Abstract
Segmentation of the vertebrate hindbrain into multiple rhombomeres is essential for proper formation of the cerebellum, cranial nerves and cranial neural crest. Paralog group 1 (PG1) hox genes are expressed early in the caudal hindbrain and are required for rhombomere formation. Accordingly, loss of PG1 hox function disrupts development of caudal rhombomeres in model organisms and causes brainstem defects, associated with cognitive impairment, in humans. In spite of this important role for PG1 hox genes, transcriptional targets of PG1 proteins are not well characterized. Here we use ectopic expression together with embryonic dissection to identify novel targets of the zebrafish PG1 gene hoxb1b. Of 100 genes up-regulated by hoxb1b, 54 were examined and 25 were found to represent novel hoxb1b regulated hindbrain genes. The ppp1r14al gene was analyzed in greater detail and our results indicate that Hoxb1b is likely to directly regulate ppp1r14al expression in rhombomere 4. Furthermore, ppp1r14al is essential for establishment of the earliest hindbrain signaling-center in rhombomere 4 by regulating expression of fgf3.
Keywords: hindbrain, zebrafish, transcription, fgf, hox, microarray
Introduction
hox genes were first identified in Drosophila genetic screens as important regulators of embryonic development (reviewed in (Lewis, 1994)). In particular, mutations in hox genes give rise to homeotic phenotypes where one body structure is transformed, more or less completely, into a different structure. Subsequently, hox genes were shown to carry out analogous functions in vertebrates (reviewed in (Krumlauf, 1994)). In vertebrates, genome duplications have produced four hox clusters, except in teleost fish that contain seven clusters as the result of an additional duplication event (Amores et al., 1998; Kuraku and Meyer, 2009). hox genes that occupy the same position in each cluster are referred to as paralogous genes (reviewed in (Alexander et al., 2009)) and their expression is co-linear with their position in the hox clusters such that 3′ genes are expressed earlier and further anteriorly than 5′ genes. Accordingly, the earliest expressed vertebrate hox genes belong to paralog group 1 (PG1).
PG1 hox genes act together with hox genes from PG2, 3 and 4 to regulate formation of the caudal hindbrain. In particular, PG1-4 hox genes act to impart distinct identities on rhombomeres 4-7. In the mouse, Hoxa1, which is expressed in the caudal hindbrain, is the earliest-acting hox gene and it is required for appropriate patterning of rhombomere (r) 4, 5 and 6 (Carpenter et al., 1993; Mark et al., 1993). Hoxa1 is also required to activate Hoxb1, which is expressed exclusively in r4. Accordingly, in Hoxb1 mutants, r4 is misidentified and takes on r2-like characteristics (Goddard et al., 1996; Studer et al., 1996). In addition, Hoxa1/Hoxb1 double mutants show a more severe phenotype than the single mutants (Gavalas et al., 1998; Gavalas et al., 2001; Rossel and Capecchi, 1999; Studer et al., 1998), indicating that these hox genes may regulate non-overlapping sets of genes. Notably, the additional genome duplication in teleosts has led to a re-shuffling of responsibilities among hox genes in zebrafish. In particular, the only zebrafish hoxa1 gene (hoxa1a) is not expressed in the hindbrain. Instead, a second hoxb1 copy (hoxb1b) has taken on the role performed by Hoxa1 in the mouse, while zebrafish hoxb1a plays the same role as murine Hoxb1 (McClintock et al., 2001; McClintock et al., 2002). Hindbrain patterning and PG1 hox genes have been implicated in developmental defects during human development. In particular, mutations in Hoxa1 have been linked to defects of the brainstem (which derives partly from the embryonic hindbrain) that are associated with some cases of autism (Bosley et al., 2007; Tischfield et al., 2005).
Secreted factors are also required for hindbrain patterning. Indeed, one of the earliest events during hindbrain patterning is the establishment of a signaling center in r4 that secretes Fgf3 and Fgf8 (Maves et al., 2002; Walshe et al., 2002). Fgf3 and Fgf8 are required for proper formation of r5 and r6, apparently by acting together with the vhnf1 gene to regulate expression of krox20 in r5 and valentino in r5/r6 (Hernandez et al., 2004; Wiellette and Sive, 2003). Nevertheless, it remains unclear how this r4 signaling center is set up and what role hox genes may play in this process.
To fully understand the role of PG1 hox genes in hindbrain development, it is necessary to identify genes regulated - directly or indirectly - by PG1 proteins. Some direct PG1 target genes are known, but many such targets are other hox genes (e.g. hoxb1, hoxa2, hoxb2; (Maconochie et al., 1997; Popperl et al., 1995; Tumpel et al., 2007)) – although there are also examples of non-hox direct targets (e.g. krox20; (Wassef et al., 2008)). In terms of indirect target genes, any gene whose expression is lost in PG1 mutants would be a candidate, but in most cases it has not been determined if such genes can actually be induced by PG1 proteins. Here we take advantage of the ease of gene misexpression and dissection in zebrafish to identify genes induced by hoxb1b. We identify 100 genes that are up-regulated more than 2-fold by hoxb1b. Subsequent expression analysis of 54 genes revealed that 28 are expressed in hindbrain-associated structures. Three of these have been previously reported as expressed in the hindbrain, while the remaining 25 are either novel genes or known genes not previously reported as expressed in the hindbrain. Furthermore, 20 of the 28 hindbrain-associated genes show rhombomere-restricted expression. One r4-restricted gene, the protein phosphatase 1 regulatory subunit ppp1r14al, was selected for detailed analysis to determine its role in hindbrain development and to confirm its regulation by Hoxb1b. We find that ppp1r14al is required for fgf3 expression in the r4 signaling center and that loss of ppp1r14al leads to defects in hindbrain patterning, as well as disruption of subsequent neurogenesis. Lastly, chromatin immunoprecipitation reveals that Hoxb1, as well as Pbx and Meis cofactors, occupy the ppp1r14al promoter in developing zebrafish embryos. Hence, our screening strategy efficiently identified bona fide hoxb1b target genes in zebrafish hindbrain development and identified a regulator of the r4-signaling center.
Materials and Methods
Zebrafish
Zebrafish and their embryos were handled and staged according to standard protocols (Kimmel et al., 1995).
Microinjections and embryo dissection
All mRNAs for microinjections were synthesized in vitro using the SP6 mMessage mMachine kit (Ambion) as previously described (Vlachakis et al., 2001). For microarray experiments, hoxb1b+meis3 (166pg each) or meis3+βgal (166pg each) were microinjected into 1-2 cell stage zebrafish embryos and raised to 14 hours post fertilization. Embryos were then manually dechorinated in fish Ringer solution on a 1% agarose-bed 35-mm culture dish. Anterior tissues were dissected and collected using a pair of forceps and were then resuspended in 750μl of Trizol Reagent (Invitrogen) and stored at -80°C. For morpholino (MO) injections, 4ng of MO targeting the translational start site of ppp1r14al was microinjected into 1-2 cell stage of embryos. For double morpholino injections, 2ng of MOfgf8 was injected solely or in combination with 4ng of MOppp1r14al. Rescue experiments were performed using 4ng MOppp1r14al+300pg ppp1r14al mRNA compared to 4ng MOppp1r14al+300pg GFP mRNA. The sequences of MO are as follows: MOppp1r14al 5′-CACCCGATTCGCAGCCATCTCCAGA-3′, MOfgf8 5′-TCAACCGTGAAGGTATGAGTCTC-3′ (Maves et al., 2002). For rescue experiments, 6 nucleotides at the 5′ end of the ppp1r14al mRNA were changed using the primer 5′-GGAATTCGATGGCCGCCAACAGAGTCGGGAGGCG-3′ to prevent targeting by MOs, while encoding the same amino acids as in wild type ppp1r14al. Pbx MOs were reported previously (Waskiewicz et al., 2002).
RNA isolation and qRT-PCR
Total RNA from dissected anterior tissues was isolated using standard protocols and dissolved in 20 ⌠L nuclease-free water (Ambion). 1 ⌠g total RNA per sample was shipped on dry ice for microarray analysis. For RT-PCR, cDNA was first synthesized using 1 ⌠g total RNA, 200 units of Superscript III reverse transcriptase (Invitrogen), and 2.5 ⌠M oligo dT primer in a 20 ⌠L reaction for 2 hours at 37°C. Quantitative PCR was performed using QuantiFast SYBR Green PCR kit (Qiagen) using 500ng of cDNA and gene specific primers in a 50 ⌠L reaction and detected in a 7300 realtime PCR system (Applied Biosystems). Sequences of PCR primers were as follows: tubulin, 5′-CTGTTGACTACGGAAAGAAGT-3′ and 5′-TATGTGGACGCTCTATGTCTA-3′; krox20, 5′-AAACGCAGGAGATGGCCTGA-3′ and 5′-GGTACTGGGAGTCGATGGAA-3′.
Microarray analysis
Microarray analysis was carried out by the Kimmel Cancer Center microarray facility at Thomas Jefferson University. Biotinylated cRNA probes were synthesized by linear amplification from total mRNA and hybridized to microarray slides containing 16,399 distinct 65-mer oligonucleotides (Compugen/Sigma-Genosys oligo set XEBLIB96), corresponding to approximately 12,500 zebrafish cDNAs. The experiments were performed in triplicate and each replicate array was hybridized with probe prepared from a separate injection and dissection. Background readings were subtracted from experimental readings, followed by normalization where each data point was divided by the 50th percentile of all data points. p-values were derived using Student's T-test. Genes up-regulated by hoxb1b+meis3 were defined as follows: 1) up-regulation by more than 2-fold by hoxb1b+meis3 as compared to meis3, and 2) a p-value lower than 0.05.
