Abstract
Genetic variation among females is likely to influence the outcome of both pre- and post-copulatory sexual selection in Drosophila melanogaster. Here we use association testing to survey natural variation in 10 candidate female genes for their effects on female reproduction. Females from 91 chromosome 2 substitution lines were scored for phenotypes affecting pre- and post-copulatory sexual selection such as mating and remating rate, propensity to use sperm from the second male to mate, and measures of fertility. There were significant genetic contributions to phenotypic variation for all the traits measured. Resequencing of the 10 candidate genes in the 91 lines yielded 68 nonsynonymous polymorphisms which were tested for associations with the measured phenotypes. Twelve significant associations (markerwise P < 0.01) were identified. Polymorphisms in the putative serine protease homolog CG9897 and the putative odorant binding protein CG11797 associated with female propensity to remate and met an experimentwise significance of P < 0.05. Several other associations, including those impacting both fertility and female remating rate suggest that sperm storage might be an important factor mitigating female influence on sexual selection.
Keywords: Sperm competition, association testing, female mating, sexual selection, genotype-phenotype
Introduction
Sexual selection can be a complex interplay of male and female influence on traits such as mating, sperm utilization and allocation of resources to current versus future matings (Zeh & Zeh 2003). The disparity between the reproductive interests of males and females can potentially lead to sexual conflict as each sex attempts to maximize its own reproductive fitness at the potential cost to members of the opposite sex (Chapman et al. 2003a; Parker 2006). Understanding how natural selection might be affecting the evolution of phenotypes influencing sexual selection requires a detailed understanding of patterns of phenotypic variation in natural populations and ideally, knowledge of the genetic polymorphisms underlying the observed variation.
In Drosophila melanogaster, females frequently mate with and store the sperm from multiple males (Milkman & Zeitler 1974; Prout & Bundgaard 1977; Harshman & Clark 1998; Imhof et al. 1998) establishing the opportunity for both pre- and post-copulatory sexual selection to operate. The male perspective and his influence on traits such as induced fidelity and sperm competition has been studied in some detail. We know that a variety of male seminal fluid proteins play a large role in mediating traits affecting sexual selection (Ravi Ram et al. 2005; Wolfner 2009) including sperm storage (Wong et al. 2008), female fidelity (Chapman et al. 2003b; Liu & Kubli 2003; Ram & Wolfner 2009), female egg laying (Herndon & Wolfner 1995) and egg hatching rates (Chapman et al. 2001). Furthermore, natural variation in these genes is associated with sperm competition phenotypes (Clark et al. 1995; Fiumera et al. 2005, 2006, 2007) suggesting that sexual selection might be driving the non-neutral patterns of evolution observed in many male reproductive genes (Clark et al. 2006).
Females are also active participants in pre- and post-copulatory sexual selection and thus can be contributing to the variation present in natural populations. Male Drosophila melanogaster cannot force copulations on adult females, and several factors are suspected to influence female mating and remating rates. It is well known that the number of sperm in storage has a large effect on female remating rate, the so called ‘sperm effect’ (Manning 1962). This is influenced by the male (Chapman et al. 2003b; Liu & Kubli 2003) but could easily be affected by the female as well. For example, sex peptide is gradually cleaved from sperm within the female storage organs, allowing for long-term remating suppression (Peng et al. 2005) and female variation in the storage environment might impact the duration of this effect. Females are capable of dumping sperm from storage, which could also play a role in regulating remating rates and sperm utilization (Snook & Hosken 2004; Manier et al. 2010). The physical environment also impacts remating rates. Females with reduced access to food are less likely to remate, although this effect disappears once their stored sperm are depleted (Harshman et al. 1988). Nutrition also impacts remating rate via differential response to sex peptide (Fricke et al. 2009). Density appears to affect female remating rate as well (Gromko & Gerhart 1984; Marks et al. 1988), although this effect may depend on the lines that are surveyed (Harshman et al. 1988). The perceived attractiveness of the males can also influence mating and remating rates (Jones & Ratterman 2009); females prefer to mate with larger males (Ewing 1961; Wilkinson 1987; Taylor & Kekic 1988; Pitnick 1991; Pitnick & Garcia-Gonzalez 2002; Friberg & Arnqvist 2003) and this may be driven to some degree by environmental factors (Zhang et al. 2008). Chemical communication through cuticular hydrocarbons and olfactory receptors has been shown to be important for species recognition (Billeter et al. 2009). Although their effects within species are still debated (Takahashi & Ting 2004; Coyne & Elwyn 2006; Greenberg et al. 2006), they could be influencing female mate choice in this species.
Currently we have only a cursory characterization of patterns of genetic variation for the female role in pre- and post-copulatory sexual selection in D. melanogaster. Clark and Begun (1998) showed that female genotype affects both remating frequency and also the propensity of doubly mated females to use sperm from either the first or second male. Clark et al. (1999) then demonstrated a strong interaction between the male and female genotype impacting sperm utilization. Females are known to vary in their ability to resist the cost of mating and much of this variation is due to differences in mating rates (Wigby & Chapman 2004; Linder & Rice 2005). Several genes such as sarah (Ejima et al. 2004), muscleblind (Juni & Yamamoto 2009), dissatisfaction (Finley et al. 1998), lozenge (Fuyama 1995), and insulin signaling genes (Wigby et al. 2010), among others, are known to impact female mating. Recently, the receptor for sex peptide, CG16752, has been identified and shown to affect female receptivity (Yapici et al. 2008), and potentially mediate male × female mating interactions (Chow et al. 2010). Furthermore, QTL mapping has identified additional regions of the genome that might be influential (Lawniczak & Begun 2005). Using an evolutionary EST approach, studies have identified novel genes expressed in the female reproductive tract (Swanson et al. 2004; Kelleher et al. 2007; Prokupek et al. 2008; Kelleher & Pennington 2009; Prokupek et al. 2009). Several studies have also used microarray data to compare gene expression in D. melanogaster females in response to some aspect of mating such as the transfer of sperm, seminal fluid proteins, and/or courtship (Lawniczak & Begun 2004; McGraw et al. 2004; Mack et al. 2006; McGraw et al. 2008). Overall, we now have a sufficiently strong set of candidate genes to directly investigate the impact of polymorphism on variation among female reproductive fitness components.
