Abstract
Co-divergence between host and parasites suggests that evolutionary processes act across similar spatial and temporal scales. Although there has been considerable work on the extent and correlates of co-divergence of RNA viruses and their mammalian hosts, relatively little is known about the extent to which virus evolution is determined by the phylogeographic history of host species. To test hypotheses related to co-divergence across a variety of spatial and temporal scales, we explored phylogenetic signatures in Andes virus (ANDV) sampled from Chile and its host rodent, Oligoryzomys longicaudatus. ANDV showed strong spatial subdivision, a phylogeographic pattern also recovered in the host using both spatial and genealogical approaches, and despite incomplete lineage sorting. Lineage structure in the virus seemed to be a response to current population dynamics in the host at the spatial scale of ecoregions. However, finer scale analyses revealed contrasting patterns of genetic structure across a latitudinal gradient. As predicted by their higher substitution rates, ANDV showed greater genealogical resolution than the rodent, with topological congruence influenced by the degree of lineage sorting within the host. However, despite these major differences in evolutionary dynamics, the geographic structure of host and virus converged across large spatial scales.
Keywords: Chile, co-divergence, hantavirus, O. longicaudatus, phylogeography, spatial genetics
Introduction
Host-parasite interactions occur across several levels of organization (Mideo et al. 2008), and are influenced by processes that act at various temporal scales (Archie et al. 2009; Pybus& Rambaut 2009). Changes in host ecology, population dynamics, and evolutionary processes are all associated with changes in parasite population structure. Among parasites, RNA viruses represent an important group of emerging human pathogens. High mutation rates, large population sizes, and short generation times mean that RNA viruses evolve extremely rapidly (Holmes 2009b), with viral phylogenies often providing information about temporal and spatial dynamics (Holmes 2004). Therefore, RNA viruses typically exhibit far greater phylogenetic resolution than that of their hosts. For example, feline immunodeficiency virus (FIV) provided a more detailed view of demographic expansion in cougars (Puma concolor), particularly in recent time, than could be inferred from host data alone (Biek et al. 2006).
Distinct evolutionary processes act to shape the pattern of host-parasite congruence at different hierarchical scales. Intimate interactions between the two organisms, combined with vertical transmission of the parasite through host generations, are key features that should increase the likelihood of shared common histories (Nieberding& Olivieri 2007). At the phylogeographic scale, founder, divergence and migration events act as major determinants of congruence/incongruence in host-parasite genealogies (Nieberding& Olivieri 2007), and generally lead to limited congruence between the phylogenetic trees of RNA viruses and their hosts (Rannala& Michalakis 2003). Indeed, thus far, co-divergence of RNA viruses and their hosts has been observed infrequently (Liu et al. 2008; Switzer et al. 2005).
Landscape features may also strongly structure populations of reservoir species involved in zoonotic emergences (Archie et al. 2009; Biek& Real 2010; Reisen 2009), thereby influencing the dynamics of host-parasite systems (Hirzel et al. 2007; Su et al. 2009; Webb et al. 2007) and ultimately the complexity of epidemiological patterns (Barton et al. 2010). The physical environment affects host-parasite interactions by influencing gene flow among host populations, and by acting as a selective force in the form variable environmental conditions (Biek& Real 2010). Indeed, the genetic structure of parasites depends heavily on the overall dispersal of the host (especially in obligatory parasites like RNA viruses) and responds to the spatial heterogeneity affecting the population dynamics of the species (Biek& Real 2010; Nadin-Davis et al. 2010). For RNA viruses, geographic discontinuities in the host may lead to strong phylogenetic signatures in the virus that can be revealed using a suite of phylogeographic methods (Biek et al. 2007; Holmes 2004; Real et al. 2005).
Globally, Hantavirus (Bunyaviridae: Hantavirus) includes several strains recognized as human pathogens (Hjelle& Torres-Pérez 2010; Jonsson et al. 2010; Schmaljohn& Hjelle 1997). Rodents of the families Muridae and Cricetidae were described as the primary zoonotic reservoirs of these viruses, but distinct hantaviruses also have been discovered in several species of shrews and moles (Soricomorpha) (Arai et al. 2007; Kang et al. 2009; Klempa et al. 2007; Yadav et al. 2007). Hantaviruses are segmented negative-strand RNA viruses, and contain small (S), medium (M), and large (L) genomic segments, encoding nucleocapsid (N), glycoproteins Gn and Gc, and RNA-dependent RNA polymerase, respectively (Jonsson& Schmaljohn 2001). Horizontal transmission by rodent-to-rodent contact is the main mechanism for hantavirus maintenance and spread (Hjelle& Yates 2001). Human infection by hantaviruses is thought to follow accidental exposure to secretions or excretions produced by infected rodents. To date, person-to-person transmission has only been documented in the case of the South American Andes virus (Enria et al. 1996; Ferres et al. 2007; Lazaro et al. 2007; Martinez et al. 2005; Padula et al. 1998; Wells et al. 1998; Wells et al. 1997). Previously, hantaviruses were hypothesized to co-diverge with their associated mammalian hosts (Plyusnin et al. 1996; Yates et al. 2002). However, studies comparing rates of nucleotide substitution and times to most recent common ancestry suggest that at least some hantaviruses have diverged far more recently than implied under the assumption of divergence such that much of their evolution is dominated by cross-species transmission (Ramsden et al. 2009; Ramsden et al. 2008). The latter strongly contrasts with the temporal scale of divergence of their mammalian hosts (Palma et al. 2010; Ramsden et al. 2009). In the southern cone of South America, the sigmodontine rodent, Oligoryzomys longicaudatus, constitutes the main reservoir for Andes virus (ANDV). In Chile, this rodent occurs in mesic habitats from 28°S to 55°S latitude (Belmar-Lucero et al. 2009; Palma et al. 2005), where > 65% of the human population resides. This area encompasses climates varying from warm semi-desert in northern Chile to cold subpolar in the extreme south. In southern Chile and Patagonia, biotic communities were structured by the dramatic glacial cycles of the Pleistocene (Harrison 2004; Lessa et al. 2010).
To date, the extent and pattern of co-divergence among RNA viruses and their hosts has generally been studied either at deeper phylogenetic (i.e. inter-specific) levels or on much shorter time-scales by way of comparisons of intra-host dynamics (Paterson& Piertney 2011). Far fewer studies have explored these processes simultaneously using both phylogeographic and population genetic approaches (Biek et al. 2006; Dekonenko et al. 2003; Nemirov et al. 2010). To determine the ecological and evolutionary processes that might be responsible for genetic diversity of hantaviruses, we undertook a detailed analysis of the spatial and temporal scale of co-divergence utilizing both contemporary spatial genetics and longer-term co-phylogeny. These data allow us to evaluate whether ANDV in Chile and the associated host follow similar genealogical patterns, and if ANDV might be thought of as a genealogical proxy that is tracking ecological processes in O. longicaudatus. The obligate parasitic nature of ANDV in O. longicaudatus (Medina et al. 2009), coupled with a broad latitudinal distribution in Chile, provides a useful model for examining processes acting to shape contemporary population structure in hosts and parasites and ultimately exploring the history and epidemiology of zoonotic diseases.
