11 – methotrexate, 1 μM; 12 – pemetrexed, 1 μM; 13 – vinorelbine, 100 nM.
We treated cells with 11 other clinically relevant chemotherapeutic drugs, including cytosine arabinoside, hydroxyurea, 5-fluorouracil, doxorubicin, methotrexate, carboplatin, etoposide, docetaxel, cisplatin, pemetrexed, and vinorelbine, and found that only gemcitabine could induce the 110 kD RRM1 (Fig. 2E). Surprisingly, cytosine arabinoside did not result in formation of the 110 kD band, suggesting a highly specific relationship between gemcitabine and the 110 kD band.
During our investigations, other laboratories reported that RNR recombinant proteins of E. Coli [19], H. Sapiens [24], and L. Leichmannii [25], when reacted with F2dCDP in vitro, showed a molecular shift in SDS-PAGE gels under non-boiling conditions only. Our in vivo cell line and animal data are consistent with these in vitro results; however, since our results were obtained under denaturing condition with boiling, the gemcitabine-induced RRM1 modification may be more stable under in vivo conditions, and experimental conditions may also contribute to this observation.
The 20 kD shift of RRM1 is stabilized by N- ethylmaleimide (NEM)
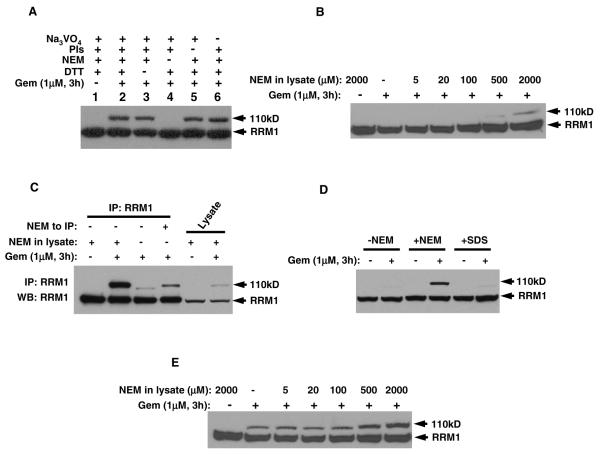
We investigated conditions that stabilize the gemcitabine-induced RRM1 shift in vivo. The lysate buffer contained components intended to preserve potential protein modifications. When using a series of lysis buffers with selectively omitted components, we found that only NEM was required to show the 110 kD RRM1 band (Fig. 3A). The minimum required NEM dose to achieve the shift was of 0.5 mM (Fig. 3B).
Fig. 3.
N-ethylmaleimide (NEM) is required to induce the 110 kD RRM1. (A) Western blots of HEK293 cells treated with or without 1 μM gemcitabine. Cell lysis buffers contained the indicated components. (B) The indicated doses of NEM were used in the lysis buffer. (C) HEK293 cell lysates prepared with or without NEM were immunoprecipitated with anti-RRM1. The non-NEM lysate immunocomplex was then treated with NEM. (D) Cells treated with gemcitabine were lysed with or without NEM or directly lysed in SDS sample buffer. (E) The experiment was carried out as in (B), except that samples were not boiled prior to gel loading.
Since NEM is a thiol-modifying agent that has been used for inhibition of cysteine proteases, we studied whether the effect is the result of enzymatic inhibition during lysate preparation rather than a direct interaction with RRM1. To address this, we prepared cell lysates directly in SDS buffer after gemcitabine treatment to suppress any enzymatic activity during lysate preparation. Under these conditions, a 110 kD band was undetectable, leading us to conclude that NEM exerts its effect independent of its protease inhibitory capability (Fig. 3D). When immunoprecipitated RRM1 from gemcitabine-treated cells was resuspended in the NEM containing buffer, the shifted RRM1 protein could also be detected (Fig. 3C), indicating that NEM directly impacts on gemcitabine-treated RRM1.
We examined NEMs influence under non-boiling conditions. Cell lysates from gemcitabine treated cells displayed a molecular shift in the absence of NEM (Fig. 3E). However, the amount of the shifted band increased with increasing amounts of NEM (Fig. 3E), similar to the dose response observed under boiling conditions (Fig. 3B). NEM alone without gemcitabine treatment could not induce a molecular shift of RRM1. These data suggest that NEM directly modifies gemcitabine-conjugated RRM1 and stabilizes this interaction.
We investigated if protein modifications, such as ubiquitination, sumoylation, or neddylation could account for the described 20 kD band shift, and we found no evidence for this using Western blotting and mass spectroscopy (data not shown).