Here, we characterized females from 91 second chromosome extraction lines for traits influencing sexual selection, including mating and remating rates, fertility and sperm utilization. We sequenced 10 candidate genes which have been shown to change in expression level due to some aspect of mating (McGraw et al. 2004) or are expressed in the female reproductive tract and are likely under positive selection (Swanson et al. 2004). In particular, we targeted odorant binding proteins, based on an a priori assumption that they would affect mating decisions. We then tested for associations between genotype and phenotype. We identified several associations between nonsynonymous polymorphisms and phenotypes affecting the female role in sexual selection. Two of the associations, both influencing female remating rate, met an experimentwise P-value < 0.05.
Materials and methods
Scoring phenotypes
Females were selected from 91 chromosome 2 substitution lines originating from a natural Drosophila melanogaster population in State College, Pennsylvania (Lazzaro et al. 2004). Each homozygous line has a unique second chromosome from nature but they have identical first, third, and fourth chromosomes. The first males to mate were Oregon-R (wild type red eyes) and the second males had a brown dominant (bwD) eye color mutation. All fly stocks were maintained at medium density on standard agar-dextrose-yeast media at room temperature (~22°C) on a 12 hour light/dark cycle with partially overlapping generations.
Virgin males and females were collected over CO2, and maintained in single sex vials of five flies until 4–7 days old. Single pair matings of a female from a chromosome extraction line and a single Oregon-R male were set up on day 1 in vial 1 (V1) starting at 0800 hours. Vials were observed for mating at no greater than 15 minute intervals until 1230 hours and the mating times were recorded to the nearest 15 minutes. Males were removed quickly after mating was completed in order to prevent additional copulations. Vials in which no mating was observed were left to mate unobserved until 1900 hours, at which time males were removed. On day 3, females were tapped into vial 2 (V2) with a pair of virgin bwD males starting at 0800 hours. Matings were observed as described above and all females were tapped to vial 3 (V3) at 1900 hours. On day 8, females were transferred to vial 4 (V4) and then discarded on day 12. Paternity was scored via progeny eye color approximately 16 days after the female was removed from each vial, such that all progeny had an opportunity to eclose. For each of the 91 lines, 10 replicate females were scored in each of two generations (blocks). Only those females that survived the entire experiment were included in the analyses.
The following phenotypes were analyzed: the proportion of virgin females mating within 30 minutes (mated-30), the proportion of females that remated (remated), female fertility in vial 1 (fertility-V1) using only those females that ultimately mated to both males, overall fertility of doubly mated females (fertility), and sperm utilization measured as the proportion of offspring sired by the second male to mate (P2). Analysis of variance (ANOVA) was used to test for significance of fertility-V1, fertility and P2 between lines (these phenotypes approximated normal distributions). Permutation tests based on chi-squared statistics were used to test for significance of mated-30 and remated (Fiumera et al. 2005). Line means were then used for association testing as described below.
Genotyping
PCR amplicons of 10 candidate genes from each line were sequenced (CG4847, CG5395, CG6641, CG8965, CG9820, CG9897, CG10363, CG11797, CG13873, CG13939, Table 1). These genes were chosen either because their regulation was altered by some aspect of mating (McGraw et al. 2004), or they are expressed in the female reproductive tract and are likely under positive selection (Swanson et al. 2004). We focused specifically on odorant binding proteins, under the a priori assumption that they would be relevant to mating decisions. PCR and sequencing primers were designed using Primer3 (Rozen & Skaletsky 2000) and we attempted to include ~1 kb upstream and downstream of the coding region. All sequencing was completed using Applied Biosystems Automated 3730 DNA Analyzer with Big Dye Terminator chemistry according to manufacturers’ protocols. Sequences were clipped to maximize regions with error rates below 0.01 using the program CodonCode Aligner (www.codoncode.com), and were then assembled by line into contigs. All sequences with fewer than 25 bases were discarded, as were sequences with fewer than 50 Phred20 bases. Contigs were aligned to reference sequences obtained from flybase.org. Polymorphism tables were exported from CodonCode Aligner.
Table 1.
Summary of the genes analyzed. The putative function according to FlyBase, the number of nonsynonymous polymorphisms (dN), the number of synonymous polymorphisms (dS) and the number of noncoding polymorphisms (noncoding) that were identified.
| Gene | Putative Function | dS | dN | noncoding |
|---|---|---|---|---|
| CG4847c | endopeptidase | 15 | 4 | 97 |
| CG5395c | ATPase activity | 8 | 3 | 32 |
| CG6641a | pheromone binding | 4 | 4 | 47 |
| CG8965c | unknown | 33 | 4 | 115 |
| CG9820a | olfactory receptor | 22 | 10 | 81 |
| CG9897b | endopeptidase | 17 | 14 | 103 |
| CG10363b | peptidase inhibitor | 26 | 20 | 44 |
| CG11797a | odorant binding | 6 | 4 | 80 |
| CG13873a | odorant binding | 8 | 4 | 100 |
| CG13939a | odorant binding | 18 | 1 | 47 |
| Total | 157 | 68 | 746 | |
Genes were identified from McGraw et al. 2004a, Swanson et al. 2004b or bothc
Association testing
Association tests between genotype and phenotype were conducted in MATLAB using permutation tests based on simple linear regression. We tested the effects of nonsynonymous polymorphisms, synonymous polymorphisms and non-coding polymorphisms independently. For each phenotype, we used 100,000 permutations based on the F-value from the linear model to generate both markerwise and experimentwise P-values (Churchill & Doerge 1994). Experimentwise P-values may represent a conservative statistic, as they compare observed P-values to permuted values across all polymorphisms. Markerwise P-values are less stringent, and do not adequately control for the large number of tests completed. In order to correct for this, we also applied a False Discovery Rate calculation (FDR) to the markerwise P-values to control for false positives (Storey & Tibshirani 2003).