Materials and Methods
Sampling and rodents sequencing
A total of 197 O. longicaudatus were collected from 22 localities in Chile ranging from 28°S to 51°S (Fig. 1, Table S1, Supporting Information). Rodents were collected in live traps (H.B. Sherman Traps, Inc., Tallahassee, FL), following established safety guidelines for rodent captures and processing (Gannon& Sikes 2007; Mills et al. 1995). Genomic DNA from liver tissue was extracted using a modified salt extraction method (Fleming& Cook 2002). A fragment of 924 bp of the mitochondrial cytochrome b (cyt b) was amplified using the polymerase chain reaction (PCR), and primers MVZ 07, MVZ 26, LBE 05 and H 15767, following the procedures described previously (Palma et al. 2005). We also sequenced the mtDNA control region (1014-nt) for 12 selected individuals used in the co-divergence analyses using primers (DLO-L) CGGAGGCCAACCAGTAGA-3′ and (DLO-H) TAAGGCCAGGACCAAACC-3′. PCR products were purified using QIAquick PCR purification kit (Qiagen Inc., Valencia, CA, USA). Cycle sequencing used flanking primers labeled with the Big Dye terminator Kit (Perkin Elmer, Norwalk, Connecticut, USA). Sequencing reactions were analyzed on an ABI Prism 3100 (Applied Biosystems) automated sequencer. Sequences were edited using the BioEdit Sequence Alignment Editor (Hall 1999), and aligned using Clustal W implemented in BioEdit.
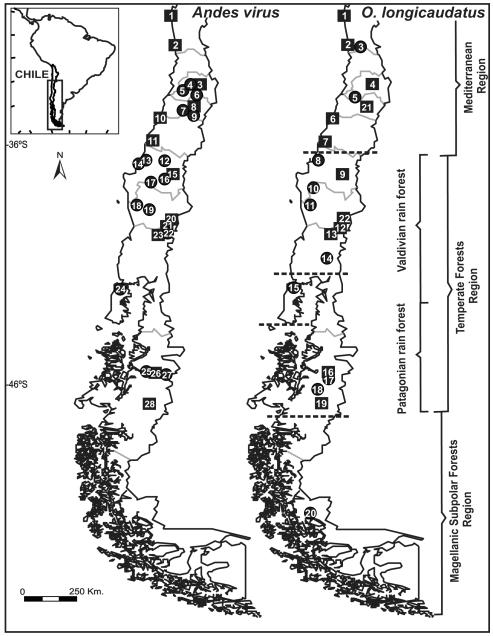
Figure 1.
Map of the sampled localities of ANDV in Chile (left) and O. longicaudatus (right). Squares represent localities used for co-divergence analyses. Dashed lines separate groups indicated from SAMOVA. Numbers are explained in Table S1 of the supplementary data.
ANDV Sequences, Recombination and Selection Analyses
Sequences from ANDV were obtained from human cases and seropositive rodents (Medina et al. 2009). We sequenced the S-segment (396-nt) in 38 samples representative of the known geographic range of the virus in Chile (Toro et al. 1998; Torres-Pérez et al. 2004) (GenBank Accession numbers EU241665-EU241702). We also included four ANDV samples taken from GenBank (Accession numbers AY228237, AF004660, AF291702, AF482712), for a total of 42 sequences. These samples were used to perform spatial genetic and molecular clock analyses. For the co-divergence analyses we used a longer S-segment (931-nt) from fewer ANDV samples (N = 12).
We screened for potential recombination in 42 S-segment sequences (396-nt) used in phylogeographic analyses (Medina et al. 2009), and the 12 S-segment (931-nt) sequences used in co-divergence analyses (see below). These analysis were performed using the Recombination Detection Program (RDP3, http://darwin.uvigo.es/rdp/rdp.html), which applies a suite of recombination detection methods (Martin et al. 2005). In all cases, analyses were performed using default detection thresholds. The Genetic Algorithm for Recombination Detection (GARD) method (Kosakovsky Pond et al. 2006) was also used to examine the occurrence of recombination events between ANDV sequences (as implemented in datamonkey.org resource). When potential recombination among sequences was detected, analyses were performed using only the regions in which recombination was not suspected.
Analyses of selection pressure were conducted on the Datamonkey server (Kosakovsky Pond& Frost 2005a) using the following methods: single-likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), random effects likelihood (REL), and PARRIS (a PARtitioning approach for Robust Inference of Selection). All these methods allow positive selection to be detected at individual nucleotide sites (Kosakovsky Pond& Frost 2005b).We also tested for selection using alternative evolutionary models under a Bayesian approach implemented in the Selecton server (Stern et al. 2007).
Rates of nucleotide substitution and time to the most recent common ancestor
We used the Bayesian Markov Chain Monte Carlo (MCMC) method available in BEAST v1.5.3 (Drummond& Rambaut 2007) to estimate the rate of nucleotide substitution and time to the most recent common ancestor (tMRCA) for ANDV in Chile. Because of the short length of the fragment analyzed and the relatively limited time span of sampling (nine years) we employed the simple HKY85 model of nucleotide substitution; more complex substitution models resulted in model over-parameterization and unreliable estimates (not shown). This analysis also employed the conservative Bayesian skyline coalescent prior and both strict and relaxed (uncorrelated lognormal) molecular clocks, and which produced similar results (see below). The MCMC chains were run for 1 × 108 steps with a discarded 10% burn-in and statistical uncertainty is depicted in values of the 95% Highest Probability Density (HPD). For O. longicaudatus, a strict molecular clock was found to be the best-fit to the data (employing the GTR model and the Bayesian skyline coalescent), and convergence of the MCMC was reached after running the BEAST program for 6 × 108 steps. The samples from two runs were combined, and convergence of the chain, sampling and mixing was confirmed by inspection of the MCMC samples using the program Tracer v.1.5 (Rambaut& Drummond 2009). All analyses were performed until parameter convergence was obtained. For our molecular clock analysis of rodent mtDNA, we assumed a sigmodontine-based rate of 2.3% substitutions per site per million years (Smith& Patton 1993). Finally, to check for temporal structure in ANDV, essential for the accurate estimation of evolutionary dynamics, the BEAST analysis described above was repeated using 20 data sets in which the year of sampling of each sequence had been randomized (and assuming an uncorrelated lognormal relaxed molecular clock). Real data containing temporal structure would be expected to produce results that are qualitatively different to those seen in the random data sets.
Spatial Genetic Analyses
Spatial analysis of molecular variance was performed in SAMOVA v.1.0 (Dupanloup et al. 2002). This analysis was previously shown to be useful in identifying latitudinal structure in O. longicaudatus in south-central Chile (Torres-Pérez et al. 2010), so we expanded the analysis by increasing sample size (195 sequences) across a larger geographic range. This approach uses a simulated annealing approach to identify partitions of geographically adjacent sampling areas (K) that are maximally differentiated by maximizing Fct (the proportion of the total genetic variance due to differences among groups of populations). The number of groups was estimated by running 500 random initial conditions and forcing the data into k groups (where k = 2 to 10).
We used an interpolation-based graphical method to generate a three-dimensional genetic landscape shape (GLS) within the program Alleles in Space (AIS) (Miller 2005). The procedure creates peaks in areas where genetic distances between individuals are high, and valleys or troughs where genetic distances between individuals are low, providing a visual perspective of the spatial distribution of genetic structure over landscapes (Miller et al. 2006). GLS analysis was performed to visually explore the congruence in the spatial pattern of genetic divergence in ANDV and O. longicaudatus. A location-based connectivity network (based on Delaunay triangulations) was created first among all sampling locations. Then, residual genetic distances between observations (or surface heights) are calculated and placed at the midpoints of each connection in the network. To infer surface heights, AIS incorporates an inverse distance weighted interpolation procedure across a uniform grid. We selected a distance weighting parameter (a) of 0.5 for ANDV and 1.0 for O. longicaudatus (we also tested a = 0.2, 0.5, 1.0, and 1.2), and a grid dimension of 50 × 50. The AIS program was also used for spatial autocorrelation analysis and calculates the statistic Ay for each of the y = 1 to Z distance classes. Analyses were performed using Z = 10 and 20 distance classes. A permutation procedure of 10,000 replicates was performed as a global test for the full data set to quantify the heterogeneity of Ay values among distance classes.