In situ hybridization
Plasmids containing zebrafish cDNA sequences were purchased from OpenBiosystems (Huntsville, AL). DIG-labeled antisense probes for hoxb1b+meis3 target genes were synthesized using PCR-amplified DNA inserts from the plasmids whose inserts had been verified by sequencing. hoxb1a, krox20, valentino, pax2, dlx2a and pea3 were described previously (Akimenko et al., 1994; Brown et al., 1998; Kiefer et al., 1996; Krauss et al., 1991; Moens et al., 1996; Oxtoby and Jowett, 1993; Prince et al., 1998). Plasmids containing fgf20a and ngn1 cDNA was purchased from OpenBiosystems. In situ hybridizations were carried out as described previously (Choe et al., 2002).
Acridine Orange staining
Acridine orange staining was performed as described previously (Kwak et al., 2006). Briefly, dechorinated embryos at desired stages were incubated in 0.2% acridine orange (Sigma) in phosphate-buffered saline (PBS) for 30 minutes at room temperature. Embryos were then washed 5 times with PBS and apoptotic cells were visualized under a UV microscope. Live images were captured using the SPOT software (version 4.6, SPOT imaging solutions).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously using antibodies to Hoxb1b/a, Pbx and Meis (Choe et al., 2009). A Hox/Pbx/Meis binding site located in the first intron of the ppp1r14al gene was assayed using primers 5′-GGTGCTAAAAAGTAACAGCCCCCACTGAGG-3′ and 5′-GGACAGTTGCAGGAGGGCTTTCTTTGTGTGAT-3′.
Results
An assay for the identification of hoxb1b target genes
Several reports have demonstrated that misexpression of paralog group 1 (PG1) hox genes drives ectopic gene expression in the anterior zebrafish embryo (Alexandre et al., 1996; McClintock et al., 2001; Vlachakis et al., 2001). In particular, hoxb1b, together with its meis and pbx cofactors, induces ectopic expression of hindbrain genes such as hoxb1a, hoxb2a, krox20 and valentino in the anterior CNS of zebrafish embryos (Fig. 1C;(Choe et al., 2002; Vlachakis et al., 2001)). In fact, the forebrain region undergoes a wholesale transformation to a hindbrain fate in these embryos - including differentiation of ectopic hindbrain Mauthner neurons (Vlachakis et al., 2001). This transformation is accompanied by morphological changes that shorten and thicken the anterior neural region into a characteristic rounded shape, as well as by the lack of eye formation. In contrast, misexpression of hoxb1b alone, or either cofactor alone, does not induce ectopic gene expression in the anterior CNS (Fig. 1B) and has no effect on morphology. Hence, we reasoned that misexpression could form the basis of a simple system to isolate novel genes acting downstream of hoxb1b in the hindbrain.
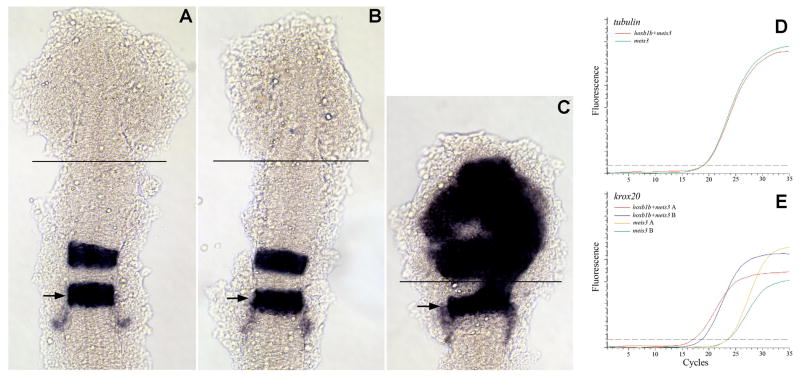
Figure 1.
An ectopic expression/dissection strategy to identify hoxb1b-regulated genes. (A-C) Uninjected (A), meis3+βgal mRNA-injected (B) or meis3+hoxb1b mRNA-injected (C) embryos were analyzed by in situ hybridization for expression of krox20. The position of r5 is indicated by arrows and the site where the anterior embryo was dissected is indicated by black lines in A-C. (D-E) Quantitative RT-PCR was used to assay expression of tubulin (D) and krox20 (E) in meis3-injected and meis3+hoxb1b-injected embryos. Two independent injections are shown in E. Embryos in A-C were flat mounted at 14hpf and are shown in dorsal view with anterior to the top.
hoxb1b-mediated induction of ectopic hindbrain gene expression is readily detected by in situ hybridization (Fig. 1C; (Vlachakis et al., 2001)). To quantify this induction, we used quantitative RT-PCR and assayed expression of three hindbrain genes (krox20, valentino and hoxa3; (Amores et al., 1998; Moens et al., 1998; Oxtoby and Jowett, 1993)), one forebrain/midbrain gene (otx2; (Li et al., 1994)) and one “house-keeping” gene (tubulin). We microinjected hoxb1b and meis3 mRNA – we have previously found that it is not necessary to co-inject pbx mRNA (Vlachakis et al., 2001) since pbx2 and pbx4 are highly expressed throughout the embryo (Vlachakis et al., 2000; Waskiewicz et al., 2002) - and raised embryos to 14hpf. Successfully injected embryos were identified based on the loss of eyes and the characteristic rounding of the head. The anteriormost region of hoxb1b/meis3-injected embryos was dissected at the line indicated in Fig. 1C. As control, we used the anterior-most region of embryos injected with meis3 alone (dissected as outlined in Fig. 1B). We find that expression of hindbrain genes is robustly induced such that krox20 transcripts are increased ∼10-fold (Fig. 1E) and valentino and hoxa3 are induced ∼2-4 fold (not shown). The forebrain/midbrain gene otx2 is repressed ∼2-fold (not shown), as expected based on the loss of forebrain fates and in accordance with our previously reported in situ hybridization data (Vlachakis et al., 2001), while expression of tubulin is unchanged (Fig. 1D). We conclude that hindbrain genes are induced 2- to 10-fold in the anterior of hoxb1b/meis3-injected embryos at the expense of forebrain/midbrain genes and that this induction is sufficiently pronounced to be detected by quantitative RT-PCR.
Expression profiling efficiently identifies hoxb1b-induced genes
To identify novel hindbrain genes acting in the hoxb1b genetic cascade, anterior tissue was dissected from embryos misexpressing hoxb1b/meis3 or meis3 alone at 14hpf, a stage when cell fate specification is ongoing in the hindbrain, as shown in Fig. 1. Probes were prepared from each tissue and hybridized to microarrays containing 65-mer oligonucleotides (Compugen/Sigma-Genosys oligo set XEBLIB96), representing ∼12,500 unique zebrafish genes, by the Kimmel Cancer Center Microarray Core Facility at Thomas Jefferson University. The experiment was carried out in triplicate where each replicate array was hybridized to probe prepared from a separate injection/dissection. Array results were analyzed as outlined in Materials and Methods and genes were considered hoxb1b targets if they were up-regulated more than 2-fold in hoxb1b/meis3-injected relative to meis3-injected controls with a p-value below 0.05 for the three experiments. Using these criteria, we find that 100 genes are up-regulated by hoxb1b at 14hpf (Table S1).
Strikingly, we find that six of the ten most highly induced genes – krox20, hoxa3a, valentino, hoxb3a, hoxa2b, hoxb2a - have been previously found to be expressed in the hindbrain and four of these (krox20, valentino, hoxa2a, hoxb2a) have already been shown to be regulated by hoxb1b/meis3 (Choe et al., 2002; Vlachakis et al., 2001; Waskiewicz et al., 2001). Given the efficient identification of known hoxb1b-regulated hindbrain genes by our assay, we reasoned that the uncharacterized genes in Table S1 might also represent novel hoxb1b-regulated genes expressed in the hindbrain. To test this directly, we selected 54 genes for further analysis. Since some known hindbrain genes showed relatively modest induction (e.g. ephA4a; 2.07-fold induction), we selected a set of genes to represent a cross-section of high and low induced genes, as well as genes with a range of p-values (including some genes that did not make the cut for Table S1 due to low fold-induction or high p-value). The genes selected for further analysis were re-annotated and are listed in Table 1. cDNA clones containing the longest insert available for each gene were purchased and initial sequencing revealed that 49 of the purchased clones corresponded to the correct gene on the microarray.