Results
A total of 1686 females were scored for propensity to mate within 30 minutes and propensity to remate. 1554 of these females mated to a second male in vial 2, and were scored for fertility in vial 1, overall fertility, and sperm utilization. There were highly significant line effects (P < 0.005) for all the traits scored (Table 2) but no significant effect of block (not shown). Fertility in vial 1 (fertility-V1) averaged 20.7 offspring and line means ranged from 1.8 to 39.5 offspring, while overall fertility averaged 85.8 offspring and lines means ranged from 39.4 to 137.8 offspring. Overall, approximately 56% of virgin females mated within 30 minutes but line means ranged from 6% up to 88%. On average, 48% of the females ultimately remated to the second male and line means ranged from 7% to 94%. Among those females that doubly mated, approximately 56% of the offspring were sired by the second male to mate and line means ranged from 8% to 89%. Fertility and fertility-V1 were strongly positively correlated (r = 0.681, P < 0.001) which is not surprising given one is a subset of the other. In addition, P2 and fertility were negatively correlated (r = −0.22, P = 0.039). We further analyzed this finding by decomposing fertility into fertility in the first and second vials (fertility-V1V2) and fertility in the third and fourth vials (fertility-V3V4), such that fertility-V1V2 corresponds almost entirely to progeny of the first male, and fertility-V3V4 corresponds to progeny from both the first and second male. Fertility-V1V2 and fertility-V3V4 were positively correlated (r = 0.576, P < 0.001). Fertility-V3V4 was strongly negatively correlated with P2 (r = −0.272, P = 0.009) but fertility-V1V2 was not correlated with P2 (r = −0.040, P = 0.708). Females that tended to use the second male’s sperm also tended to have reduced overall fertility and this was due to reduced fertility after the second mating.
Table 2.
Summary of phenotypic distributions.
| Mean of Line Means |
Range of Line Means |
Standard Error for Line Means |
|||
|---|---|---|---|---|---|
| Phenotype | Statistic | P-value | |||
| Fertility-V1 | 20.66 | 1.8 – 39.5 | 0.87 | F90,549 = 3.32 | P <0.0001 |
| Fertility-overall | 85.76 | 34.9 – 137.8 | 2.69 | F90,549 = 3.27 | P < 0.0001 |
| Mated-30 | 0.56 | 0.06 – 0.88 | 0.02 | X2 = 91.42 | P < 0.0001a |
| Remating | 0.48 | 0.07 – 0.94 | 0.02 | X2 = 99.75 | P < 0.0001a |
| P2 | 0.56 | 0.08 – 0.89 | 0.02 | F90,549 = 1.48 | P = 0.005 |
P-value determined via permutation
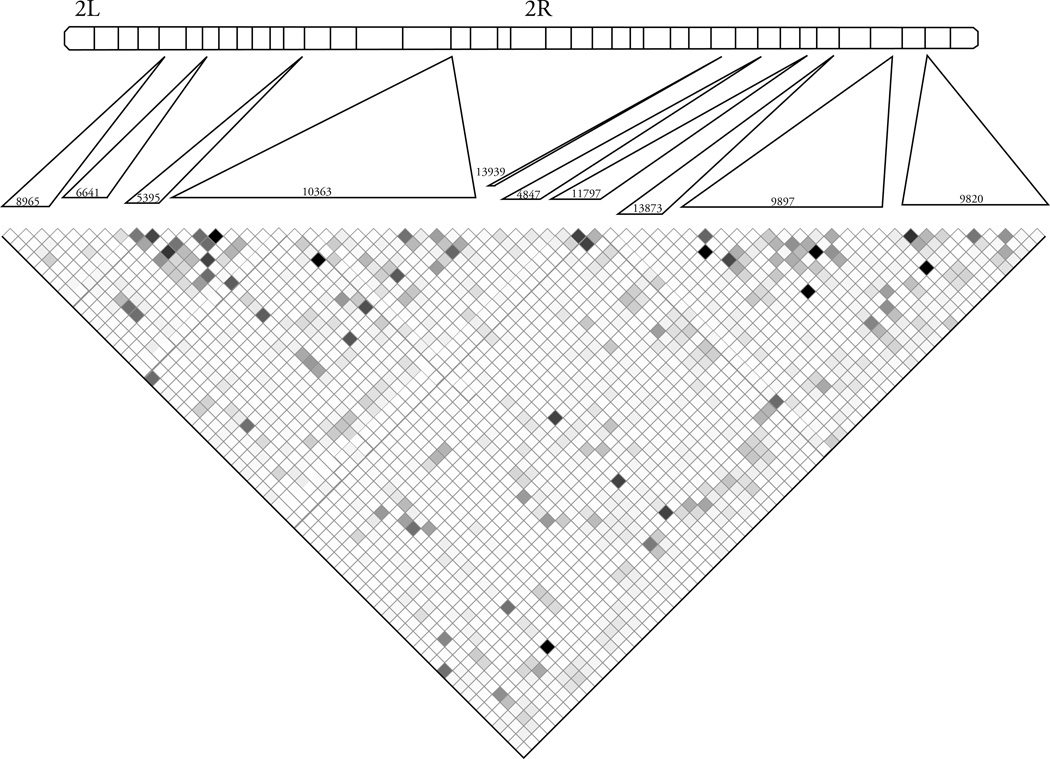
The analyzed genes were highly polymorphic (Table 1). We excluded 494 singletons and 6 polymorphisms with less than 8 lines successfully scored. This left 68 nonsynonymous polymorphisms, 157 synonymous polymorphisms and 746 polymorphisms in noncoding regions. The pattern of linkage disequilibrium among the 68 nonsynonymous polymorphisms is shown in Figure 1.
Figure 1.
Patterns of linkage disequilibrium for the nonsynonymous polymorphisms. Markers are arranged in order along chromosome 2 and the approximate location of each gene on chromosome 2 is shown. Darker colors indicate higher levels of linkage disequilibrium (r2 = 1 shown in black, 1 > r2 > 0 shown with shades of gray, r2 = 0 shown in white).
Association Testing
We identified twelve (12) significant associations (markerwise P < 0.01; q-value = 0.22) at nonsynonymous polymorphisms. These associations included five different genes and four phenotypes. Associations were identified between mated-30 and CG9820, remated and CG9820, CG9897 (4 polymorphisms), CG10363 and CG11797 (2 polymorphisms), fertility-V1 and CG9897 and CG10363, and fertility and CG11797 (Table 3). Some of these markers are in high linkage disequilibrium (Figure 1) with each other and therefore may not actually represent 12 independent effects.