Phylogenetic Analyses and Host-parasite Co-divergence
We used a selected number of sequences (pruned data set) to test the hypothesis of congruence (co-divergence) in the O. longicaudatus and ANDV tree topologies. ANDV may eventually be found in rodent species other than O. longicaudatus due to “spillover” (Toro et al. 1998; Torres-Pérez et al. 2004). However, ANDV prevalence in these non-reservoir species is always significantly lower than in O. longicaudatus (Medina et al. 2009; Toro et al. 1998). Also, ANDV segregates in Chile by geographic location of the sampling site, and does not segregate by host source (Medina et al. 2009). To prevent bias due to non-reservoir sources, co-phylogenetic analyses only used ANDV sequenced from O. longicaudatus, and both sets (mitochondrial and S-segment sequences) were obtained from the same O. longicaudatus specimens. Analyses of ANDV were performed using 931-nt of the S-segment, while analyses of O. longicaudatus were performed using concatenated cyt b (927-nt) and d-loop (1014-nt) sequences. In all cases maximum likelihood (ML) phylogenetic trees were inferred using the PAUP* package (Swofford 2002). jModeltest (Posada 2008) identified TVM + I and HKY + I as the best-fit models of sequence evolution for ANDV and O. longicaudatus, respectively. Node support was evaluated with 1,000 non-parametric bootstrap pseudoreplicates (Felsenstein 1985) using the settings obtained from jModeltest.
In addition, we employed a Bayesian MCMC procedure using BayesPhylogenies (Pagel& Meade 2004) to infer a posterior distribution of phylogenetic trees and assuming the general time-reversible (GTR) model of gene-sequence evolution with gamma distribution of rate variation among sites. From random starting trees, 5 × 106 generations were run with four Metropolis-coupled chains, with the resulting trees sampled every 1,000 generations. The first 500 trees of the sample were removed to avoid including trees sampled before convergence of the Markov Chain, and the last 4,500 trees were used to compute a 50% majority rule consensus tree. A posterior probability of P ≥ 95% was considered as evidence for significant support for any individual node (Alfaro et al. 2003).
The genealogical sorting index (Cummings et al. 2008) was used to estimate the degree of exclusive ancestry of individuals in specific groups on a rooted tree. Here, a tree topology is used to quantify coalescent events uniting a group. Under the null hypothesis that the degree of exclusive ancestry of branch tips observed is that which might be observed at random, this index assesses the statistical significance by holding the tree constant with subsequent permutation of the group labels assigned to the tips of the tree, thus randomizing the common ancestry of members of the groups. Analyses were performed in O. longicaudatus using 82 samples (plus 2 outgroups) randomly selected, and assigning samples (Mediterranean N = 31, Valdivian N = 34, Patagonian N = 17) to groups as derived from SAMOVA analysis. Significance was evaluated under 10,000 permutations.
To test for co-divergence of O. longicaudatus-ANDV, we used both topology-based and distance-based approaches. The congruence index (Icong) is a topology-based approach based on the maximum agreement subtree (MAST) between two trees (de Vienne et al. 2007). Icong tests the null hypothesis that two trees are not more congruent topologically than expected by chance. Rejecting the null hypothesis indicates that evolution in one group is dependent to some extent on evolution in the other (de Vienne et al. 2009). For the distance-based approach, we used the CopyCat program (Meier-Kolthoff et al. 2007), which incorporates a wrapper for the program Parafit (Legendre et al. 2002). Parafit uses matrices of raw or patristic distances (summed branch lengths along a phylogenetic tree) transformed into principle coordinates. The program assesses the fit between host and parasite phylogenetic distances, through a matrix representing host-parasite associations. Statistical significance was evaluated by performing 9,999 permutations to test the null hypothesis of no co-divergence between ANDV and O. longicaudatus populations.
We compared the results of the above tests with those from two other programs commonly used in cophylogenetic studies. Specifically, TreeMap 1.0 and TreeMap 2.0β are topology-based approaches that reconcile two trees by introducing four types of events (cospeciation, duplication, lineage sorting, and host switching). TreeMap 1.0 incorporates multiple reconstructions that attempt to produce the fewest possible number of events and to maximize the number of co-divergence events. A randomization test (10,000 permutations) was performed on each reconstruction to assess if both phylogenies are more similar to each other than expected by chance. TreeMap 2.0β uses jungles (instead of parsimony), which is a directed graph of all possible mappings of one tree onto another. When resolved, all potentially optimal solutions are reported (POpt) (Charleston 1998). The optimal reconstruction is that which minimizes the global cost. To evaluate the statistical significance of any congruence observed, a null hypothesis that the extent of co-divergence between trees is no more than that expected by chance is tested. Significance was then tested by comparing the jungle with 100 randomized virus trees.
Results
Genetic diversity and analyses of selection pressures
Estimations of allele, haplotype and nucleotide diversity (Table 1) revealed similar levels of genetic diversity for O. longicaudatus mtDNA sequences sampled from the Mediterranean and Valdivian rain forests regions, but lower levels in Patagonia. For ANDV, allele diversity was similar across the three ecoregions, although higher nucleotide diversity was observed in those viruses sampled from the Mediterranean region. An analysis of selection pressures in ANDV using a variety maximum likelihood approaches revealed no evidence for positive selection. Similar results were obtained using a Bayesian approach, and in some revealed a dominance of purifying selection on the viral S segment.
Table 1.
Descriptive statistics of genetic variation of ANDV S segment (397-nt) and O. longicaudatus cyt b (927-nt) sequences in Chile. N: Number of individuals; Nh: Number of alleles/haplotypes; S: Number of segregating sites; Hd: Allele/haplotype diversity; π: Nucleotide diversity; SD: Standard deviation.