Table 1. Genes selected for analysis1.
| Genbank2 | Fold Change3 | p-value4 | Description (gene name)5 | Unigene6 | Sequence7 | Probe8 | ISH Signal9 | Expression10 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AI601442 | 14.32 | 0.025 | si:dkeyp-95d10.1; Similar to DUSP23 | Dr.78360 | X | X | X | lateral to hindbrain |
| 2 | Y13944 | 10.78 | 0.008 | Homeo box B3a (hoxB3a) | Dr.132502 | X | X | X | rhombomere 5/6 |
| 3 | AW184433 | 8.86 | 0.001 | Membrane-spanning 4-domains, subfamily A, member 17A.11 | Dr.40434 | X | X | ||
| 4 | AW281753 | 8.68 | 0.004 | Zwilling (zwi) | Dr.81187 | X | X | X | rhombomere 3/5 |
| 5 | AI667023 | 8.41 | 0.009 | Myelin protein zero | Dr.78005 | X | X | ||
| 6 | BI671376 | 7.29 | 0.012 | Sb:cb1035 | Dr.148642 | X | X | ||
| 7 | AI641757 | 6.99 | 0.014 | Wu:fc23f06 – similar to HNF1-beta-like or HNF1 gamma | Dr.15418 | X | X | X | rhombomere 5 |
| 8 | AI957504 | 6.83 | 0.008 | Wu:fd06c09 | Dr.122018 | X | X | X | rhombomere 5 |
| 9 | AF052249 | 5.82 | 0.032 | Forkhead box D3 | Dr.75816 | X | X | X | lateral to hindbrain |
| 10 | BM024206 | 5.45 | 0.000 | cytochrome P450, family 26, subfamily b, polypeptide 1 (Cyp26b1) | Dr.76359 | X | |||
| 11 | AI942951 | 5.44 | 0.001 | Wu:fc79c11 | Dr.106214 | X | X | X | rhombomere 2/4 |
| 12 | AI878761 | 5.41 | 0.025 | EGF-like-domain, multiple 6 | Dr.79266 | X | X | X | rhombomere 2/4 |
| 13 | AI793592 | 4.96 | 0.003 | Wu:fc51e09 | Dr.150442 | X | X | X | rhombomere 3/5 |
| 14 | AI461323 | 4.86 | 0.024 | PR domain containing 12 | Dr.23693 | X | X | X | multiple rhombomeres |
| 15 | AW567515 | 4.17 | 0.006 | Tissue inhibitor of metalloproteinase 2b | Dr.81512 | ||||
| 16 | BI534285 | 3.81 | 0.021 | Zgc:55558 | Dr.83658 | ||||
| 17 | AW279672 | 3.72 | 0.011 | retinol dehydrogenase 12 (all-trans and 9-cis) RDH12 | Dr.32031 | X | |||
| 18 | BI430048 | 3.53 | 0.001 | Zgc:92288 | Dr.85849 | X | X | ||
| 19 | AI721660 | 3.28 | 0.005 | Wu:fc30f10 | Dr.150323 | X | X | X | rhombomere boundaries |
| 20 | BI845046 | 3.27 | 0.010 | Wu:fj78f01 (Si:dkeyp-84a8.1); similar to Ngfi-A binding protein 1 | Dr.83971 | X | X | X | rhombomere 3/5 |
| 21 | AI965251 | 3.13 | 0.020 | Wu:fc65f09 | Dr.79031 | X | X | X | rhombomere 2/4/6 |
| 22 | AI957828 | 3.10 | 0.004 | Rho family GTPase 3b | Dr.32839 | X | X | ||
| 23 | BI890305 | 3.03 | 0.005 | Kin of IRRE like 3 | Dr.75543 | X | X | X | multiple rhombomeres |
| 24 | BI880801 | 2.75 | 0.005 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | Dr.83871 | X | X | ||
| 25 | AY017309 | 2.73 | 0.009 | Iroquois homeobox protein 5a (irx5a) | Dr.83684 | X | X | X | rhombomere |
| 26 | AI722745 | 2.72 | 0.002 | Transcription factor AP-2 alpha (tfap2a) | Dr.8506 | X | X | X | lateral to hindbrain |
| 27 | AW115765 | 2.56 | 0.008 | Zgc:55283 - similar to growth factor-receptor-binding protein 4 (grb4) Nck2 | Dr.80366 | X | X | X | rhombomere 3/5 |
| 28 | AA495100 | 2.50 | 0.004 | fa06f01 ICRFzfls cDNA clone | None | ||||
| 29 | BG304232 | 2.46 | 0.008 | Zgc:66107 - similar to Acheron (La ribonucleoprotein domain family, member 6) | Dr.6975 | ||||
| 30 | BI672091 | 2.43 | 0.026 | ft34d03; similar to cytoglobin | Dr.29018 | X | X | ||
| 31 | AI416207 | 2.43 | 0.026 | v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived b | Dr.74192 | X | X | X | multiple rhombomeres |
| 32 | AI601770 | 2.40 | 0.006 | Wu:fc11e07 | Dr.78014 | X | X | X | lateral to hindbrain |
| 33 | BG728637 | 2.37 | 0.006 | Wu:fj66h02 | X | X | |||
| 34 | AF229448 | 2.37 | 0.010 | Jagged 1a | Dr.83677 | X | |||
| 35 | BM071271 | 2.31 | 0.006 | Potassium channel tetramerisation domain containing 12.2 | Dr.84702 | X | X | X | rhombomere 4 |
| 36 | BG891906 | 2.31 | 0.000 | Actin filament associated protein 1-like 1 | Dr.16264 | X | X | X | lateral to hindbrain |
| 37 | BM026691 | 2.28 | 0.048 | Zgc:65997 | Dr.86139 | X | X | ||
| 38 | BM182277 | 2.20 | 0.020 | Matrix metalloproteinase 2 (mmp2) | Dr.76397 | X | X | X | rhombomere 5 |
| 39 | BI710499 | 2.09 | 0.003 | Cellular retinoic acid binding protein 1 (crabp1a) | Dr.83594 | X | X | X | hindbrain vesicle? |
| 40 | BG727376 | 2.08 | 0.014 | Wu:fa10d10 | Dr.73730 | X | X | X | lateral mesoderm |
| 41 | U89380 | 2.07 | 0.014 | Ephrin type-A receptor 4a (epha4a) | Dr.47585 | X | X | X | rhombomere 3/5 |
| 42 | AF277097 | 2.05 | 0.001 | SRY-box containing gene 9b | Dr.114501 | X | |||
| 43 | BI897419 | 2.05 | 0.043 | Limb and neural patterns (lnpa) | Dr.6104 | X | X | ||
| 44 | BG883367 | 2.02 | 0.006 | Transcribed locus; similar to tox3 | Dr.85688 | X | X | X | rhombomere 3/5 |
| 45 | BE605613 | 1.99 | 0.027 | Wu:fj20a04 | Dr.80832 | X | X | ||
| 46 | AI794528 | 1.97 | 0.012 | Zinc finger, CCHC domain containing 24 | Dr.76547 | X | X | ||
| 47 | BG985478 | 1.94 | 0.007 | V-ets erythroblastosis virus E26 oncogene homolog 1a (ets1a) | Dr.98888 | X | X | X | lateral to hindbrain |
| 48 | BI864920 | 1.92 | 0.064 | Wu:fc17g06; similar to ankyrin 2,3/unc44 | Dr.105556 | X | X | X | broad expression |
| 49 | AW420407 | 1.85 | 0.005 | Transcribed locus | Dr.2211 | ||||
| 50 | BM102082 | 1.81 | 0.036 | Sulfatase FP2a | Dr.12108 | X | X | X | broadly in hindbrain |
| 51 | AW077128 | 1.71 | 0.017 | Growth arrest-specific 6 (gas6) | Dr.80253 | X | X | X | rhombomere 5 |
| 52 | BI980238 | 1.65 | 0.179 | Transcribed locus - similar to spectrin | Dr.121661 | X | X | ||
| 53 | AA494787 | 1.34 | 0.412 | Zgc:174862 | Dr.75213 | X | X | ||
| 54 | BI890937 | 1.28 | 0.062 | Zgc:73377; similar to protein phosphatase 1 subunit 14A | Dr.82617 | X | X | X | rhombomere 4 |
Each gene selected for analysis is listed in Table 1.
Genbank number for each clone analyzed.
Average fold increase for each gene across three replicates of meis3 versus meis3+hoxb1b injected embryos.
p-value for the fold increase given in column 3.
Gene name from Genbank and/or ZFIN.
Unigene number for each clone analyzed.
Purchased clones whose sequence matched the sequence deposited on the array are indicated by an X. Other clones were not pursued.
Clones for which in situ probes were successfully synthesized are indicated by an X. Other clones were not pursued.
Clones for which detectable expression patterns were identified are indicated by an X.
Brief description of expression pattern for each clone. Detailed expression patterns are shown in figure 2.