Table 3.
Associations with nonsynonymous polymorphisms. Shown is the gene with the location of the polymorphism.
| Phenotype | Gene (snp) | Polymorphism | r2 |
|---|---|---|---|
| Mated-30 | CG9820 | Lys31Asn* | 0.08 |
| Remated | CG9820 | Glu113Lys** | 0.11 |
| CG9897 | Arg76Ser**, a | 0.15 | |
| CG9897 | Gly83Asp**, a | 0.14 | |
| CG9897 | Ile88Asn*** | 0.19 | |
| CG9897 | Ala89Ser** | 0.16 | |
| CG10363 | Asp1491Glu* | 0.08 | |
| CG11797 | Ala32Val*** | 0.17 | |
| CG11797 | Lys33Stop**, a | 0.09 | |
| Fertility-V1 | CG9897 | Gly83Asp** | 0.10 |
| CG10363 | Leu955Pro** | 0.11 | |
| Fertility | CG11797 | Thr15Ala* | 0.09 |
experimentwise P< 0.05,
markerwise P < 0.005,
markerwise P < 0.01
no longer significant (P < 0.05) after inclusion of linked markers into ANOVA.
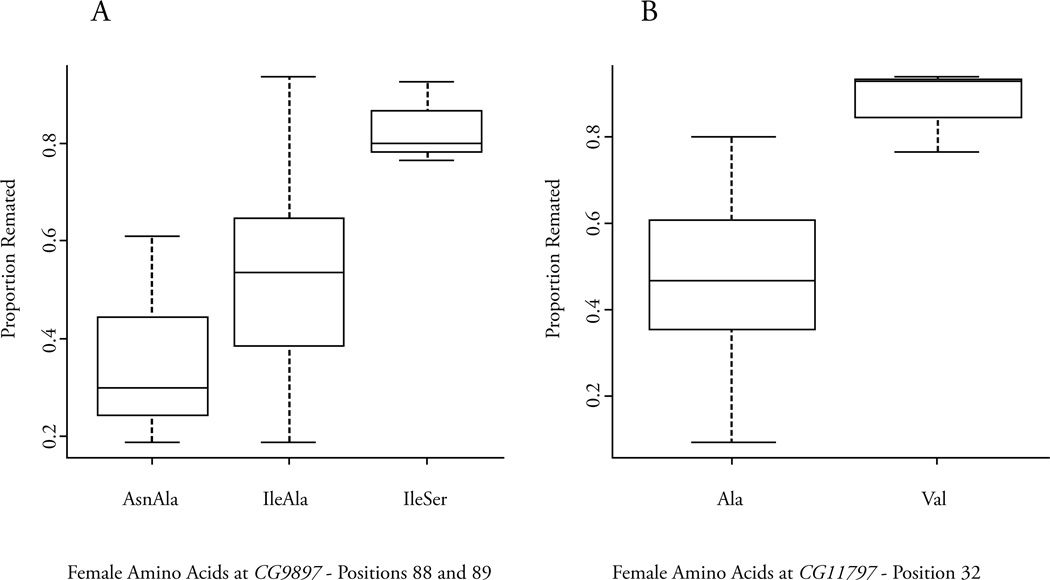
Two of these associations were significant at an experimentwise P < 0.05 (Figure 2), both affected the likelihood a female would remate (remated) and interestingly both were in regions with several linked amino acid polymorphisms. The first is an isoleucine to asparagine change at position 88 of the serine protease homolog, CG9897. This polymorphism was flanked by 3 other nonsynonymous changes all within 13 amino acids; arginine to serine at position 76, glycine to aspartic acid at position 83, and alanine to serine at position 89. All of these nonsynonymous polymorphisms associated at markerwise P < 0.005 although a synonymous change within this region did not (P = 0.59). Because these polymorphisms are in linkage disequilibrium and closely linked, we attempted to parse the individual effects by examining all four polymorphisms simultaneously using an ANOVA model. The isoleucine to asparagine polymorphism at position 88 and the alanine to serine polymorphism at position 89 retained significance at P < 0.05. Using both of these markers to form a haplotype also resulted in a significant association (P = 1.9 × 10−5) and all pair wise comparisons between the three different haplotypes were significant (Figure 2A). The other experimentwise association with remating rate is an alanine to valine change at position 32 of the putative odorant binding protein CG11797 (Figure 2B). Interestingly, there is a lysine to a premature stop codon at position 33, just one amino acid downstream. Both of these nonsynonymous polymorphisms were associated at a markerwise P < 0.005. When both were included in an ANOVA model only the alanine to valine change at position 32 had a significant effect (P < 0.05), the premature stop codon did not.
Figure 2.
Examples of associations with female influence on sexual selection. Box plots showing the median, upper and lower quartiles and the range for the different amino acid polymorphisms that associate with the proportion of females that remated at CG9897 (A) and CG11797 (B).
Several genes showed evidence of pleiotropy (Table 3). The two genes mentioned above, CG9897 and CG11797 also associated with fertility-V1 and fertility, respectively. Two different nonsynonymous polymorphisms in the putative olfactory receptor CG9820 (Lys31Asn and Glu113Lysin) associated with female mating when the females were virgins (mated-30) and after they had already mated once (remated), respectively. Finally, a lysine to proline change at position 955 in the putative peptidase inhibitor CG10363 was associated with fertility-V1, while an aspartic acid to glutamic acid change at position 1491 was associated with female remating rate (remated).
Among synonymous and noncoding polymorphisms, even the most significant associations (markerwise P < 0.005) still had a false discovery rate of around q = 0.5. These can be viewed in Supplementary Table 1. Although half these associations could be biologically relevant, the nonsynonymous polymorphisms highlighted above show the most promise to be causative or linked to the traits of interest. All line means were normally distributed except for mated-30 (P=0.019), which showed some evidence of a bimodal distribution.