| Eco-region | N | Nh | S | Hd ± SD | Π ± SD | |
|---|---|---|---|---|---|---|
| An | 1.Mediterranean | 17 | 14 | 69 | 0.971 ± 0.032 | 0.056 ± 0.003 |
| 2.Valdivian rain forests | 16 | 14 | 51 | 0.983 ± 0.028 | 0.032 ± 0.004 | |
| 3.Patagonian rain forests | 9 | 8 | 42 | 0.972 ± 0.064 | 0.030 ± 0.006 | |
| Total | 42 | 36 | 107 | 0.992 ± 0.007 | 0.076 ± 0.003 | |
|
| ||||||
| O. longicaudatus | 1.Mediterranean | 69 | 34 | 49 | 0.941 ± 0.017 | 0.004 ± 0.0004 |
| 2. Valdivian rain forests | 76 | 35 | 40 | 0.955 ± 0.011 | 0.004 ± 0.0002 | |
| 3. Patagonian rain forests | 38 | 11 | 16 | 0.839 ± 0.036 | 0.036 ± 0.0005 | |
| Total* | 183 | 83 | 0.973 ± 0.004 | 0.006 ± 0.0002 | ||
Does not include Magellanic Subpolar forests region (N = 12)
Evolutionary Rates and Times to Common Ancestry
We estimated the overall nucleotide substitution rate for the S segment of ANDV to be between 0.17 – 2.06 × 10−3 nucleotide substitutions per site, per year (subs/site/year) under a relaxed molecular clock and 0.19 – 1.96 × 10−3 subs/site/year assuming a strict clock. These rates are similar to those seen in a broad array of RNA viruses (Duffy et al. 2008; Holmes 2008) and may be > 5 orders of magnitude higher than those reported for the mitochondrial genome of rodents (Lessa& Cook 1998; Smith& Patton 1993). In addition, the mean evolutionary rate estimated under the relaxed molecular clock (1.15 × 10−3 subs/site/year) fell outside of the range of substitution rates estimated in 20 randomized data sets (range of 95% HPD values = 1.07 × 10−3 to 2.28 × 10−8 subs/site/year) indicating that they have been drawn from different distributions and that there is temporal structure in these data. In addition, the lower 95% HPD values in the randomized data tended toward a zero substitution rate (range 8.28 × 10−5 to 2.28 × 10−8 subs/site/year) as expected given a lack of temporal structure. However, that there is some overlap between the 95% HPD values of the real and randomized data indicates that more sequence data are required to obtain a precise estimate of evolutionary dynamics in ANDV. Similarly, the estimated tMRCA values for ANDV in Chile fell between 28 – 229 and 26 – 224 years before the most recent sample (2004) for the strict and relaxed clocks, respectively. In marked contrast, the tMRCA for O. longicaudatus in Chile was estimated to be between 134,000 – 346,300 years before the present (ybp) (95% HPD values; mean of 231,400 ybp). Overall, these results show that scale of evolutionary change in the genomes of ANDV and associated hosts differs dramatically.
Spatial Genetics
We assessed spatial genetic substructure within the host (O. longicaudatus, 195 samples) populations using SAMOVA (Spatial Analysis of Molecular Variance). FCT values ranged from 0.427 to 0.478, with the group structure maximized at k = 5. Collection sites from Fray Jorge to Los Ruiles (localities 1 to 7 in Fig. 1) formed the first group, Tome to Puyehue (localities 8 to 14) clustered into a second group, four localities (16 to 19) clustered into a third group, and Torres del Paine (locality 20) was separated as the fourth group (Fig. 1; Table S1). Four groups correspond to previously identified ecological regions in Chile: Mediterranean, Valdivian rain forests, North Patagonian rain forests and Magellanic subpolar forests (Armesto et al. 2007; Olson et al. 2001; Veblen 2007) (Fig. 1). Chiloé (locality 15), an isolated island, was separated as the fifth group but it is closely allied to the Valdivian rain forest ecoregion (Veblen 2007). Unlike O. longicaudatus, SAMOVA was unable to segregate discrete groups with higher FCT maximized values obtained as more groups (k) of ANDV sequences were added.
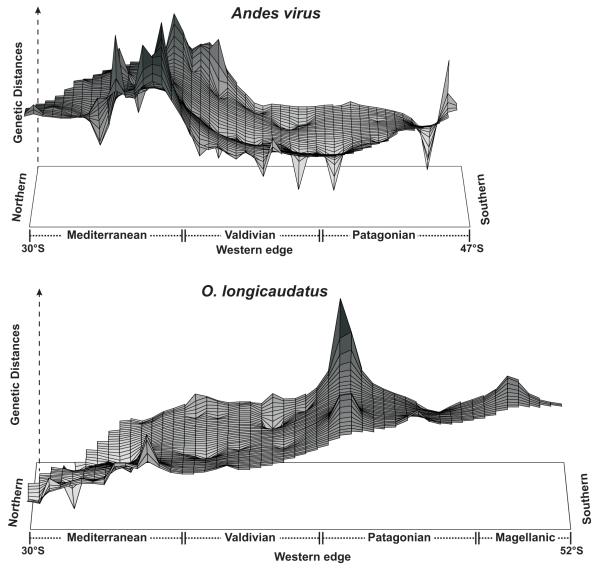
When determining the spatial distribution of alleles/haplotypes in the North – South latitudinal gradient, the genetic landscape shape interpolation analysis (Fig. 2) differed between ANDV and its host. The highest levels of pairwise genetic distance were observed between 34° – 37° S (the limit between Mediterranean and Valdivian rain forests regions) for ANDV, while the highest values for the host were observed in the south (ca. 41°- 43°S). However, ANDV and O. longicaudatus were congruent in showing higher genetic heterogeneity within the Mediterranean ecoregion, with both higher and lower peaks observed especially between virus samples (Fig. S1). Peaks and valleys (areas of high and low genetic distance between individuals) occurred both along the coast (western edge) and in the Andes. Grid size and distance weighting parameters did not affect landscape shape.
Figure 2.
A graphical interpolation-based representation of genetic structure made using a 50 × 50 grid and a distance weighting parameter of 0.5 for ANDV and 1.0 for O. longicaudatus. X- and y- axes represent geographic coordinates. Surface heights along the Z-axis indicate genetic distances. Gray scales are indicative of areas with high (black) or low (white) pairwise genetic distance between individuals. All samples (195 sequences) were included for O. longicaudautus. Scale at the bottom represents latitude in degrees and minutes.
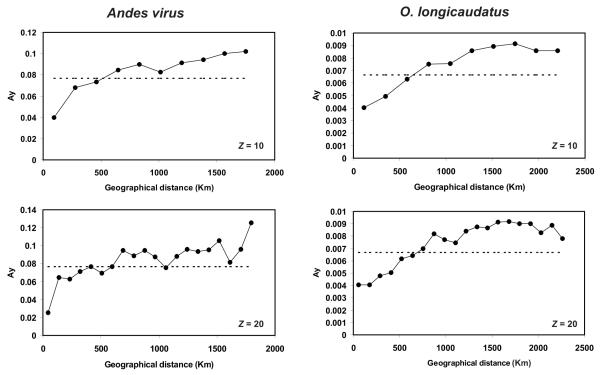
Spatial autocorrelation analyses can be used to detect geographic regions with different (high or low) genetic similarity between samples (Diniz-Filho& De Campos Telles 2002). Our analyses indicated significant geographic structure in ANDV and O. longicaudatus (P < 0.0001). For ANDV, spatial autocorrelation analyses performed using Z = 10 (10 distance classes) and Z = 20 (Fig. 3) were similar with significant autocorrelation ending about 500 km. For O. longicaudatus, autocorrelation ceased between the distance classes 3 to 4 (for Z=10) and 6 to 7 (for Z = 20) (Fig. 3). These analyses suggest that spatial phylogeographic structure in ANDV can be detected at around 400-600 km, while for O. longicaudatus structure does not appear until about 600-800 km.
Figure 3.
Results of spatial autocorrelation analyses of ANDV (S segment) and O. longicaudatus in Chile. Analyses were performed using Z = 10 and 20 distinct distance classes. Ay: Average pairwise genetic distances of haplotypes that fall within the boundaries specified for distance class y. Horizontal dotted lines indicate the average pairwise genetic distance from full data set.