In situ hybridization analysis of candidate genes up-regulated by Hoxb1b
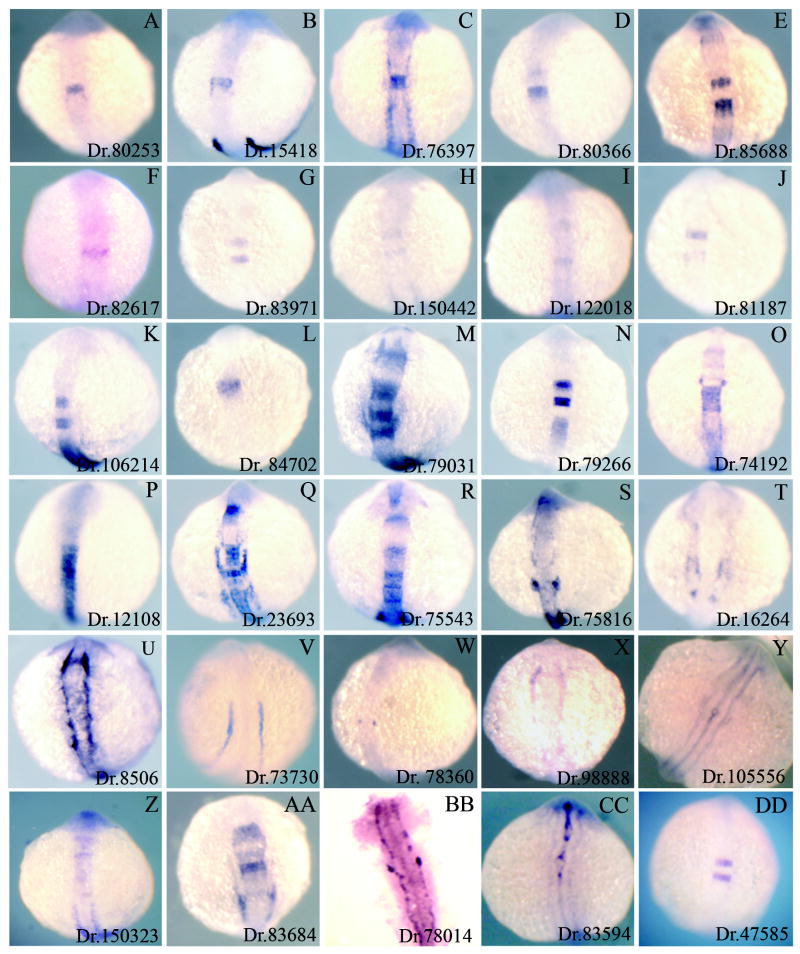
We successfully generated in situ probes from 45 clones and detectable expression patterns were observed for 31 of these, with the remaining 14 showing either no signal or high levels of background staining. We cannot distinguish if the failed in situ hybridizations are due to poor probes, or to low expression levels of the targeted genes, but all failed probes were re-synthesized at least twice. Fig. 2 shows all expression patterns obtained, with the exception of Dr.132502 (hoxb3a), for which the expression pattern is known. We note that at least 21 of the 30 genes in Fig. 2 show expression in the hindbrain (sometimes accompanied by expression elsewhere). Twenty of these genes show expression that is restricted to one, or several, rhombomeres, while one is broadly expressed in the hindbrain (Table 1; Fig. 2P). Of the remaining nine genes, at least seven are expressed next to the neural tube at the level of the hindbrain in structures that may correspond to neural crest or sensory placodes (e.g. Fig. 2S). Hence, since hoxb1b is normally expressed throughout the caudal region of the developing CNS, 28 of the 30 genes for which we have expression data are expressed in a pattern that is compatible with direct or indirect induction by hoxb1b. Only two genes in Fig. 2, Dr.73730 (Fig. 2V) and Dr.105556 (Fig. 2Y), have expression patterns that appear incompatible with either direct or indirect induction by hoxb1b, although we note that both genes are expressed near the hindbrain. We also note that of the 28 hindbrain associated genes shown in Fig. 2, only three (ngf1a/nab, irx5a and ephA4; (Mechta-Grigoriou et al., 2000; Wang et al., 2001; Xu et al., 1994)) have been previously identified as being expressed in the hindbrain, the remaining 25 genes are either novel genes whose expression has not been reported in the published literature, or known genes not previously reported to have hindbrain associated expression. Thus, approximately one half (25/54) of the genes we picked turned out to represent genes not previously known to be hindbrain associated.
Figure 2.
Expression patterns of hoxb1b-regulated genes. 30 genes identified as up-regulated by hoxb1b (see Table 1 and Table S1) were assayed by in situ hybridization to determine their expression patterns. All embryos are at 14hpf and are shown as whole mounts in dorsal view with anterior to the top. The Unigene ID for each gene is given in the lower right of each panel and corresponds to the Unigene IDs in Table 1 and Table S1.
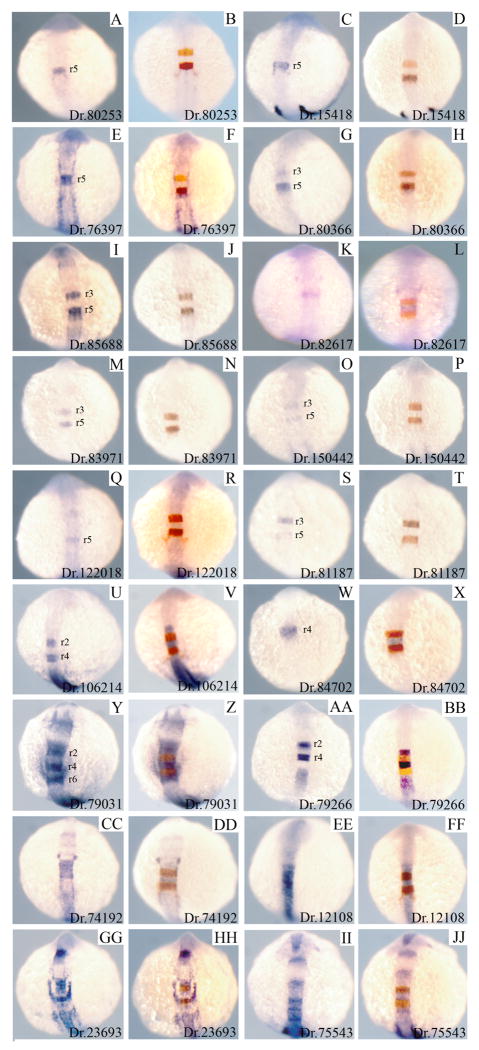
We next examined the detailed expression pattern of genes expressed within the hindbrain. The expression patterns for 18 of the 21 hindbrain genes is shown in Fig. 3 -two known hindbrain genes (iro5/Dr.83684 and ephA4/Dr.47585), as well as one gene whose in situ signal was too weak to permit analysis by double in situ hybridization (Dr.150323) were not analyzed. Of these 18 genes, we find that four are expressed primarily in r5 (Dr. 80253, Dr.15418, Dr.76397, Dr.122018), five primarily in r3/r5 (Dr.80366, Dr.85688, Dr.83971, Dr.150422, Dr.81187), two primarily in r4 (Dr.82617, Dr.84702), two primarily in r2/4 (Dr.106214, Dr.79266) and one primarily in r2/r4/r6 (Dr.79031). The remaining four genes are expressed in multiple adjacent rhombomeres (Dr.74192, Dr.23693, Dr.75543) or throughout the hindbrain (Dr.12108). In summary, at least 1/3 (20/54) of the hoxb1b-induced genes selected for analysis are expressed in rhombomere-restricted patterns during the stage when hindbrain formation takes place.
Figure 3.
Hindbrain expression patterns of hoxb1b-regulated genes. 18 hoxb1b-regulated genes identified as expressed in the hindbrain were assayed by themselves (1st and 3rd column) or as double in situ hybridization together with krox20 (2nd and 4th column; krox20 is expressed in r3 and r5 and is detected in red). Based on the double in situ hybridizations, it was determined which rhombomeres express each of the novel genes and this is indicated in each panel in columns 1 and 3. All embryos are at 14hpf and are shown as whole mounts in dorsal view with anterior to the top. In situ panels in columns 1 and 3 are duplicated from Fig. 2 for ease of comparison.
Many hoxb1b-induced r5/r6 genes act downstream of vhnf1 and valentino
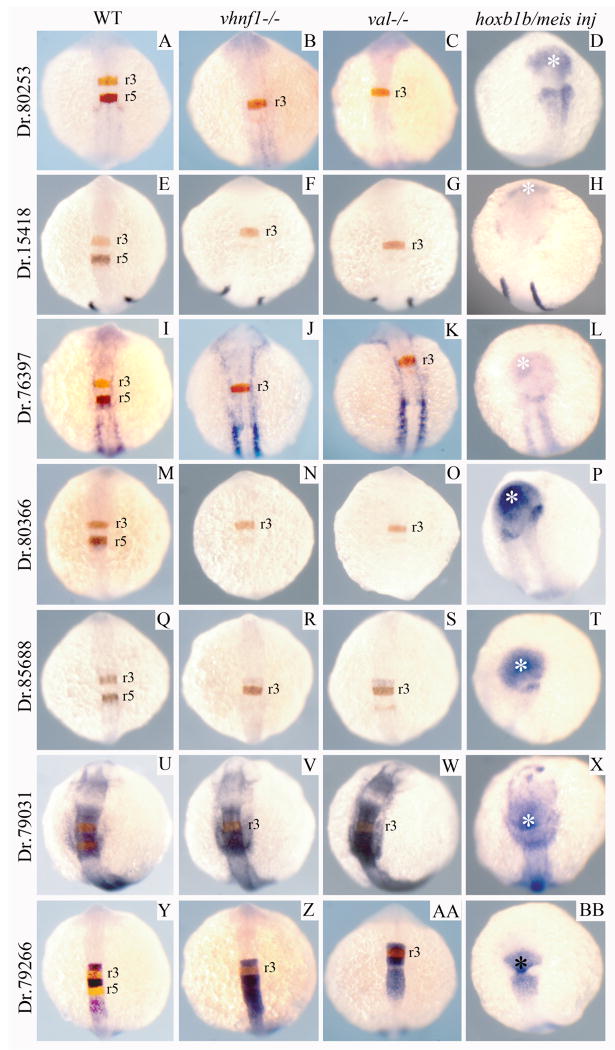
We next made use of existing mutant lines to determine where the newly identified genes act in the genetic hierarchy controlling hindbrain development. Previous analyses have indicated that hoxb1b is required for expression of vhnf1 in r5/r6 (Choe and Sagerstrom, 2004) and that vhnf1 is in turn required for r5/r6 expression of val (Hernandez et al., 2004; Sun and Hopkins, 2001; Wiellette and Sive, 2003). We therefore examined the expression of several of the newly identified genes in vhnf1 and val mutant embryos. We find that three genes expressed in r5 (Dr.80253, Dr.15418, Dr.76397; Fig. 4A-L) and two genes expressed in r3/r5 (Dr. 80366, Dr. 85688; Fig 4M-T) are not expressed in r5 of vhnf1 and val mutants. This finding indicates that these five genes act downstream of vhnf1 and val.
Figure 4.
Expression of hoxb1b-regulated genes in vhnf1 mutant, valentino mutant and hoxb1b/meis3-injected embryos. Seven hoxb1b-regulated genes were analyzed for their expression in wild type (A, E, I, M, Q, U, Y), vhnf1 mutant (B, F, J, N, R, V, Z), valentino mutant (C, G, K, O, S, W, AA) or hoxb1b+meis3-injected (D, H, L, P, T, X, BB) embryos. Genes were analyzed as double in situ hybridizations with krox20 (A-C, E-G, I-K, M-O, Q-S, U-W, Y-AA; krox20 detected in red) or as single in situ hybridizations (D, H, L, P, T, X, BB). All embryos are at 14hpf and are shown as whole mounts in dorsal view with anterior to the top. In situ panels in the left-hand column are duplicated from Fig. 3 for ease of comparison.