Discussion
Here we used association testing to survey natural variation and study the genetic basis to female influence on pre- and post-copulatory sexual selection in D. melanogaster. We sequenced 10 candidate genes in 91 chromosome extraction lines and scored females from these lines for mating rate, remating rate, propensity to use the sperm from the second male and fertility. There was a significant genetic basis to variation for all the traits studied. By independently testing nonsynonymous polymorphisms, we identified two associations with remating rate that met a stringent experimentwise P < 0.05 (CG9897 and CG11797) while 10 additional associations (across a variety of genes and phenotypes) met a more liberal markerwise P < 0.01.
CG9897 is a putative serine endopeptidase homolog (Ross et al. 2003). It was identified as a candidate gene due to its expression in female reproductive tracts (Swanson et al. 2004), and recent work demonstrates that it is expressed in the sperm storage organs (Prokupek et al. 2009). An isoleucine to asparagine change at position 88 of CG9897 associated with female remating rate (experimentwise P < 0.05). Three other nonsynonymous polymorphisms in this gene (at positions 76, 83 and 89) were also associated with remating rate at a markerwise P < 0.01. When considering these markers in an ANOVA simultaneously, only the polymorphisms at amino acid positions 88 and 89 remained significant, but it is interesting to observe so many amino acid polymorphisms in this small region. Although speculative, the expression of CG9897 in the sperm storage organs may suggest some influence on how sperm behaves in storage, and thus relate to the so called ‘sperm effect’, whereby females without properly stored sperm are more likely to remate (Manning 1962). We also identified an association between a glycine to aspartic acid change at position 83 in CG9897 and female fertility (markerwise P < 0.01). This could also be driven through an effect on sperm storage but could also be the result of pleiotropy as we do not see a correlation between female fertility and remating rate as might be expected if both were being driven via this gene’s impact on sperm storage. Interestingly, CG9897 shows evidence for elevated levels of DNA polymorphism and signatures of balancing selection (Panhuis & Swanson 2006) and may be interacting with male reproductive genes that are also known to exhibit high levels of polymorphism (Swanson & Vacquier 2002; Clark et al. 2006). Serine endopeptidase homologs are thought to lack proteolytic activity, but have been implicated in mediating protein interactions and immune responses (Kawabata et al. 1996; Asgari et al. 2003; Ross et al. 2003; Yu et al. 2003). Clearly, more work needs to be done to understand the mechanisms and selective forces acting on CG9897, but association tests in another population support these findings and analysis of the RNAi knockdown is ongoing (Chow, Wolfner and Clark, unpublished data).
The second association meeting the experimentwise threshold was an alanine to valine polymorphism at position 32 of CG11797. A lysine to premature stop codon at position 33 also was associated (markerwise P < 0.005), but this association was not significant in an ANOVA model testing the simultaneous effects of both markers. CG11797, also known as Obp56a, codes for an odorant binding protein. D. melanogaster encode 51 odorant binding proteins, which aid in the solution and transfer of odorants to specific receptors (Hekmat-Scafe et al. 2002). These odorant binding proteins are potentially of great interest in the context of female mating decisions. Species and gender recognition in D. melanogaster rely on both the production and assessment of cuticular hydrocarbons (Billeter et al. 2009; Lacaille et al. 2009). Changes in cuticular hydrocarbon profiles, specifically polymorphism at the desaturase 2 gene, have also been implicated in sexual isolation (Fang et al. 2002; Greenberg et al. 2003). These results suggest that polymorphisms in odorant binding proteins or odorant receptors may play a role in female mating decisions. Odorant binding proteins are particularly noteworthy, as they have recently been found among male seminal fluid proteins, indicating a possible role in male induced post-copulatory phenotypes in addition to female pre-copulatory decision making (Findlay et al. 2008). Interestingly, CG11797 is likely under positive selection, one of only two genes in the family to show this pattern of selection (Wang et al. 2007). A threonine to alanine polymorphism at position 15 in CG11797 was also associated with female fertility (markerwise P < 0.01). As both CG9897 and CG11797 were associated with both fertility and female remating propensity, this raises the possibility that there may be some relationship between these phenotypes, although we do not see a genetic correlation between these phenotypes in the lines assayed (r = −0.11, P = 0.28).
A lysine to asparagine change at position 31 and a glutamate to lysine change at position 113 in another odorant binding protein, CG9820, associated with mating rate when the females were virgins (mated-30) and remating rate (remated), respectively. Although CG9820 appears to affect both mating and remating rates, no single polymorphism was associated with both traits and we did not observe a genetic correlation between these phenotypes (r = 0.14, P = 0.18). It is interesting to note that mating rate when the females are virgins appears to be independent of the propensity of these females to remate. It would be very exciting if these two traits were independently controlled, but our observation could be generated by a strong male by female interaction (Clark et al. 1999; Chow et al. 2010). Under such a scenario, the interaction term between the male and female genotype swamps the marginal effects of the female genotype when scoring mating and remating rate using only two different male genotypes. Although it would demand substantial added effort, a common genetic basis underlying mating and remating rate might be observed if this experiment were repeated using the same males but in the reciprocal order.
Surprisingly, we did not identify associations for a female’s propensity to use sperm from the second male, despite significant differences in P2 among the surveyed lines in this study (P = 0.005) and previous evidence that female genotype influences sperm utilization (Clark & Begun 1998). It is possible that we lack the statistical power to identify true associations because many genes of very small effect impact P2. It is also possible that the ten genes we chose to survey may not be important in regulating sperm utilization patterns. We did not observe associations with any of the noncoding or synonymous polymorphisms that we scored; even the most significant of these yielded a false discovery rate of about 50% (see supplemental table 1). This is surprising since some of these genes were selected based on changes in expression level after mating (McGraw et al. 2004). The lack of association may be due to the large number of tests (and the requirement to control for these tests) that were completed for noncoding and synonymous polymorphisms as compared to the nonsynonymous changes. It would be interesting to look more closely at the effects of polymorphism within promoter regions for these genes, but these regions are not yet well defined.