Phylogenetic Relationships and Virus-Host Co-divergence
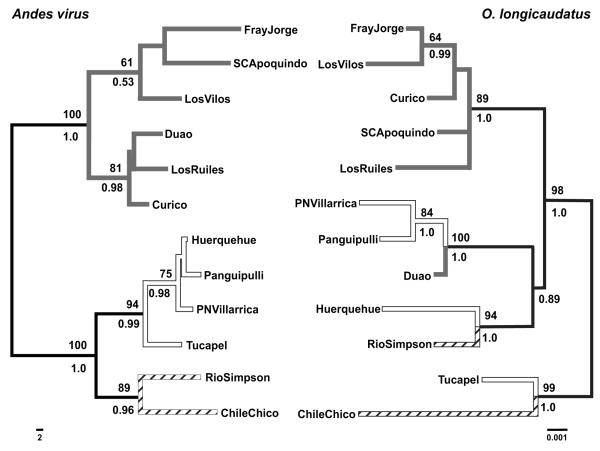
Phylogenetic analyses for ANDV (S-segment, 931-nt) and O. longicaudatus (concatenated cyt b and control region, 1942-nt) sequences were based on a reduced subset of 12 samples (Fig. 4) with both mitochondrial and S-segment sequences obtained from the same O. longicaudatus specimens. Topologies based on maximum likelihood (ML) and Bayesian MCMC did not show significant differences using the Shimodaira-Hasegawa (SH) test for either ANDV (−lnLML = 2969.668, −lnLBMCMC= 2965.829, P = 0.275) or O. longicaudatus (−lnLML = 3263.962, −lnLBMCMC= 3263.962, P = 0.523).
Figure 4.
Maximum Likelihood phylogenetic trees from pruned data sets of 12 samples using S-segment sequences (943-nt) for ANDV and concatenated cytochrome b and control region for O. longicaudatus (1,942-nt). Numbers above and below branches indicate bootstrap values obtained with maximum likelihood and posterior probabilities, respectively. Gray, white and dashed branches represent samples from Mediterranean, Valdivian rain forests and Patagonian rain forests regions, respectively. Bars indicate the number of nucleotide substitutions per site.
For ANDV, the ML (midpoint rooted) topology identified two primary clades. All samples in the Mediterranean ecoregion (Fray Jorge to Los Ruiles; Fig. 1) clustered into a well-supported clade (maximum likelihood bootstrap values = 100 and posterior probabilities = 1.0 using BMCMC). Within the second clade, two subclades were recovered, a first group with all samples belonging to the Valdivian rain forest region (MLB = 94, PP = 0.99), and a second group with the two samples from Patagonia (MLB = 89, PP = 0.96). For ANDV host, the clades identified had a few samples intermingled among ecoregions. Most Mediterranean samples (except Duao) were in a well-supported clade (MLB = 89, PP = 1.0). Samples from Valdivian rain forest region grouped together with either Duao (Mediterranean) or Rio Simpson (Patagonian) samples. Tucapel (a Valdivian rain forest sample) and Chile Chico (Patagonian sample) were in a highly supported clade (MLB = 99, PP = 1.0). Overall, and as expected given the disparity in evolutionary rates, ANDV sequences provide greater genealogical resolution than the associated host data. Incomplete lineage sorting was observed in populations of O. longicaudatus in the Mediterranean (Duao), Valdivian Temperate Forests (Tucapel) and Patagonian (Rio Simpson) regions. Based on the assumption of no lineage sorting in the O. longicaudatus samples, we estimated the degree of exclusive ancestry of individuals in specific groups using the genealogical sorting index (GSI) (Cummings et al. 2008). For the three groups tested, the index showed highly significant values (GSIMediterranean = 0.2729, P < 0.001; GSIValdivian = 0.3011, P < 0.001; GSIPatagonia = 0.5871, P < 0.001), suggesting that placement of O. longicaudatus samples into the three respective groups is greater than expected by chance. Incomplete lineage sorting was not observed for ANDV.
Using ML topologies, we performed co-divergence tests with 12 data sets. The index of congruence (Icong) indicates that evolution of ANDV is likely independent of that O. longicaudatus (P > 0.05). The ParaFit Global test of cospeciation revealed a significant association between ANDV and O. longicaudatus samples (P < 0.05), although only two out of twelve host-parasite links were significant. Importantly, both TreeMap 1.0 and TreeMap 2.0β randomization tests suggested that the observed number of co-divergence events in the parasite tree was not significantly different from that observed in random associations. Therefore, with the exception of the ParaFit Global test, all analyses revealed that the phylogenetic trees of ANDV and their O. longicaudatus rodent hosts are no more congruent than expected by chance alone.
Discussion
The extent of evolutionary congruence between host and parasite depends on processes that may not only act at different spatial and temporal scales, but also at different hierarchical scales; genes, individuals, populations, or species (Pybus& Rambaut 2009; Rannala& Michalakis 2003). For example, if viral evolutionary dynamics are coupled with ecological processes acting in female-host populations, congruence in spatial pattern may be revealed using a mitochondrial gene. An example is provided by rabies viruses sampled from British Columbia and which show a spatial correlation with cytochrome oxidase I gene from their bat host, suggesting a sex-biased pattern of viral transmission (Nadin-Davis et al. 2010). We evaluated current and historical population structure using spatial genetics and co-phylogeographic approaches in a virus – rodent system.
Previous work showed that nucleotide substitution rates in some rodent hantaviruses are significantly higher than their host, which leads to substantial differences in divergence times that appear incompatible with a history of codivergence (Ramsden et al. 2009). Indeed, estimates of the tMRCA of hantaviruses and their rodent hosts are similarly discordant (Ramsden et al. 2009; Steppan et al. 2004). At the phylogeographic scale of our study, the evolutionary rates and tMRCA estimated for ANDV and O. longicaudatus also differed substantially, although with a very large variance in the case of the virus. Specifically, the tMRCA of ANDV in Chile was under 250 ybp, while that of O. longicaudatus was about 223,000 ybp. Although inferences of tMRCA for RNA viruses using a Bayesian MCMC approach may be strongly biased (i.e. too recent) due to unsampled extant or extinct basal viral lineages or an inability to correct infer the number of mutational changes (Holmes 2009a), our results indicate that the evolution of ANDV and its rodent host is independent at the phylogenetic scale.
Both host and virus segregated across a latitudinal gradient corresponding to three major ecoregions, Mediterranean, Temperate Forests (Valdivian and Patagonian rain forests) and Magellanic Subpolar Forest (Armesto et al. 2007; Veblen 2007). Spatial genetic autocorrelation disappears at the boundaries of these ecoregions, with more rapidly evolving ANDV lineages more strongly segregated than O. longicaudatus. Some clades of the host share haplotypes across ecoregion borders (Palma et al. 2005; Torres-Pérez et al. 2010). Nonetheless, both SAMOVA and the genealogical sorting index recovered three groups that correspond to ecoregions despite incomplete lineage sorting in the host (Maddison 1997; Pamilo& Nei 1988), also congruent with three previously proposed O. longicaudatus subspecies (Osgood 1943). Ecoregions are defined by environmental characteristics of the landscape in aggregating areas with similar properties (relative homogeneity of ecosystems) (Loveland& Merchant 2004; Wilson et al. 2007), which may reflect differences in vegetation composition, soil type, climate, and geology (Bailey 1998; Olson et al. 2001). Complex geographical, historical, and/or contemporary evolutionary processes (e.g., differential selective regimes) acting on populations may result in geographic lineages that are partitioned across ecoregions. In this particular virus/mammal system there are no obvious geographic barriers that would reduce gene flow between ecoregions, nor was positive selection detected in ANDV (Table S2). Alternatively, partitioning of viral variants across ecoregions may be explained by an isolation-by-distance model (Real et al. 2005). These and other models should be fully explored in the future when evaluating divergence of ANDV lineages. Populations that are widely separated or that have arisen through recent colonization (as those occurring in previously glaciated areas within the Valdivian region), may bias gene flow estimation as an equilibrium between genetic drift and gene flow has not been reached (Crispo& Hendry 2005). ANDV follows a phylogeographic pattern of strong spatial subdivision (Holmes 2008) that was nearly recovered in O. longicaudatus, suggesting that ANDV may be a genealogical proxy of its host.