We also examined the expression of Dr.79031 (that is expressed in r2/r4/r6) and find a contiguous domain of Dr.79031 expression caudal to r3 in vhnf1 and val mutant embryos (Fig. 4U-W). This domain appears larger than just r4, suggesting that r6 expression of Dr.79031 persists in vhnf1 and val mutants. Similarly, expression of Dr.79266 (that is expressed in r2/r4) is unaffected in r2/r4 in vhnf1 and val mutants, but expression appears to expand caudally in both mutants (Fig. 4Y-BB).
Lastly, we confirmed that the newly identified genes are regulated by hoxb1b. We injected hoxb1b/meis3 mRNA and detect ectopic expression in the anterior embryo of all genes tested (Fig. 4D, H, L, P, T, X, BB). In addition, disrupting the activity of Hox cofactors from the Pbx and Meis families prevents expression of the newly identified hindbrain genes (Fig. S1B, D, F, H). Taken together, these results indicate that the genes identified in our screen are indeed hoxb1b regulated.
The ppp1r14al gene regulates formation of the caudal hindbrain
We next set out to determine if the novel hindbrain genes identified in our expressionprofiling screen are required for hindbrain development. We selected the r4-restricted Dr.82617 gene, since its expression in a single rhombomere simplifies functional analysis and since a full-length clone was readily available. Analysis of the Dr.82617 sequence revealed similarity to Protein phosphatase 1 regulatory subunit 14a. Protein phosphatase 1 (PPP1) is one of the main serine/threonine phosphatases in eukaryotic cells (reviewed in (Cohen, 2002)). While there are four PPP1 isoforms in most mammals, these are broadly expressed and show high sequence identity, suggesting that the various PPP1 proteins display similar substrate specificities. Accordingly, all five Ppp1 genes identified in zebrafish to date (ppp1caa, ppp1cab, ppp1cb, ppp1cbl and ppp1cc) are broadly expressed (ppp1 expression data were retrieved from the Zebrafish Information Network (ZFIN), University of Oregon, Eugene, OR 97403-5274; World Wide Web URL: http://zfin.org/; 5/17/10). Instead, substrate specificity is provided when a single PPP1 catalytic subunit (PPP1c) associates with one of a large number of available regulatory (PPP1r) subunits (reviewed in (Cohen, 2002)). Ppp1r14a is one such regulatory subunit that was initially identified as CPI-17 with the ability to inhibit PPP1c activity in smooth muscle. Notably, the inhibitory activity of Ppp1r14a is further enhanced by PKC-mediated phosphorylation of Ppp1r14a itself (Eto et al., 1995; Eto et al., 1997).
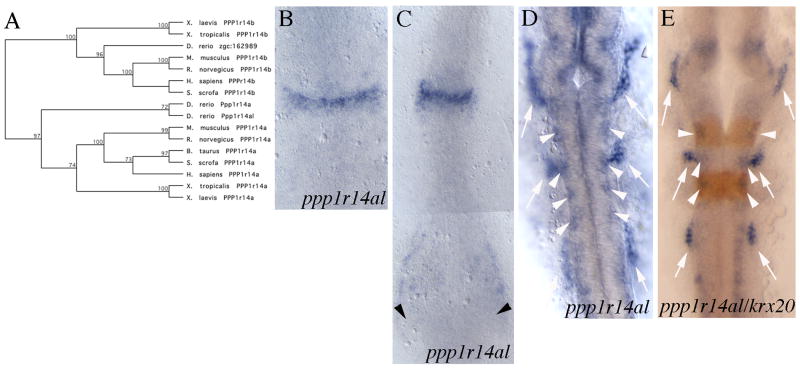
Although our analysis identified Dr.82617 as Ppp1r14a, a distinct zebrafish gene (Unigene Dr.14203) had been previously designated Ppp1r14a. Phylogenetic analysis indicates that the two genes cluster together with Ppp1r14a proteins from other species (Fig. 5A), suggesting that they may have resulted from a previously reported duplication of the zebrafish genome. As a result, we have designated Dr.82617 as ppp1r14a-like (ppp1r14al).
Figure 5.
Sequence and expression analysis of the ppp1r14al gene. (A) Phylogenetic tree demonstrating that zebrafish ppp1r14al groups with other ppp1r14a genes, not with ppp1r14b genes. (B-E) ppp1r14al expression pattern at 9hpf (B), 11hpf (C) and 20hpf (D, E). Arrows indicate cranial placodes (D, E) and arrowheads indicate staining in ventral mesoderm (C) and in the lateral neural tube (D, E). E is a double in situ hybridization with krox20 detected in red. Embryos in B-E are shown as flat mounts in dorsal view with anterior to the top.
Our initial analysis of genes from the expression screen identified ppp1r14al as being expressed in r4 (Fig. 2, 3). A more detailed analysis confirmed this finding at early somitogenesis stages (12-14hpf; Fig. 5B, C) and also revealed expression in ventral mesoderm (arrowheads in Fig. 5C). Thereafter, ppp1r14al expression is no longer r4-specific, but persists in the lateral neural tube (arrowheads in Fig. 5D, E) and also becomes detectable in cranial ganglia (arrows in Fig. 5D, E). In contrast, ppp1r14a (Dr.14203) does not show restricted gene expression at early stages of development and is not expressed in the hindbrain (ppp1r14a expression data were retrieved from the Zebrafish Information Network (ZFIN), University of Oregon, Eugene, OR 97403-5274; World Wide Web URL: http://zfin.org/; 5/17/10), but is expressed in intestinal smooth muscle by 3dpf (Georgijevic et al., 2007).
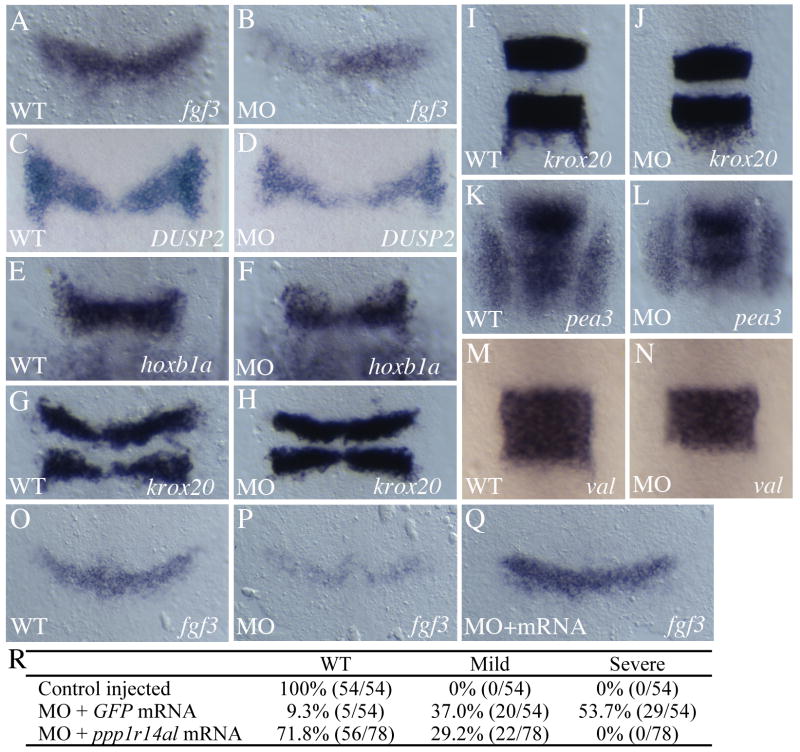
To examine the role of ppp1r14al in zebrafish development, we made use of MOs targeting the ppp1r14al translation initiation site. We find that disrupting ppp1r14al function leads to reduced fgf3 expression in r4 at the end of gastrulation (10.5hpf; Fig. 6A, B). We also observe modest reductions in the r4 expression of dual specificity phosphatase 2 (DUSP2; Fig. 6C, D) and hoxb1a (Fig 6E, F), but not of ephrinB2, cyp26b or irx7 (Fig. S2A-F). Expression levels of krox20 in r3 and r5 are not affected, although the r3 and r5 expression domains may be somewhat closer together in MO-injected embryos (Fig. 6G-J). Since fgf3 appears to be the earliest gene affected upon interfering with ppp1r14al function, we examined expression of pea3, a known Fgf-target (Raible and Brand, 2001). We find that the size of the pea3 expression domain is reduced somewhat (Fig. 6K, L), consistent with impaired Fgf-signaling in MO-injected embryos. We also observe a slight reduction in the size of the val domain in r5/r6 (Fig. 6M, N), consistent with previous reports that val expression is Fgf-dependent (Hernandez et al., 2004; Wiellette and Sive, 2003). Lastly, we co-injected MOs with ppp1r14al mRNA mutated to be refractory to the MOs. We find that ppp1r14al mRNA efficiently rescues the MO phenotype at 10.5hpf (Fig. 6O-R), confirming the specificity of the MOs. In particular, less than 10% of MO-injected embryos show normal fgf3 expression and over 50% are severely affected (Fig. 6P, R). This situation is reversed when ppp1r14al mRNA is co-injected with the MO, leading to no embryos with a severe phenotype and more than 70% with strong fgf3 expression (Fig. 6Q, R).
Figure 6.