In addition to the correlations between fertility measures, we observed a negative relationship between fertility and the propensity of a female to use sperm from the second male (P2). One explanation for this correlation between P2 and fertility is variation in male quality. Females are known to vary in the extent to which they use sperm from the second male to mate (Clark & Begun 1998). If certain female genotypes are inclined to use sperm from the second male regardless of quality, via mechanisms such as sperm dumping or ejection (Snook & Hosken 2004; Manier et al. 2010), she may find herself in the unfortunate position of reducing her overall fertility in the instance of a low quality second mating partner. To explore this possibility, we further decomposed fertility into fertility in vials 1 and 2 (fertility-V1V2) and fertility in vials 3 and 4 (fertility-V3V4), which are positively correlated. We found that fertility–V1V2, when the female had very little chance to lay eggs sired by the second male, was uncorrelated with P2 (r = −0.040, P = 0.708) however fertility-V3V4 showed a strong negative correlation with P2 (r = −0.272, P = 0.009). This is consistent with the hypothesis of sperm dumping by the female coupled with mating to a low quality second partner.
The negative correlation between P2 and fertility could also be explained by effects of variation in female reproductive tract environment on sperm longevity. Under this scenario, female genotypes causing sperm to degrade in quality quickly could result in reduced hatchability of eggs sired by the first male with increasing time after mating. If the female continues laying eggs sired by the first male despite this loss of hatchability, this would lead to an inflated P2 value and could also lead to decreased fertility. Previous work has identified an effect of male genotype on sperm longevity in storage (Chapman et al. 2001; Civetta et al. 2008) and has shown that seminal fluid has a protective effect on sperm in the female reproductive tract (Holman 2009), but no effect of female genotype on sperm longevity was identified. However, this does not preclude such a possibility, as neither study attempted to assess variation across a wide variety of female backgrounds. As mentioned above with the effect of CG9897 on remating rate, it is possible that much of the variation in female influence on postcopulatory sexual selection is driven through sperm storage (or ejection) rates, a potentially productive area for future research.
Another important problem is to quantify the degree to which the female influence on postcopulatory sexual selection impacts her overall fitness. The effects of female fertility are obvious, but the fitness benefits of female choice remain an active area of research (Jennions & Petrie 2000; Zeh & Zeh 2003; Hettyey et al. 2010). Understanding the benefits of female choice can be further complicated when there are strong male by female interactions (Clark et al. 1999) potentially influencing parentage through cryptic female choice (Eberhard 1996). Females do have some control over how sperm is utilized (Qazi & Hogdal 2010), although much of the observed dynamics may fit a ‘fair raffle’ (Manier et al. 2010). Further research characterizing the fitness consequences of female influence on postcopulatory sexual selection will greatly enhance our understanding of reproductive outcomes.
Caveats of association testing
It is important to recognize that association testing does not imply causality and the potential for false positives is well recognized (Cowperthwaite et al. 2010). By focusing on nonsynonymous polymorphisms, we identified twelve significant associations between genotype and phenotype at a markerwise P < 0.01 and a false discovery rate calculation using Q-value (Storey & Tibshirani 2003) suggests that perhaps two of these associations are false positives. Several factors, however, indicate that false positives are not the sole driving force underlying the identified associations. First, the genes in this study were selected a priori based on the biological assumption that they could be affecting female influence on sexual selection. We also tested for associations between our female phenotypes and polymorphisms in male reproductive genes (Fiumera et al. 2005) or immunity genes (Lazzaro et al. 2004) that had been genotyped in these same lines. No markers from these data sets met an experimentwise P < 0.05. In addition, female reproductive genes had significantly more associations meeting a markerwise P < 0.01 as compared to either male reproductive genes (P = 0.007) or immunity genes (P = 0.015) indicating that long distance, unobserved linkage disequilibrium is not driving the observed associations. Furthermore, we observed the strongest associations with amino acid polymorphisms as compared to synonymous or arbitrary noncoding polymorphisms that were identified.
In D. melanogaster, and other non-human species, the goal of association testing may not be to identify markers for diagnostics but to provide a detailed genetic screen that can inform future studies using technologies such as RNAi. For example, Fiumera et al. (2005) identified a weak association between Acp29Ab and the proportion of offspring sired by the first male to mate and this was subsequently verified using a null mutation (Wong et al. 2008). As such, it is often acceptable to allow a slightly liberal false discovery rate to prevent missing true associations. The synonymous and noncoding associations had much higher false discovery rates; the Q-value was greater than 0.5 for associations with a markerwise P< 0.005. This makes it less likely that these represent true associations (Supplemental Table 1), but these results certainly motivate direct testing of RNAi knockdowns to assess their impact on the phenotypes measured here.
In summary, we have shown that extensive genetic variation exists for female influence on postcopulatory sexual in D. melanogaster. Furthermore, we have used association testing to identify twelve associations of polymorphisms in four genes with mating rate, remating rate and measures of female fertility. Two different genes related to olfactory systems influence either mating or remating rate suggesting that pheromonal communication may be important within this species. In addition, a serine protease homolog associated with both fertility and remating rate and this may be influencing sperm storage or ejection. If, as we believe, these genes are crucial determinants of female post-mating behavior, then further research may answer questions about the nature of antagonistic sexual coevolution in D. melanogaster, and help to explain the dynamics of male × female interactions in determining mating outcomes.
Supplementary Material
Acknowledgements
We thank J. Belote, C. Chow, M. Manier, S. Pitnick, M. Wolfner, and several anonymous reviewers for useful discussions and comments. S. Ryan assisted with the figures. This work was supported by NSF grant DEB-0743125 to A.C.F and A.C.G. and NIH R01 HD059060 to A.C.G.
Footnotes
Data Accessibility
Phenotype data is available as Supplemental Table 2. DNA sequences are available under the Genbank accession numbers JN162918-JN163851 and the genotype file is available as Supplemental Table 3.