Landscape structure is likely an important determinant of the genetic structure of ANDV. Transmission in obligate parasites depends on host dispersal and population dynamics, which may be strongly affected by spatial heterogeneity (Biek& Real 2010). For example, O. longicaudatus shows a heterogeneous population genetic structure across the latitudinal span of Chile. Within the Mediterranean ecoregion, populations of O. longicaudatus have higher genetic divergence than either Valdivian or Patagonian rain forest populations (Torres-Pérez et al. 2010). This finding is congruent with a pattern of higher genetic distances among northern ANDV samples as revealed by the genetic landscape shape analysis. The Mediterranean region is characterized by a heterogeneous vegetation mosaic that transitions between desert and mixed deciduous evergreen forests (Amigo& Ramirez 1998; Armesto et al. 2007; Veblen 2007). This highly fragmented area supports a large number of threatened endemics (Wilson et al. 2007) and 65% of the Chilean human population. For mesic species such as O. longicaudatus, this dry, fragmented environment should lead to greater population subdivision, reduced gene flow and strong effects on genetic diversity due to drift (Hartl& Clark 2007; Pilot et al. 2006; Sacks et al. 2008; Wright 1931). Hence, differences in processes such as fragmentation and drift in host populations likely impacted genetic structure of ANDV, with rapid evolutionary rates in the virus resulting in higher genealogical resolution and structuring than the host.
Fahrenholz’s rule assumes that host and associated parasite trees will be congruent due to a shared history (Fahrenholz 1913). However, a growing number of molecular phylogenetic studies have challenged this idea (Jackson& Charleston 2004; Koehler et al. 2009; Ramsden et al. 2009). At the phylogeographic level, several factors likely contribute to decreased host-parasite congruence between O. longicaudatus and ANDV: i) Higher nucleotide substitution rates in ANDV suggest that evolution occurs at different temporal scales; ii) Hantaviruses are reported to be horizontally transmitted in the wild, although host-parasite co-divergence is most likely to occur under vertical transmission (see Nieberding & Olivieri 2007, and references therein); iii) Weak genealogical structure in O. longicaudatus contrasts with the increased sorting of the parasite; iv) Genealogical reconstructions of O. longicaudatus based on a female-inherited molecular marker such as mtDNA may not accurately reflect the history of ANDV transmission, because ANDV is thought to be primarily transmitted by males (Padula et al. 2004; Torres-Pérez et al. 2004). However, there is no evidence of differences in dispersal patterns between males and females of O. longicaudatus (Murúa et al. 1986), suggesting that our genealogical reconstructions likely reflect the history of the species.
Both current and historical processes have acted to produce major differences in the genetic structure of ANDV and O. longicaudatus populations. Globally, our results suggest that despite contrasting evolutionary rates, geographic structure of both the host and RNA virus may converge across large spatial scales. Host structure is related to historic (e.g. effects of glacial cycles during Pleistocene; Palma et al. submitted) and contemporary (e.g. reduced gene flow among ecoregions) dynamics of its populations. Although ANDV does not co-diverge with O. longicaudatus, strong lineage structure in the virus seems to be a response to current population dynamics in the host at the scale of ecoregions.
Supplementary Material
Acknowledgments
We thank the Ministry of Health in Chile and local health services for providing useful information on sampling sites, and the Servicio Agrícola y Ganadero (SAG) and Corporación Nacional Forestal (CONAF) for trapping permits. Support from Life Science Informatics at the University of Alaska is also greatly appreciated (#P20RR016466 National Center for Research Resources). We also thank the hantavirus field crew, M. Acuña and the Centro de Investigaciones Médicas (PUC) for fieldwork or lab support. Financial support was provided by the Fogarty International Center Research Grant # D43 TW007131, the NIH-ICIDR Chilean Hantavirus Grant 1 U19 AI45452–01, FONDECYT 1070331, 1100558, 1110664, and Fondecyt CASEB 1501-0001 Program 2. ECH was supported in part by NIH grant GM080533-04. F. Balloux (Subject Editor) and three anonymous reviewers provided insightful comments.
Footnotes
Data accessibility DNA sequences: GenBank accessions GQ282502-GQ282603; JN034440-JN034536; AF 346566; AY275693; AY275692; AY275694; AY275696; AY275699; AY452197; AY452198; AF346568; AY275690. (For an individual-by-individual listing, please see the online supplemental material.)
Final DNA sequence assembly uploaded as online supplemental material
REFERENCES
- Alfaro ME, Zoller S, Lutzoni F. Bayes or Bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molecular Biology and Evolution. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- Amigo J, Ramirez C. A bioclimatic classification of Chile: woodland communities in the temperate zone. Plant Ecology. 1998;136:9–26. [Google Scholar]
- Arai S, Song JW, Sumibcay L, et al. Hantavirus in northern short-tailed shrew, United States. Emerging Infectious Diseases. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology & Evolution. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Armesto JJ, Arroyo MTK, Hinojosa LF, et al. The Mediterranean Environment of Central Chile. In: Veblen TT, Orme AR, Young KR, editors. The Physical Geography of South America. Oxford University Press; Oxford: 2007. pp. 184–199. [Google Scholar]
- Bailey RG. Ecoregions: the Ecosystem Geography of the Oceans and Continents. Springer; New York: 1998. [Google Scholar]
- Barton HD, Gregory AJ, Davis R, Hanlon CA, Wisely SM. Contrasting landscape epidemiology of two sympatric rabies virus strains. Molecular Ecology. 2010;19:2725–2738. doi: 10.1111/j.1365-294X.2010.04668.x. [DOI] [PubMed] [Google Scholar]
- Belmar-Lucero S, Godoy P, Ferres M, Vial P, Palma RE. Range expansion of Oligoryzomys longicaudatus (Rodentia, Sigmodontinae) in Patagonian Chile, and first record of Hantavirus in the region. Revista Chilena de Historia Natural. 2009;82:265–275. [Google Scholar]
- Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R, Real LA. The landscape genetics of infectious disease emergence and spread. Molecular Ecology. 2010;19:3515–3531. doi: 10.1111/j.1365-294X.2010.04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charleston MA. Jungles: a new solution to the host/parasite phylogeny reconciliation problem. Mathematical Biosciences. 1998;149:191–223. doi: 10.1016/s0025-5564(97)10012-8. [DOI] [PubMed] [Google Scholar]
- Crispo E, Hendry A. Does time since colonization influence isolation by distance? A meta-analysis. Conservation Genetics. 2005;6:665–682. [Google Scholar]
- Cummings MP, Neel MC, Shaw KL. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62:2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Giraud T, Martin OC. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Giraud T, Martin OC. In response to comment on ‘A congruence index for testing topological similarity between trees’. Bioinformatics. 2009;25:150–151. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- Dekonenko A, Yakimenko V, Ivanov A, et al. Genetic similarity of Puumala viruses found in Finland and western Siberia and of the mitochondrial DNA of their rodent hosts suggests a common evolutionary origin. Infection, Genetics and Evolution. 2003;3:245–257. doi: 10.1016/s1567-1348(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, De Campos Telles MP. Spatial Autocorrelation Analysis and the Identification of Operational Units for Conservation in Continuous Populations. Conservation Biology. 2002;16:924–935. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nature Reviews: Genetics. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Enria D, Padula P, Segura EL, et al. Hantavirus pulmonary syndrome in Argentina - Possibility of person to person transmission. Medicina-Buenos Aires. 1996;56:709–711. [PubMed] [Google Scholar]