It has been reported that F2CDP covalently links to the RNR substrate binding site [2] resulting in a conformational change that is sufficient to cause a 20 kD band shift in vitro [24]. Our results demonstrate this also happens to endogenous RNR in cells treated with gemcitabine at doses as low as 5 nM. This conformational change can be stabilized by NEM, resulting in a shifted RRM1 even under boiling conditions.
RRM1 mutants respond to Gemcitabine/NEM treatment
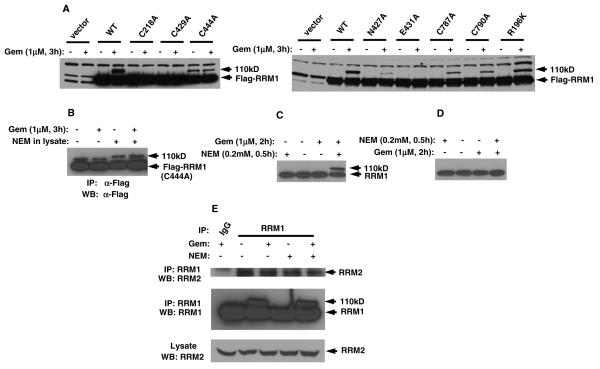
The detectable 110 kD of RRM1 stabilized by NEM is indicative of a gemcitabine effect on RRM1. To understand how endogenous RRM1 is modified by gemcitabine and further stabilized by NEM, we asked which residues are essential for the appearance of the 110 kD RRM1. There are seven amino acids that are reported to be critical for RRM1 activity. They are Cys218, Asn427, Cys429, Glu431, and Cys444 from the active site and Cys787 and Cys790 from the C-terminal. We mutated these residues to alanine with a Flag-tag at the N-terminal. We also mutated an arbitrary residue outside of the active site, R196, as a negative control. The mutants were transfected into HEK293 cells, and lysates were prepared with buffer containing NEM. Western blotting was carried out with rabbit-anti Flag. Results showed that, using boiling conditions, mutants C218A, C429A, and E431A had no detectable 110 kD signal, and mutants N427A, C787A, and C790A had a diminished 110 kD signal. Surprisingly, although the band intensity was low, mutant C444A showed a 110 kD band even without gemcitabine treatment. By contrast, the control mutant R196K did not show any changes compared to wildtype RRM1 (Fig. 4A). This was confirmed by IP with and without NEM in the lysis buffer (Fig. 4B).
Fig. 4.
RRM1 mutations and NEM treatment of cells. (A) HEK293 cells with the indicated RRM1 mutations were treated with 1 μM of gemcitabine and lysates were prepared with NEM. Transfected RRM1 was visualized in Western blots using anti-Flag antibody. (B) C444A-mutant RRM1, transfected into HEK293 cells, was immunoprecipitated with mouse-anti Flag antibody. Western blot was carried out using rabbit-anti Flag antibody. (C) H23 cells were treated with 1 μM of gemcitabine for 2 h followed by 0.2 mM NEM for 0.5 h. Cell lysates were prepared without NEM. (D) H23 cells were treated with NEM for 0.5 h followed by 2 h with gemcitabine. (E) HEK293 cells were treated with or without 1 μM of gemcitabine for 3 h. Endogenous RRM1 was immunoprecipitated with anti-RRM1 with NEM in the lysis buffer. RRM2 was detected in immunoprecipitates and lysates.
These results indicate that Cys218, Cys429, and Glu431 are essential for the conformational change induced by gemcitabine. Wang [24] and Artin [19] proposed that it is the covalent modification of Cys218, Cys429, Glu431, Cys787, and Cys790, but not Cys444, by F2CDP that causes the conformational change, resulting in the molecular shift in vitro, which is consistent with our in vivo results. The amount of covalently bound sugar corresponds roughly to the proportion of band shifting, and the absence of the shifted band in a mutant is indicative that the site is covalently modified. Our result indicating that cytosine arabinoside does not result in a 110 kD RRM1 band, is consistent with the proposed covalent binding of gemcitabine to RRM1 through its ribose sugar moiety.
Consistent with these published data, our in vivo results confirmed that Cys218, Glu431, and Cys429 are essential to the covalent interaction with F2CDP and that the C-terminal Cys787 and Cys790 are partially involved. In addition, we found that another residue in the active site, Asn427, is also essential to the interaction. Moreover, Cys444 is unique since its alanine substitution showed a shifted band even without gemcitabine treatment, suggesting this mutation has the same conformation as F2CDP-labled wildtype RRM1. This highlights the importance of Cys444 to the activity of RRM1 and explains the results by Wang [24] mentioned above. Although it is known that Cys218 and Cys444 form a disulfide bond to become cystine, which is essential to the normal function of RRM1 [26], the in vitro and our in vivo data demonstrated that these two cysteines play different roles with regard to maintaining the functional conformation of RRM1.