References
- Asgari S, Zhang GM, Zareie R, Schmidt O. A serine proteinase homolog venom protein from an endoparasitoid wasp inhibits melanization of the host hemolymph. Insect Biochemistry and Molecular Biology. 2003;33:1017–1024. doi: 10.1016/s0965-1748(03)00116-4. [DOI] [PubMed] [Google Scholar]
- Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends in Ecology and Evolution. 2003a;18:41–47. [Google Scholar]
- Chapman T, Bangham J, Vinti G, et al. The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2003b;100:9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Herndon LA, Heifetz Y, Partridge L, Wolfner MF. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2001;268:1647–1654. doi: 10.1098/rspb.2001.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Wolfner MF, Clark AG. The genetic basis for male × female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics. 2010;186:1355–1365. doi: 10.1534/genetics.110.123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Rosing KR, Fisher JH. Differences in sperm competition and sperm competition avoidance in Drosophila melanogaster. Animal Behaviour. 2008;75:1739–1746. [Google Scholar]
- Clark AG, Aguade M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory-gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ. Female genotypes affect sperm displacement in Drosophila. Genetics. 1998;149:1487–1493. doi: 10.1093/genetics/149.3.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. Female × male interactions in Drosophila sperm competition. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Cowperthwaite MC, Mohanty D, Burnett MG. Genome-wide association studies: a powerful tool for neurogenomics. Neurosurgical Focus. 2010;28:E2. doi: 10.3171/2010.10.FOCUS09186. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Elwyn S. Does the desaturase-2 locus in Drosophila melanogaster cause adaptation and sexual isolation? Evolution. 2006;60:279–291. [PubMed] [Google Scholar]
- Eberhard WG. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press; 1996. [Google Scholar]
- Ejima A, Tsuda M, Takeo S, et al. Expression level of sarah, a Homolog of DSCR1, is critical for ovulation and female courtship behavior in Drosophila melanogaster. Genetics. 2004;168:2077–2087. doi: 10.1534/genetics.104.029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AW. Body size and courtship behaviour in Drosophila melanogaster. Animal Behaviour. 1961;9:93–99. [Google Scholar]
- Fang S, Takahashi A, Wu CI. A mutation in the promoter of desaturase 2 is correlated with sexual isolation between Drosophila behavioral races. Genetics. 2002;162:781–784. doi: 10.1093/genetics/162.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi XH, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biology. 2008;6:1417–1426. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Foss M, et al. dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron. 1998;21:1363–1374. doi: 10.1016/s0896-6273(00)80655-8. [DOI] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Natural variation in male-induced 'cost-of-mating' and allele-specific association with male reproductive genes in Drosophila melanogaster. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 2006;361:355–361. doi: 10.1098/rstb.2005.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 2007;176:1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg U, Arnqvist G. Fitness effects of female mate choice: preferred males are detrimental for Drosophila melanogaster females. Journal of Evolutionary Biology. 2003;16:797–811. doi: 10.1046/j.1420-9101.2003.00597.x. [DOI] [PubMed] [Google Scholar]
- Fricke C, Wigby S, Hobbs R, Chapman T. The benefits of male ejaculate sex peptide transfer in Drosophila melanogaster. Journal of Evolutionary Biology. 2009;22:275–286. doi: 10.1111/j.1420-9101.2008.01638.x. [DOI] [PubMed] [Google Scholar]
- Fuyama Y. Genetic-evidence that ovulation reduces sexual receptivity in Drosophila melanogaster females. Behavior Genetics. 1995;25:581–587. doi: 10.1007/BF02327581. [DOI] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Coyne JA, Wu CI. Ecological adaptation during incipient speciation revealed by precise gene replacement. Science. 2003;302:1754–1757. doi: 10.1126/science.1090432. [DOI] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Fang S, Wu CI. Adaptive loss of an old duplicated gene during incipient speciation. Molecular Biology and Evolution. 2006;23:401–410. doi: 10.1093/molbev/msj045. [DOI] [PubMed] [Google Scholar]
- Gromko MH, Gerhart PD. Increased density does not increase remating frequency in laboratory populations of Drosophila melanogaster. Evolution. 1984;38:451–455. doi: 10.1111/j.1558-5646.1984.tb00305.x. [DOI] [PubMed] [Google Scholar]
- Harshman LG, Clark AG. Inference of sperm competition from broods of field-caught Drosophila. Evolution. 1998;52:1334–1341. doi: 10.1111/j.1558-5646.1998.tb02015.x. [DOI] [PubMed] [Google Scholar]
- Harshman LG, Hoffmann AA, Prout T. Environmental-Effects on Remating in Drosophila-Melanogaster. Evolution. 1988;42:312–321. doi: 10.1111/j.1558-5646.1988.tb04135.x. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Research. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Wolfner MF. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg-laying in females for 1 day after mating. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10114–10118. doi: 10.1073/pnas.92.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettyey A, Hegyi G, Puurtinen M, et al. Mate Choice for Genetic Benefits: Time to Put the Pieces Together. Ethology. 2010;116:1–9. [Google Scholar]
- Holman L. Drosophila melanogaster seminal fluid can protect the sperm of other males. Functional Ecology. 2009;23:180–186. [Google Scholar]
- Imhof M, Harr B, Brem G, Schlotterer C. Multiple mating in wild Drosophila melanogaster revisited by microsatellite analysis. Molecular Ecology. 1998;7:915–917. doi: 10.1046/j.1365-294x.1998.00382.x. [DOI] [PubMed] [Google Scholar]
- Jennions MD, Petrie M. Why do females mate multiply? A review of the genetic benefits. Biological Reviews. 2000;75:21–64. doi: 10.1017/s0006323199005423. [DOI] [PubMed] [Google Scholar]
- Jones AG, Ratterman NL. Mate choice and sexual selection: What have we learned since Darwin? Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10001–10008. doi: 10.1073/pnas.0901129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni N, Yamamoto D. Genetic analysis of chaste, a new mutation of Drosophila melanogaster characterized by extremely low female sexual receptivity. Journal of Neurogenetics. 2009;23:329–340. doi: 10.1080/01677060802471601. [DOI] [PubMed] [Google Scholar]