- Fahrenholz H. Ectoparasiten und Abstammungslehre. Zoologischer Anzeiger. 1913;41:371–374. [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies - an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferres M, Vial P, Marco C, et al. Prospective evaluation of household contacts of persons with hantavirus cardiopulmonary syndrome in Chile. Journal of Infectious Diseases. 2007;195:1563–1571. doi: 10.1086/516786. [DOI] [PubMed] [Google Scholar]
- Fleming MA, Cook JA. Phylogeography of endemic ermine (Mustela erminea) in southeast Alaska. Molecular Ecology. 2002;11:795–807. doi: 10.1046/j.1365-294x.2002.01472.x. [DOI] [PubMed] [Google Scholar]
- Gannon WL, Sikes RS. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy. 2007;88:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harrison S. The Pleistocene Glaciations in Chile. Part III: South America, Asia, Africa, Australia, Antarctica. In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations - Extent and Chronology. Elsevier Science; 2004. pp. 89–104. [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 4th edn Sinauer Associates, Inc.; Sunderland, MA: 2007. [Google Scholar]
- Hirzel AH, Nisbet RM, Murdoch WW. Host-parasitoid spatial dynamics in heterogeneous landscapes. Oikos. 2007;116:2082–2096. [Google Scholar]
- Hjelle B, Torres-Pérez F. Hantaviruses in the Americas and their role as emerging pathogens. Viruses. 2010;2:2559–2586. doi: 10.3390/v2122559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelle B, Yates T. Modeling hantavirus maintenance and transmission in rodent communities. Current Topics in Microbiology and Immunology. 2001;256:77–90. doi: 10.1007/978-3-642-56753-7_5. [DOI] [PubMed] [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Molecular Ecology. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Evolutionary history and phylogeography of human viruses. Annual Review of Microbiology. 2008;62:307–328. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- Holmes EC. The Evolution and Emergence of RNA Viruses. Oxford University Press; Oxford: 2009a. [Google Scholar]
- Holmes EC. The Evolutionary Genetics of Emerging Viruses. Annual Review of Ecology, Evolution, and Systematics. 2009b;40:353–372. [Google Scholar]
- Jackson AP, Charleston MA. A cophylogenetic perspective of RNA-virus evolution. Molecular Biology and Evolution. 2004;21:45–57. doi: 10.1093/molbev/msg232. [DOI] [PubMed] [Google Scholar]
- Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clinical Microbiology Reviews. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CB, Schmaljohn CS. Replication of hantaviruses. Current Topics in Microbiology and Immunology. 2001;256:15–32. doi: 10.1007/978-3-642-56753-7_2. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Sumibcay L, et al. Evolutionary Insights from a Genetically Divergent Hantavirus Harbored by the European Common Mole (Talpa europaea) PLoS ONE. 2009;4:e6149. doi: 10.1371/journal.pone.0006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B, Fichet-Calvet E, Lecompte E, et al. Novel hantavirus sequences in Shrew, Guinea. Emerging Infectious Diseases. 2007;13:520–522. doi: 10.3201/eid1303.061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler AVA, Hoberg EP, Dokuchaev NE, et al. Phylogeography of a Holarctic nematode, Soboliphyme baturini, among mustelids: climate change, episodic colonization, and diversification in a complex host–parasite system. Biological Journal of the Linnean Society. 2009;96:651–663. [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005a;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular Biology and Evolution. 2005b;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Molecular Biology and Evolution. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- Lazaro ME, Cantoni GE, Calanni LM, et al. Clusters of hantavirus infection, southern Argentina. Emerging Infectious Diseases. 2007;13:104–110. doi: 10.3201/eid1301.060404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Desdevises Y, Bazin E. A statistical test for host-parasite coevolution. Systematic Biology. 2002;51:217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- Lessa EP, Cook JA. The molecular phylogenetics of tuco-tucos (genus Ctenomys, Rodentia : Octodontidae) suggests an early burst of speciation. Molecular Phylogenetics and Evolution. 1998;9:88–99. doi: 10.1006/mpev.1997.0445. [DOI] [PubMed] [Google Scholar]
- Lessa EP, D’Elia G, Pardinas UF. Genetic footprints of late Quaternary climate change in the diversity of Patagonian-Fueguian rodents. Molecular Ecology. 2010;19:3031–3037. doi: 10.1111/j.1365-294X.2010.04734.x. [DOI] [PubMed] [Google Scholar]
- Liu W, Worobey M, Li Y, et al. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 2008;4:e1000097. doi: 10.1371/journal.ppat.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland TR, Merchant JM. Ecoregions and ecoregionalization: Geographical and ecological perspectives. Environmental Management. 2004;34:S1–S13. doi: 10.1007/s00267-003-5181-x. [DOI] [PubMed] [Google Scholar]
- Maddison WP. Gene trees in species trees. Systematic Biology. 1997;46:523–536. [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Martinez VP, Bellomo C, Juan J San, et al. Person-to-person transmission of Andes virus. Emerging Infectious Diseases. 2005;11:1848–1853. doi: 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RA, Torres-Pérez F, Galeno H, et al. Ecology, genetic diversity, and phylogeographic structure of Andes virus in humans and rodents in Chile. Journal of Virology. 2009;83:2446–2459. doi: 10.1128/JVI.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Huson DH, Goker M. COPYCAT: cophylogenetic analysis tool. Bioinformatics. 2007;23:898–900. doi: 10.1093/bioinformatics/btm027. [DOI] [PubMed] [Google Scholar]
- Mideo N, Alizon S, Day T. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends in Ecology & Evolution. 2008;23:511–517. doi: 10.1016/j.tree.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Miller MP. Alleles In Space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. Journal of Heredity. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- Miller MP, Bellinger MR, Forsman ED, Haig SM. Effects of historical climate change, habitat connectivity, and vicariance on genetic structure and diversity across the range of the red tree vole (Phenacomys longicaudus) in the Pacific Northwestern United States. Molecular Ecology. 2006;15:145–159. doi: 10.1111/j.1365-294X.2005.02765.x. [DOI] [PubMed] [Google Scholar]
- Mills JN, Yates TL, Childs JE, et al. Guidelines for working with rodents potentially infected with hantavirus. Journal of Mammalogy. 1995;76:716–722. [Google Scholar]
- Murúa R, Gonzales LA, Meserve PL. Population ecology of Oryzomys longicaudatus Philippii (Rodentia, Cricetidae) in southern Chile. Journal of Animal Ecology. 1986;55:281–293. [Google Scholar]
- Nadin-Davis SA, Feng Y, Mousse D, Wandeler AI, Aris-Brosou S. Spatial and temporal dynamics of rabies virus variants in big brown bat populations across Canada: footprints of an emerging zoonosis. Molecular Ecology. 2010;19:2120–2136. doi: 10.1111/j.1365-294X.2010.04630.x. [DOI] [PubMed] [Google Scholar]