Disruption of ppp1r14al leads to loss of fgf3 expression. (A-N) Control (A, C, E, G, I, K, M) and ppp1r14al MO-injected (B, D, F, H, J, L, N) embryos were assayed for expression of fgf3 at 10.5hpf (A, B), DUSP2 at 10.5 hpf (C, D), hoxb1a at 11.5 hpf (E, F) krox20 at 10.5hpf (G, H) and at 12hpf (I, J), pea3 at 12hpf (K, L) and valentino at 12.5hpf (M, N). (O-R) Analysis of fgf3 expression in control (O), ppp1r14al MO-injected (P), ppp1r14al MO + ppp1r14al mRNA injected (Q; note that mRNA is mutated to resist MO effect) reveals rescue of the ppp1r14al MO phenotype at 10.5hpf. Actual data is given in R. Phenotypes were scored as severe (similar to panel P), or mild (similar to panel B). Embryos in A-Q are shown as flat mounts in dorsal view with anterior to the top.
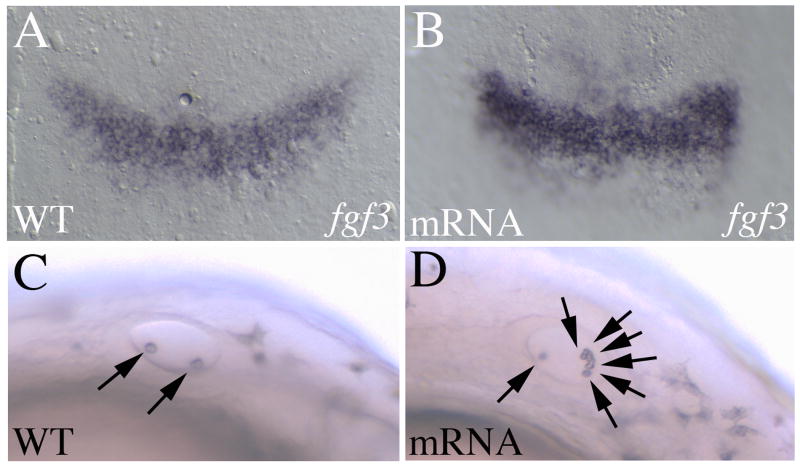
We next examined the effect of ppp1r14al overexpression and observe elevated fgf3 expression in r4 (Fig. 7A, B), further supporting a role for ppp1r14al in the regulation of fgf3 expression. Notably, ppp1r14al overexpression also affects formation of the otic vesicle, detected as an increased number of otoliths (Fig. 7C, D), consistent with the known role for fgf3 in otic vesicle formation (Kwak et al., 2002; Maroon et al., 2002; Phillips et al., 2001).
Figure 7.
Overexpression of ppp1r14al leads to elevated fgf3 expression. Control (A, C) or ppp1r14al mRNA-injected (B, D) embryos were assayed for fgf3 expression (A, B), or observed under brightfield for the presence of otoliths (C, D). A, B are shown as flat mounts in dorsal view with anterior to the top. C, D are live embryos in lateral view with anterior to the left. Arrows in C, D indicate otoliths.
Since ppp1r14al is expressed in lateral cells in several rhombomeres (arrowheads in Fig. 5D, E), as well as in cranial ganglia (arrows in Fig. 5D, E), at later stages, we also examined gene expression in those tissues in later-stage MO-injected embryos. We find that expression of ascl1a (zash1a), which is regulated by fgf3 in the forebrain (Walshe and Mason, 2003), is down regulated in MO-injected embryos already by mid-somitogenesis (13hpf; Fig. S3A, B). Similarly, ngn1, fgf20 and pax2a (Krauss et al., 1991; Liao et al., 1999; Whitehead et al., 2005), three genes expressed laterally within the neural tube, are down regulated in MO-injected embryos by late somitogenesis (22-24hpf; Fig. S3C-H). This effect appears to be specific to the hindbrain, since ngn1 and pax2 expression at the MHB is largely unaffected (asterisks in Fig. S3C, D, G, H). Notably, dlx2 expression in the cranial ganglia (arrows in Fig. S3I, J) is down regulated only modestly. While it is possible that this role for ppp1r14al is independent of its role in r4, a recent lineage tracing analysis in the mouse (Makki and Capecchi, 2010) revealed that numerous structures outside the hindbrain (including the cranial ganglia) derive from cells that initially express hoxa1 (the murine hoxb1b homolog). Hence, hoxb1b may regulate ppp1r14al expression at most of the embryonic sites that we observe in this analysis.
When observing MO-injected embryos at later stages of development, we also noticed increased cell death in the neural tube (Fig. S4). Acridine orange staining confirmed increased cell death in the hindbrain and anterior neural tube of MO-injected embryos at 12-13hpf (Fig. S4B). Co-injection of MOs targeted to p53 does not rescue this cell-death (not shown), demonstrating that it is not due to p53-mediated MO-toxicity. In contrast, using the same rescue strategy as in Fig. 6, we find that ppp1r14al mRNA rescues the cell death (Fig. S4C). Notably, acridine orange staining of 9hpf MO-injected embryos was indistinguishable from that of control embryos (not shown), indicating that the reduction in fgf3 expression precedes the increased cell death and suggesting that cell death is a later consequence of loss of ppp1r14al and/or fgf3 activity.
Simultaneous disruption of ppp1r14al and fgf8 interferes with r5/r6 formation
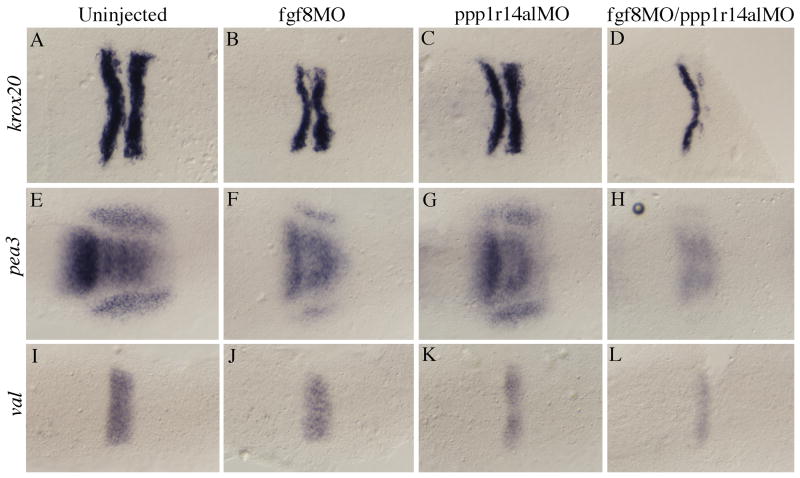
The experiments in Fig. 6 reveal a robust effect of ppp1r14al on fgf3 expression, but modest effects on the expression of other hindbrain genes. This observation suggests that reduced fgf3 expression does not have profound effects on overall hindbrain development. Indeed, it has been previously reported that fgf3 acts redundantly with fgf8 in a r4 signaling center to regulate formation of r5/r6 (Maves et al., 2002; Walshe et al., 2002). In particular, individual disruption of fgf8 or fgf3 function (using MOs and/or the acerebellar line that carries a fgf8 mutation) produces modest phenotypes, while simultaneous disruption of fgf8 and fgf3 function leads to significant loss of krox20 expression in r5 and valentino expression in r5/r6 (Maves et al., 2002; Walshe et al., 2002).
Since fgf3 expression is reduced (Fig. 6), but fgf8 expression is unaffected (not shows) in embryos injected with ppp1r14l MO, we reasoned that simultaneous disruption of ppp1r14al and fgf8 might produce a phenotype similar to simultaneous disruption of fgf3 and fgf8. As reported (Maves et al., 2002; Walshe et al., 2002), we find that fgf8 disruption has a modest effect on pea3, krox20 and val expression (Fig. 8B, F, J). This effect is similar to that observed upon disruption of ppp1r14al (Fig. 6; Fig. 8C, G, K). In contrast, co-injection of ppp1r14al and fgf8 MOs causes pronounced reduction in the expression of pea3, krox20 and val (Fig. 8D, H, L). This latter phenotype is similar to that reported for simultaneous loss of fgf3 and fgf8 (Maves et al., 2002; Walshe et al., 2002), supporting the notion that ppp1r14al acts by regulating fgf3 expression.
Figure 8.
Combined loss of ppp1r14al and fgf8 function leads to disruption of the r4 signaling center. Uninjected (A, E, I), fgf8 MO-injected (B, F, J), ppp1r14al MO-injected (C, G, K) or fgf8 + ppp1r14al MO-injected embryos (D, H, L) were assayed for expression of krox20 (A-D), pea3 (E-H) or valentino (I-L) by in situ hybridization. All embryos are dorsal views at 11hpf with anterior to the left.
ppp1r14al expression is regulated by Hoxb1b
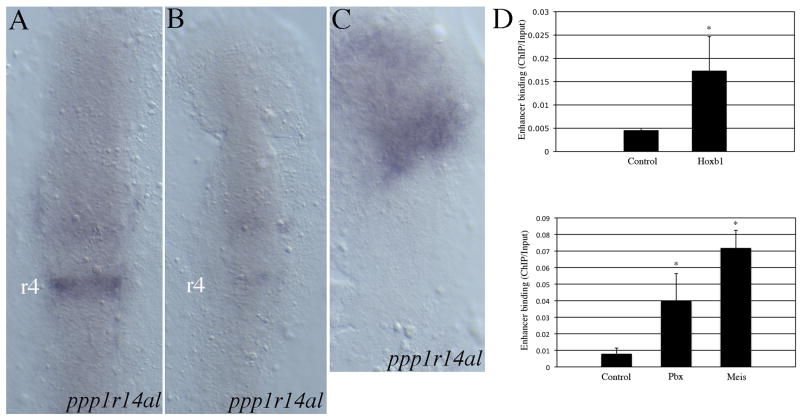
We next determined if ppp1r14al expression requires Hoxb1b activity. As discussed, Hoxb1b functions in a complex that also includes Meis and Pbx cofactors and we have found that such complexes cannot form in the absence of Meis proteins. Hence, we used a dominant negative Meis construct (Choe et al., 2002) to disrupt Meis/Pbx/Hoxb1b complexes during zebrafish embryogenesis and find that ppp1r14al expression is lost (Fig. 9B). We also find that injection of meis3, pbx4 and hoxb1b mRNA induces ectopic expression of ppp1r14al in the anterior embryo (Fig. 9C), as expected based on the design of our screen. Lastly, we identified a Meis/Pbx/Hox binding element in the first intron of the ppp1r14al gene and, using ChIP analysis, we find that endogenous Meis, Pbx and Hoxb1 proteins occupy this element in wild type embryos (Fig. 9D).