- Kawabata S, Tokunaga F, Kugi Y, et al. Limulus factor D, a 43-kDa protein isolated from horseshoe crab hemocytes, is a serine protease homologue with antimicrobial activity. Febs Letters. 1996;398:146–150. doi: 10.1016/s0014-5793(96)01224-0. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Pennington JE. Protease gene duplication and proteolytic activity in Drosophila female reproductive tracts. Molecular Biology and Evolution. 2009;26:2125–2134. doi: 10.1093/molbev/msp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. Plos Genetics. 2007;3:1541–1549. doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaille F, Everaerts C, Ferveur JF. Feminization and alteration of Drosophila taste neurons induce reciprocal effects on male avoidance behavior. Behavior Genetics. 2009;39:554–563. doi: 10.1007/s10519-009-9286-8. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. [DOI] [PubMed] [Google Scholar]
- Lawniczak MKN, Begun DJ. A QTL analysis of female variation contributing to refractoriness and sperm competition in Drosophila melanogaster. Genetical Research. 2005;86:107–114. doi: 10.1017/S0016672305007755. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- Linder JE, Rice WR. Natural selection and genetic variation for female resistance to harm from males. Journal of Evolutionary Biology. 2005;18:568–575. doi: 10.1111/j.1420-9101.2004.00872.x. [DOI] [PubMed] [Google Scholar]
- Liu HF, Kubli E. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9929–9933. doi: 10.1073/pnas.1631700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack PD, Kapelnikov A, Heifetz Y, Bender M. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10358–10363. doi: 10.1073/pnas.0604046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier MK, Belote JM, Berben KS, et al. Resolving mechanisms of competitive fertilization success in Drosophila melanogaster. Science. 2010;328:354–357. doi: 10.1126/science.1187096. [DOI] [PubMed] [Google Scholar]
- Manning A. Sperm factor affecting receptivity of Drosophila melanogaster females. Nature. 1962;194:252–&. [Google Scholar]
- Marks RW, Seager RD, Barr LG. Local ecology and multiple mating in a natural population of Drosophila melanogaster. American Naturalist. 1988;131:918–923. [Google Scholar]
- McGraw LA, Clark AG, Wolfner MF. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics. 2008;179:1395–1408. doi: 10.1534/genetics.108.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Current Biology. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Milkman R, Zeitler RR. Concurrent multiple paternity in natural and laboratory populations of Drosophila melanogaster. Genetics. 1974;78:1191–1193. doi: 10.1093/genetics/78.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis TM, Swanson WJ. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics. 2006;173:2039–2047. doi: 10.1534/genetics.105.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GA. Sexual conflict over mating and fertilization: an overview. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 2006;361:235–259. doi: 10.1098/rstb.2005.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Chen S, Busser S, et al. Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Current Biology. 2005;15:207–213. doi: 10.1016/j.cub.2005.01.034. [DOI] [PubMed] [Google Scholar]
- Pitnick S. Male Size Influences Mate Fecundity and Remating Interval in Drosophila-Melanogaster. Animal Behaviour. 1991;41:735–745. [Google Scholar]
- Pitnick S, Garcia-Gonzalez F. Harm to females increases with male body size in Drosophila melanogaster. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2002;269:1821–1828. doi: 10.1098/rspb.2002.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokupek A, Hoffmann F, Eyun SI, et al. An Evolutionary Expressed Sequence Tag Analysis of Drosophila Spermatheca Genes. Evolution. 2008;62:2936–2947. doi: 10.1111/j.1558-5646.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- Prokupek AM, Kachman SD, Ladunga I, Harshman LG. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Molecular Biology. 2009;18:465–475. doi: 10.1111/j.1365-2583.2009.00887.x. [DOI] [PubMed] [Google Scholar]
- Prout T, Bundgaard J. The population genetics of sperm displacement. Genetics. 1977;85:95–124. doi: 10.1093/genetics/85.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi MCB, Hogdal L. Hold on: Females modulate sperm depletion from storage sites in the fly Drosophila melanogaster. Journal of Insect Physiology. 2010;56:1332–1340. doi: 10.1016/j.jinsphys.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Ram KR, Wolfner MF. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15384–15389. doi: 10.1073/pnas.0902923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi Ram K, Ji S, Wolfner MF. Fates and targets of male accessory gland proteins in mated female Drosophila melanogaster. Insect Biochemistry and Molecular Biology. 2005;35:1059–1071. doi: 10.1016/j.ibmb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Snook RR, Hosken DJ. Sperm death and dumping in Drosophila. Nature. 2004;428:939–941. doi: 10.1038/nature02455. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nature Reviews Genetics. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Ting CT. Genetic basis of sexual isolation in Drosophila melanogaster. Genetica. 2004;120:273–284. doi: 10.1023/b:gene.0000017649.51782.5b. [DOI] [PubMed] [Google Scholar]
- Taylor CE, Kekic V. Sexual selection in a natural population of Drosophila melanogaster. Evolution. 1988;42:197–199. doi: 10.1111/j.1558-5646.1988.tb04120.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Shabalina SA, Mackay TFC, Anholt RRH. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics. 2007;177:1655–1665. doi: 10.1534/genetics.107.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S, Chapman T. Female resistance to male harm evolves in response to manipulation of sexual conflict. Evolution. 2004;58:1028–1037. doi: 10.1111/j.0014-3820.2004.tb00436.x. [DOI] [PubMed] [Google Scholar]
- Wigby S, Slack C, Gronke S, et al. Insulin signalling regulates remating in female Drosophila. Proceedings of the Royal Society B-Biological Sciences. 2010 doi: 10.1098/rspb.2010.1390. eprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GS. Equilibrium-analysis of sexual selection in Drosophila melanogaster. Evolution. 1987;41:11–21. doi: 10.1111/j.1558-5646.1987.tb05767.x. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. Battle and ballet: Molecular interactions between the sexes in Drosophila. Journal of Heredity. 2009;100:399–410. doi: 10.1093/jhered/esp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Albright SN, Giebel JD, et al. A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics. 2008;180:921–931. doi: 10.1534/genetics.108.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N, Kim YJ, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451:33–38. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Jiang HB, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochemistry and Molecular Biology. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- Zeh JA, Zeh DW. Toward a new sexual selection paradigm: Polyandry, conflict and incompatibility (Invited article) Ethology. 2003;109:929–950. [Google Scholar]
- Zhang R, Amah L, Fiumera AC. Autosomal variation for male body size and sperm competition phenotypes is uncorrelated in Drosophila melanogaster. Biology Letters. 2008;4:500–503. doi: 10.1098/rsbl.2008.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.