The second pair of cysteines that form cystine is Cys787 and Cys790. It can serve to rereduce the Cys218/Cys444 disulfide to complete the turnover of active sites in cells [9]. The identical and weaker intensity of the molecular shift from the mutation of Cys787 and Cys790 further implies this pair of cysteines is indeed not in the essential active site, but it is still involved in RRM1’s function and inactivation by gemcitabine.
Our in vivo data and the in vitro work by others suggest that the altered conformation is likely due to the formation of an intramolecular cross-link within RRM1, rather than an intermolecular cross-link between RRM1 subunits. The cross-link that causes the conformational change is likely formed between C218 and C787 or C218 and C790 [24]. Our data further suggest that F2CDP-labeling of C218 may lead to the disruption of disulfide bonding between C218 and C444, which may facilitate the formation of cross-links between C218 and C787 or C790; i.e., the altered conformation that results in the 110 kD RRM1. The mutation of C444 to alanine may mimic the disruption of a disulfide bond between C218 and C444. The roles of E431 and N427, both essential to this cross-link formation, remain unclear.
NEM also works in cells
To explore if NEM has any effect in vivo on the gemcitabine-RRM1 interaction, we studied if treatment of cells with gemcitabine and NEM would show a conformational change of RRM1. H23 cells were treated with 1 μM gemcitabine for 2.5 h followed by 0.2 mM NEM for 30 min. The cells were lysed in buffer without NEM. Western blots showed the 110 kD RRM1, indicating that NEM also modified the conformational change of RRM1 induced by gemcitabine in vivo (Fig. 4C). However, when we switched the treatment sequence; i.e. NEM followed by gemcitabine, the shifted RRM1 was not detectable (Fig. 4D). This implies that the NEM-modified RRM1 is no longer a target of gemcitabine, further confirming that cysteine residues are the targets of gemcitabine modification. It also implies that NEM, as a thiol-alkylator of cysteine, may release C444 and facilitate the cross-link between C218 and one of the cysteines in the C-terminus (C787 and C790). This may explain why NEM can stabilize the conformational change induced by gemcitabine.
Finally, we investigated if NEM and gemcitabine could affect the RRM1/RRM2 interaction in cells. Cells were treated with a combination of gemcitabine and NEM (1 μM gemcitabine for 2.5 h, followed by 0.2 mM NEM for 30 min). RRM1 was immunoprecipitated in the presence of 0.5 mM ATP in the lysis buffer [20]. Western blots showed that RRM2 co-immunoprecipitated under all conditions (Fig. 4E), implying that the modification in the active sites of RRM1 by gemcitabine or NEM does not affect the formation of the holoenzyme.
We sought to investigate potential synergism between NEM and gemcitabine cytotoxicity. However, due to the overwhelming non-specific cytotoxicity of NEM at concentrations that stabilize the gemcitabine-induced RRM1 conformation, we were unable to obtain meaningful results using cell proliferation-based assays.
Taken together, our findings confirm that gemcitabine covalently modifies RRM1 in the active site in cell line and animal models. This modification leads to a conformational change that can be stabilized by the thiol alkylator NEM. Site-directed mutagenesis revealed Cys218, Cys429, and Glu431 as essential for the conformational change. C444A was unique in that it caused the conformational change without gemcitabine induction. These results suggest that the 110 kD RRM1 may be of utility in the development of drugs that target RRM1 and for monitoring of gemcitabine efficacy.
Contextual Overview/Highlights.