Figure 9.
ppp1r14al is regulated by Hoxb1b together with Pbx and Meis cofactors. (A-C) ppp1r14al expression was analyzed in control (A), dominant negative Meis-injected (B) and hoxb1b/pbx4/meis3-injected (C) embryos. Embryos in A-C are flat mounts at 11hpf in dorsal view with anterior to the top. (D) Chromatin immunoprecipitation from 11 hpf zebrafish embryos revealed binding of Hoxb1b (top panel), Meis and Pbx (bottom panel) proteins to the regulatory region of ppp1r14al. Asterisks indicate values that are statistically different (p < 0.05) from the control value based on Student's T-test.
Discussion
In this report we use ectopic expression of hoxb1b/meis3 followed by dissection and microarray analysis to isolate novel hoxb1b-regulated hindbrain genes. A total of 100 genes fulfilled our criteria of at least a 2-fold induction with a p-value under 0.05 over three independent experiments. 54 genes were characterized further and we find that 25 of these represent novel genes or known genes not previously reported as expressed in the hindbrain. This fraction of novel hindbrain genes (25/54) may in fact be an underestimate since only 31 of the 54 genes gave detectable expression patterns by in situ hybridization. Specifically, while some of the genes that failed to show an expression pattern are likely to represent true false positives (i.e. genes identified as up-regulated by hoxb1b when they are really not), other genes may have failed to show an expression pattern for technical reasons (such as poor probe synthesis) and may in fact be regulated by hoxb1b. We also note that there is little correlation between the fold induction of a gene by hoxb1b in our assay and the likelihood that the gene will be expressed in the hindbrain. This is clear from Table 1, which shows that 12 of the genes induced by more than 3-fold display hindbrain specific expression and 10 of the genes induced by only 2-3 fold are expressed in the hindbrain. This lack of correlation is also illustrated by the fact that ephA4a, a known gene expressed in r1, r3 and r5, was induced only 2.071-fold by ectopic hoxb1b/meis3 expression. Lastly, since the 54 genes we analyzed were not biased to the most highly induced ones, but were selected to represent the full range of observed fold induction and p-values, it is likely that genes in Table S1 that were not analyzed will harbor a similar fraction (∼50%) of hindbrain expressed genes.
Two recent studies used microarray analysis to isolate PG1 hox-regulated genes. Rohrschneider et al compared gene expression between wild type and hoxb1a-depleted rhombomere 4 (Rohrschneider et al., 2007) and successfully identified 12 genes regulated by hoxb1a in r4. There are several differences between our study and the Rohrschneider study. First, they focused on a different PG1 hox gene (hoxb1a versus hoxb1b). Second, our study used gain of function (ectopic hoxb1b expression), while Rohrschneider used loss of function (hoxb1a depletion by morpholino injection). Third, our study would identify any gene activated by hoxb1b in the caudal hindbrain (r4-r7), while Rohrschneider focused on hoxb1a-regulated genes in r4. In spite of these differences, our screen identified two of the 12 genes from the Rohrschneider screen (collagen VIIa and calretinin). We conclude that, taken together, these two screens provide a comprehensive analysis of PG1 target genes in the hindbrain. In a distinct study, van den Akker et al used ectopic hoxb1b expression to identify target genes (van den Akker et al., 2010). However, this study analyzed embryos prior to gastrulation (40% epiboly) - well before the onset of neural development - and did not co-express hox cofactors. As a result, none of the highest-scoring hoxb1b-regulated genes identified by van den Akker et al appear in our dataset.
In order to determine if the genes identified in our screen are important for hindbrain development, we analyzed the ppp1r14al gene in detail. ppp1r14al encodes a regulatory subunit that associates with protein phosphatase 1 (PPP1) to modulate its activity, but ppp1r14a was originally identified as inhibiting PPP1 activity in smooth muscle (Eto et al., 1995) and has not been previously implicated in hindbrain development. Given the ubiquitous expression and broad substrate specificity of PPP1 proteins, it is difficult to identify a likely substrate for the PPP1/Ppp1r14al complex in the hindbrain and we instead examined a panel of r4 genes to determine if they were affected by loss of ppp1r14al. Since ppp1r14al is predicted to act downstream of PG1 hox genes, it is not surprising that we see only modest effects on the expression of hoxb1a or the size of r4 in ppp1r14al-depleted embryos. In contrast, we observe robust reduction of fgf3 expression in r4 of pp1r14al-depleted embryos. This is in agreement with previous reports indicating that fgf3 acts downstream of PG1 hox genes in r4 (Waskiewicz et al., 2002). We note that, while fgf signaling is known to regulate formation of r5 and r6 (Maves et al., 2002; Walshe et al., 2002), we observe only minor effects on valentino expression in r5 of ppp1r14al-depleted embryos. This is likely due to the fact that fgf8, which acts together with fgf3 to regulate r5/r6, is unaffected in ppp1r14al-depleted embryos. Indeed, apart from the robust loss of fgf3 expression, the ppp1r14al loss of function phenotype closely resembles the subtle effect of loss of fgf3 reported by Walshe et al (Walshe et al., 2002). By simultaneously disrupting both fgf8 and ppp1r14al, we observe a more pronounced phenotype with clear loss of krox20, pea3 and val expression – similar to the phenotype reported for simultaneous disruption of fgf3 and fgf8 (Maves et al., 2002; Walshe et al., 2002). We also observe increased cell death at later stages in ppp1r14al MO-injected embryos. Since the cell death occurs subsequent to the loss of fgf3 expression and since co-injection of ppp1r14al mRNA (which restores fgf3 expression) rescues the cell death, our data are consistent with the cell death being due to loss of fgf3, although we can not formally rule out the possibility that increased r4 expression of survival factors other than fgf3 may also be induced in the rescue experiments. Taken together, our data suggest a role for ppp1r14al upstream of fgf3 in the establishment of the key r4 signaling center during early hindbrain development. Notably, while we demonstrate that ppp1r14al is likely directly regulated by Hoxb1, Meis and Pbx, we do not know how ppp1r14al regulates fgf3 expression. Intriguingly, fgf3 expression is regulated by GATA4 in other systems (Murakami et al., 1999) and GATA4 activity is modulated by phosphorylation on Ser-105 (Kitta et al., 2003), making GATA4 a potential substrate for Ppp1r14al.
Lastly, we note that our screen does not select specifically for genes that are directly regulated by Hoxb1b. Accordingly, r5 expression of a number of genes identified in the screen is lost in vhnf1 and val mutant embryos. Since vhnf1 and val both act downstream of PG1 hox genes (Choe and Sagerstrom, 2004; Choe et al., 2002; Waskiewicz et al., 2001; Waskiewicz et al., 2002), this finding suggests that such genes are indirectly regulated by Hoxb1b, although it remains possible that they receive combinatorial input from Hoxb1b and vHnf1 and/or Val.
Supplementary Material
Figure S1. Disruption of Pbx and Meis prevents expression of four novel hindbrain genes. Control embryos (A, C, E, G) and embryos co-injected with a dominant negative Meis construct (PBCAB) together with anti-Pbx MOs (B, D, F, H) were analyzed for expression of Dr. 15418, Dr. 85688, Dr.156272 and Dr.80253. Embryos are shown as flat mounts in dorsal view with anterior to the left.
Figure S2. Expression of several r4 genes is independent of ppp1r14al. Control (A, C, E) and ppp1r14al MO-injected (B, D, F) embryos were assayed for expression of ephrinB2 (A, B), cyp26b (C, D) and irx7 (E, F). Embryos are shown as flat mounts in dorsal view with anterior to the top. Arrowheads point to r4.
Figure S3. ppp1r14al regulates neurogenesis. (A-J). Control (A, C, E, G, I) or ppp1r14al MO-injected (B, D, F, H, J) embryos were analyzed for expression of ascl1a (A, B), ngn1 (C, D), fgf20 (E, F), pax2a (G, H) and dlx2 (I, J). Embryos are shown as flat mounts in dorsal view with anterior to the top. Asterisks in C, D, G, H indicate the midbrain-hindbrain boundary. Arrows in I, J indicate cranial ganglia.
Figure S4. Cell death is observed in later stage ppp1r14al MO-injected embryos. Acridine orange staining of control (A), ppp1r14al MO-injected (B) and ppp1r14al MO + ppp1r14al mRNA-injected (C) embryos revealed rescue of ppp1r14al MO-mediated cell death. Live embryos are shown in lateral view with anterior to the left. White lines indicate the approximate position of the hindbrain.
Research Highlights.