- Nemirov K, Leirs H, Lundkvist A, Olsson GE. Puumala hantavirus and Myodes glareolus in northern Europe: no evidence of co-divergence between genetic lineages of virus and host. Journal of General Virology. 2010;91:1262–1274. doi: 10.1099/vir.0.016618-0. [DOI] [PubMed] [Google Scholar]
- Nieberding CM, Olivieri I. Parasites: proxies for host genealogy and ecology? Trends in Ecology & Evolution. 2007;22:156–165. doi: 10.1016/j.tree.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, et al. Terrestrial ecoregions of the worlds: A new map of life on Earth. Bioscience. 2001;51:933–938. [Google Scholar]
- Osgood WH. The mammals of Chile. Field Museum of Natural History; Chicago: 1943. (Zoology Series). [Google Scholar]
- Padula P, Figueroa R, Navarrete M, et al. Transmission study of Andes hantavirus infection in wild sigmodontine rodents. Journal of Virology. 2004;78:11972–11979. doi: 10.1128/JVI.78.21.11972-11979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula PJ, Edelstein A, Miguel SDL, et al. Hantavirus Pulmonary Syndrome outbreak in Argentina: Molecular evidence for person-to-person transmission of Andes virus. Virology. 1998;241:323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Systematic Biology. 2004;53:571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- Palma RE, Rivera-Milla E, Salazar-Bravo J, et al. Phylogeography of Oligoryzomys longicaudatus (Rodentia: Sigmodontinae) in temperate South America. Journal of Mammalogy. 2005;86:191–200. [Google Scholar]
- Palma RE, Rodríguez-Serrano E, Rivera-Milla E, et al. Phylogenetic relationships of the pygmy rice rats of the genus Oligoryzomys Bangs, 1900 (Rodentia, Sigmodontinae) Zoological Journal of the Linnean Society. 2010;160:551–566. [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Molecular Biology and Evolution. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Paterson S, Piertney SB. Frontiers in host-parasite ecology and evolution. Molecular Ecology. 2011;20:869–871. doi: 10.1111/j.1365-294X.2010.04991.x. [DOI] [PubMed] [Google Scholar]
- Pilot M, Jedrzejewski W, Branicki W, et al. Ecological factors influence population genetic structure of European grey wolves. Molecular Ecology. 2006;15:4533–4553. doi: 10.1111/j.1365-294X.2006.03110.x. [DOI] [PubMed] [Google Scholar]
- Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. Journal of General Virology. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A. Modelling: Evolutionary analysis of the dynamics of viral infectious disease. Nature Reviews: Genetics. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.5. 2009 Available from http://beast.bio.ed.ac.uk/Tracer.
- Ramsden C, Holmes EC, Charleston MA. Hantavirus Evolution in Relation to its Rodent and Insectivore Hosts: No Evidence for Co-divergence. Molecular Biology and Evolution. 2009;26:143–153. doi: 10.1093/molbev/msn234. [DOI] [PubMed] [Google Scholar]
- Ramsden C, Melo FL, Figueiredo LM, Holmes EC, Zanotto PM. High Rates of Molecular Evolution in Hantaviruses. Molecular Biology and Evolution. 2008;25:1488–1492. doi: 10.1093/molbev/msn093. [DOI] [PubMed] [Google Scholar]
- Rannala B, Michalakis Y. Population Genetics and Cospeciation: From Process to Pattern. In: Page RDM, editor. Tangled Trees: Phylogeny, Cospeciation, and Coevolution. University Of Chicago Press; Chicago: 2003. pp. 120–143. [Google Scholar]
- Real LA, Henderson JC, Biek R, et al. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12107–12111. doi: 10.1073/pnas.0500057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen WK. Landscape Epidemiology of Vector-Borne Diseases. Annual Review of Entomology. 2009;55:461–483. doi: 10.1146/annurev-ento-112408-085419. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Bannasch DL, Chomel BB, Ernest HB. Coyotes demonstrate how habitat specialization by individuals of a generalist species can diversify populations in a heterogeneous ecoregion. Molecular Biology and Evolution. 2008;25:1384–1394. doi: 10.1093/molbev/msn082. [DOI] [PubMed] [Google Scholar]
- Schmaljohn C, Hjelle B. Hantaviruses: A global disease problem. Emerging Infectious Diseases. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MF, Patton JL. The diversification of South American murid rodents: evidence from mitochondrial DNA sequence data for the akodontine tribe. Biological Journal of the Linnean Society. 1993;50:149–177. [Google Scholar]
- Steppan SJ, Adkins RM, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Systematic Biology. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Stern A, Doron-Faigenboim A, Erez E, et al. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Research. 2007;35:W506–511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Li W, Li Z, Zhang F, Hui C. The effect of landscape heterogeneity on host–parasite dynamics. Ecological Research. 2009;24:889–896. [Google Scholar]
- Switzer WM, Salemi M, Shanmugam V, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Beta Version. Phylogenetic Analysis Using Parsimony (* and other methods) Sinauer Associated; Sunderland, MA, USA: 2002. [Google Scholar]
- Toro J, Vega JD, Khan AS, et al. An outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerging Infectious Diseases. 1998;4:687–694. doi: 10.3201/eid0404.980425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Pérez F, Navarrete-Droguett J, Aldunate R, et al. Peridomestic small mammals associated with confirmed cases of human hantavirus disease on southcentral Chile. American Journal of Tropical Medicine and Hygiene. 2004;70:305–309. [PubMed] [Google Scholar]
- Torres-Pérez F, Palma RE, Hjelle B, Ferres M, Cook JA. Andes virus infections in the rodent reservoir and in humans vary across contrasting landscapes in Chile. Infection, Genetics and Evolution. 2010;10:820–825. doi: 10.1016/j.meegid.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veblen TT. Temperate Forests of the Southern Andean Region. In: Veblen TT, Orme AR, Young KR, editors. The Physical Geography of South America. Oxford University Press; Oxford: 2007. pp. 217–231. [Google Scholar]
- Webb SD, Keeling MJ, Boots M. Host-parasite interactions between the local and the mean-field: how and when does spatial population structure matter? Journal of Theoretical Biology. 2007;249:140–152. doi: 10.1016/j.jtbi.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Wells RM, Estani SS, Yadon ZE, et al. Seroprevalence of antibodies to hantavirus in health care workers and other residents of southern Argentina. Clinical Infectious Diseases. 1998;27:895–896. doi: 10.1086/517161. [DOI] [PubMed] [Google Scholar]
- Wells RM, Estani SS, Yadon ZE, et al. An unusual hantavirus outbreak in southern Argentina: Person-to-person transmission? Emerging Infectious Diseases. 1997;3:171–174. doi: 10.3201/eid0302.970210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, Underwood EC, Morrison SA, et al. Conserving Biodiversity Efficiently: What to Do, Where, and When. PLoS Biology. 2007;5:e223. doi: 10.1371/journal.pbio.0050223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian Populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Vincent MJ, Nichol ST. Thottapalayam virus is genetically distant to the rodent-borne hantaviruses, consistent with its isolation from the Asian house shrew (Suncus murinus) Virology Journal. 2007;4:80. doi: 10.1186/1743-422X-4-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates TL, Mills JN, Parmenter CA, et al. The ecology and evolutionary history of an emergent disease: Hantavirus pulmonary syndrome. Bioscience. 2002;52:989–998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.