Gemcitabine induces a RRM1 conformational change in tumor cell lines and xenografts The 110 kD RRM1 is unique to gemcitabine interaction among 12 cytotoxic agents The 110 kD RRM1 can be stabilized by the thiol alkylator N-ethylmaleimide C218A, C429A, and E431A mutations in RRM1 abolished the conformational change The 110 kD RRM1 may be specific biomarker of gemcitabine’s therapeutic efficacy
ACKNOWLEDGEMENTS
This work was supported by a grant (NCI-CA129343) from the National Cancer Institute
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- [2].Stubbe J, van der Donk WA. Ribonucleotide reductases: radical enzymes with suicidal tendencies. Chem Biol. 1995;2:793–801. doi: 10.1016/1074-5521(95)90084-5. [DOI] [PubMed] [Google Scholar]
- [3].Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci USA. 1997;94:13181–13186. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- [5].Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- [6].Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- [7].Persson AL, Eriksson M, Katterle B, Potsch S, Sahlin M, Sjoberg BM. A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase. J Biol Chem. 1997;272:31533–41. doi: 10.1074/jbc.272.50.31533. [DOI] [PubMed] [Google Scholar]
- [8].Kasrayan A, Persson AL, Sahlin M, Sjoberg BM. The conserved active site asparagine in class I ribonucleotide reductase is essential for catalysis. J Biol Chem. 2002;277:5749–55. doi: 10.1074/jbc.M106538200. [DOI] [PubMed] [Google Scholar]
- [9].Mao SS, Holler TP, Yu GX, Bollinger JM, Booker S, Johnston MI, Stubbe J. A model for the role of multiple cysteine residues involved in ribonucleotide reducation: amazing and still confusing. Biochemistry. 1992;31:9733–43. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- [10].Rosell R, Danenberg K, Alberola V, Bepler G, Sanchez JJ, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A. Ribonucleotide reductase mRNA expression and survival in gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- [11].Bepler G, Kusmartseva I, Sharma S, Gautam A, Cantor A, Sharma A, Simon G. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–7. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- [12].Akita H, Zheng Z, Takeda Y, Chiwan K, Kittaka N, Kobayashi S, Marubashi S, Takemasa I, Nagano H, Dono K, Nkamori S, Monden M, Mori M, Doki Y, Bepler G. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. 2009;28:2903–2909. doi: 10.1038/onc.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bepler G, Sharma S, Cantor A, Gautam A, Haura E, Simon G, Sharma A, Sommers E, Robinson L. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- [14].Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. The DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- [15].Danesi R, Altavilla G, Giovannetti E, Rosell R. Pharmacogenomics of gemcitabine in non-small-cell lung cancer and other solid tumors. Pharmacogenomics. 2009;10:69–80. doi: 10.2217/14622416.10.1.69. [DOI] [PubMed] [Google Scholar]
- [16].Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanism of action. Semin Oncol. 1996;23:3–15. [PubMed] [Google Scholar]
- [17].Pereira S, Fernandez PA, Ramos MJ. Mechanism for ribonucleotide reductase inactivation by the anticancer drug gemcitabine. J Comput Chem. 2004;25:1286–94. doi: 10.1002/jcc.20054. [DOI] [PubMed] [Google Scholar]
- [18].Lohman GJ, Stubbe J. Inactivation of Lactobacillus leichmannii ribonucleotide reductase by 2′,2′-difluoro-2′-deoxycytidine 5′-triphosphate: covalent modification. Biochemistry. 2010;49:1404–17. doi: 10.1021/bi902132u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Artin E, Wang J, Lohman GJ, Yokoyama K, Yu G, Griffin RG, Bar G, Stubbe J. Insight into the mechanism of inactivation of ribonucleotide reductase by gemcitabine 5′-diphosphate in the presence or absence of reductant. Biochemistry. 2009;48:11622–9. doi: 10.1021/bi901590q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibits ribonucleotide reductases. Proc Natl Acad Sci U S A. 2007;104:14324–9. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Smyth DG, Blumenfeld OO, Konigsberg W. Reactions of N-ethylmaleimide with peptides and amino acids. Biochem J. 1964;91:589–95. doi: 10.1042/bj0910589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yellaturu CR, Bhanoori M, Neeli I, Rao GN. N-Ethylmaleimide inhibits platelet-derived growth factor BB-stimulated Akt phosphorylation via activation of protein phosphatase 2A. J Biol Chem. 2002;277:40148–55. doi: 10.1074/jbc.M206376200. [DOI] [PubMed] [Google Scholar]
- [23].Humphries KM, Deal MS, Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–8. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- [24].Wang J, Lohman GJ, Stubbe J. Mechanism of inactivation of human ribonucleotide reductase with p53R2 by gemcitabine 5′-diphosphate. Biochemistry. 2009;48:11612–21. doi: 10.1021/bi901588z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lohman GJ, Gerfen GJ, Stubbe J. Inactivation of Lactobacillus leichmannii ribonucleotide reductase by 2′,2′-difluoro-2′-deoxycytidine 5′-triphosphate: adenosylcobalamin destruction and formation of a neucleotide-based radical. Biochemistry. 2010;49:1396–403. doi: 10.1021/bi9021318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Erickson HK. Formation of the cystine between cysteine 225 and cysteine 462 from ribonucleoside diphosphate reductase is kinetically competent. Biochemistry. 2000;39:9241–50. doi: 10.1021/bi992820y. [DOI] [PubMed] [Google Scholar]