> hox genes are required for hindbrain formation, but hox targets are largely unknown > We identify 100 genes up-regulated by zebrafish hoxb1b > Of 54 genes analyzed further, 25 represent novel hindbrain genes > Expression of the ppp1r14al gene is directly regulated by hoxb1b in rhombomere 4 > ppp1r14al encodes a protein phosphatase subunit and regulates fgf3 expression
Acknowledgments
We are grateful to Rahul Bharadway, Brian Johnston and Joe Boyd for their initial assistance with the analysis of hoxb1b-induced genes and to Drs. P. diIorio, C. Moens and T. Piotrowski for plasmids. We wish to acknowledge Letitiah Etheridge for technical assistance and expert animal care. This work was supported by grant R01 NS038183 to CGS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Alexandre D, Clarke JD, Oxtoby E, Yan YL, Jowett T, Holder N. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid-induced phenotype. Development. 1996;122:735–746. doi: 10.1242/dev.122.3.735. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Bosley TM, Salih MA, Alorainy IA, Oystreck DT, Nester M, Abu-Amero KK, Tischfield MA, Engle EC. Clinical characterization of the HOXA1 syndrome BSAS variant. Neurology. 2007;69:1245–1253. doi: 10.1212/01.wnl.0000276947.59704.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LA, Amores A, Schilling TF, Jowett T, Baert JL, de Launoit Y, Sharrocks AD. Molecular characterization of the zebrafish PEA3 ETS-domain transcription factor. Oncogene. 1998;17:93–104. doi: 10.1038/sj.onc.1201911. [DOI] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR. Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development. 1993;118:1063–1075. doi: 10.1242/dev.118.4.1063. [DOI] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerström CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Choe SK, Sagerstrom CG. Paralog group 1 hox genes regulate rhombomere 5/6 expression of vnhf1, a repressor of rostral hindbrain fates, in a meis-dependent manner. Developmental Biology. 2004;271:350–361. doi: 10.1016/j.ydbio.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Eto M, Ohmori T, Suzuki M, Furuya K, Morita F. A novel protein phosphatase-1 inhibitory protein potentiated by protein kinase C. Isolation from porcine aorta media and characterization. J Biochem. 1995;118:1104–1107. doi: 10.1093/oxfordjournals.jbchem.a124993. [DOI] [PubMed] [Google Scholar]
- Eto M, Senba S, Morita F, Yazawa M. Molecular cloning of a novel phosphorylation-dependent inhibitory protein of protein phosphatase-1 (CPI17) in smooth muscle: its specific localization in smooth muscle. FEBS Lett. 1997;410:356–360. doi: 10.1016/s0014-5793(97)00657-1. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128:3017–3027. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- Georgijevic S, Subramanian Y, Rollins EL, Starovic-Subota O, Tang AC, Childs SJ. Spatiotemporal expression of smooth muscle markers in developing zebrafish gut. Dev Dyn. 2007;236:1623–1632. doi: 10.1002/dvdy.21165. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Rossel M, Manley NR, Capecchi MR. Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VII nerve. Development. 1996;122:3217–3226. doi: 10.1242/dev.122.10.3217. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Kiefer P, Strahle U, Dickson C. The zebrafish Fgf-3 gene: cDNA sequence, transcript structure and genomic organization. Gene. 1996;168:211–215. doi: 10.1016/0378-1119(95)00736-9. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullman B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kitta K, Day RM, Kim Y, Torregroza I, Evans T, Suzuki YJ. Hepatocyte growth factor induces GATA-4 phosphorylation and cell survival in cardiac muscle cells. The Journal of biological chemistry. 2003;278:4705–4712. doi: 10.1074/jbc.M211616200. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A. The evolution and maintenance of Hox gene clusters in vertebrates and the teleost-specific genome duplication. Int J Dev Biol. 2009;53:765–773. doi: 10.1387/ijdb.072533km. [DOI] [PubMed] [Google Scholar]
- Kwak SJ, Phillips BT, Heck R, Riley BB. An expanded domain of fgf3 expression in the hindbrain of zebrafish valentino mutants results in mis-patterning of the otic vesicle. Development. 2002;129:5279–5287. doi: 10.1242/dev.129.22.5279. [DOI] [PubMed] [Google Scholar]
- Kwak SJ, Vemaraju S, Moorman SJ, Zeddies D, Popper AN, Riley BB. Zebrafish pax5 regulates development of the utricular macula and vestibular function. Dev Dyn. 2006;235:3026–3038. doi: 10.1002/dvdy.20961. [DOI] [PubMed] [Google Scholar]
- Lewis EB. Homeosis: the first 100 years. Trends Genet. 1994;10:341–343. doi: 10.1016/0168-9525(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Li Y, Allende ML, Finkelstein R, Weinberg ES. Expression of two zebrafish orthodenticle-related genes in the embryonic brain. Mech Dev. 1994;48:229–244. doi: 10.1016/0925-4773(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Liao J, He J, Yan T, Korzh V, Gong Z. A class of neuroD-related basic helix-loop-helix transcription factors expressed in developing central nervous system in zebrafish. DNA Cell Biol. 1999;18:333–344. doi: 10.1089/104454999315394. [DOI] [PubMed] [Google Scholar]
- Maconochie MK, Nonchev S, Studer M, Chan SK, Popperl H, Sham MH, Mann RS, Krumlauf R. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- Makki N, Capecchi MR. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, Gorry P, Lumsden A, Chambon P. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–338. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–2108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–2484. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F, Garel S, Charnay P. Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development. 2000;127:119–128. doi: 10.1242/dev.127.1.119. [DOI] [PubMed] [Google Scholar]
- Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- Moens CB, Yan YL, Appel B, Force AG, Kimmel CB. valentino: a zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–3990. doi: 10.1242/dev.122.12.3981. [DOI] [PubMed] [Google Scholar]
- Murakami A, Thurlow J, Dickson C. Retinoic acid-regulated expression of fibroblast growth factor 3 requires the interaction between a novel transcription factor and GATA-4. The Journal of biological chemistry. 1999;274:17242–17248. doi: 10.1074/jbc.274.24.17242. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx20) and its expression during development. Nucl Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes:expression in the hindbrain region of wild-type and mutants of the segmentation gene valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–5040. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2005;37:1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Tumpel S, Cambronero F, Ferretti E, Blasi F, Wiedemann LM, Krumlauf R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol. 2007;302:646–660. doi: 10.1016/j.ydbio.2006.10.029. [DOI] [PubMed] [Google Scholar]
- van den Akker WM, Durston AJ, Spaink HP. Identification of hoxb1b downstream genes: hoxb1b as a regulatory factor controlling transcriptional networks and cell movement during zebrafish gastrulation. Int J Dev Biol. 2010;54:55–62. doi: 10.1387/ijdb.082678wv. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Choe SK, Sagerström CG. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Ellstrom DR, Sagerström CG. A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev Dyn. 2000;217:109–119. doi: 10.1002/(SICI)1097-0177(200001)217:1<109::AID-DVDY10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of Hindbrain Segmental Identity Requires Signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/s0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- Walshe J, Mason I. Unique and combinatorial functions of Fgf3 and Fgf8 during zebrafish forebrain development. Development. 2003;130:4337–4349. doi: 10.1242/dev.00660. [DOI] [PubMed] [Google Scholar]
- Wang X, Emelyanov A, Sleptsova-Friedrich I, Korzh V, Gong Z. Expression of two novel zebrafish iroquois homologues (ziro1 and ziro5) during early development of axial structures and central nervous system. Mech Dev. 2001;105:191–195. doi: 10.1016/s0925-4773(01)00400-2. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3:723–733. doi: 10.1016/s1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Wassef MA, Chomette D, Pouilhe M, Stedman A, Havis E, Desmarquet-Trin Dinh C, Schneider-Maunoury S, Gilardi-Hebenstreit P, Charnay P, Ghislain J. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development. 2008;135:3369–3378. doi: 10.1242/dev.023614. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wiellette EL, Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130:3821–3829. doi: 10.1242/dev.00572. [DOI] [PubMed] [Google Scholar]
- Xu Q, Holder N, Patient R, Wilson SW. Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development. 1994;120:287–299. doi: 10.1242/dev.120.2.287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Disruption of Pbx and Meis prevents expression of four novel hindbrain genes. Control embryos (A, C, E, G) and embryos co-injected with a dominant negative Meis construct (PBCAB) together with anti-Pbx MOs (B, D, F, H) were analyzed for expression of Dr. 15418, Dr. 85688, Dr.156272 and Dr.80253. Embryos are shown as flat mounts in dorsal view with anterior to the left.
Figure S2. Expression of several r4 genes is independent of ppp1r14al. Control (A, C, E) and ppp1r14al MO-injected (B, D, F) embryos were assayed for expression of ephrinB2 (A, B), cyp26b (C, D) and irx7 (E, F). Embryos are shown as flat mounts in dorsal view with anterior to the top. Arrowheads point to r4.
Figure S3. ppp1r14al regulates neurogenesis. (A-J). Control (A, C, E, G, I) or ppp1r14al MO-injected (B, D, F, H, J) embryos were analyzed for expression of ascl1a (A, B), ngn1 (C, D), fgf20 (E, F), pax2a (G, H) and dlx2 (I, J). Embryos are shown as flat mounts in dorsal view with anterior to the top. Asterisks in C, D, G, H indicate the midbrain-hindbrain boundary. Arrows in I, J indicate cranial ganglia.
Figure S4. Cell death is observed in later stage ppp1r14al MO-injected embryos. Acridine orange staining of control (A), ppp1r14al MO-injected (B) and ppp1r14al MO + ppp1r14al mRNA-injected (C) embryos revealed rescue of ppp1r14al MO-mediated cell death. Live embryos are shown in lateral view with anterior to the left. White lines indicate the approximate position of the hindbrain.