Abstract
The curated CSAR-NRC benchmark sets provide valuable opportunity for testing or comparing the performance of both existing and novel scoring functions. We apply two different scoring functions, both independently and in combination, to predict binding affinity of ligands in the CSAR-NRC datasets. One, reported here for the first time, employs multiple chemical-geometrical descriptors of the protein-ligand interface to develop Quantitative Structure – Binding Affinity Relationships (QSBAR) models; these models are then used to predict binding affinity of ligands in the external dataset. Second is a physical force field-based scoring function, MedusaScore. We show that both individual scoring functions achieve statistically significant prediction accuracies with the squared correlation coefficient (R2) between actual and predicted binding affinity of 0.44/0.53 (Set1/Set2) with QSBAR models and 0.34/0.47 (Set1/Set2) with MedusaScore. Importantly, we find that the combination of QSBAR models and MedusaScore into consensus scoring function affords higher prediction accuracy than any of the contributing methods achieving R2 of 0.45/0.58 (Set1/Set2). Furthermore, we identify several chemical features and non-covalent interactions that may be responsible for the inaccurate prediction of binding affinity for several ligands by the scoring functions employed in this study.
Introduction
Scoring functions play a critical role in structure-based virtual screening.1, 2 An ideal scoring function can guide docking programs to generate and identify native-like docking poses. Based on the correct docking models, an ideal scoring function can also predict the binding affinity and correctly rank all compounds in the virtual screening library. Still, despite extensive research over many years, the accuracy of scoring functions remains a major bottleneck in structure-based virtual screening.3, 4
The binding affinity is defined by free energy of the protein-ligand binding. Direct calculation of free energy requires extensive sampling in the conformational space, which is generally infeasible except in a few special cases. Given the computational inefficiency of conformational sampling, certain approximations or assumptions are often made to estimate the binding free energy using physical force field models that sometime also account for implicit solvation.5, 6 With the improvement of the underlying force fields and increased computational power of modern computers, the performance of binding affinity calculations using physical methods is expected to improve gradually. On the other hand, there are alternative approaches that take advantage of the rapidly growing data on the experimental binding affinity of many compounds. These experimental databases are used to derive empirical scoring functions or statistical models to predict the binding affinity.7, 8 Such knowledge-based scoring functions may capture certain factors that are often ignored or difficult to describe explicitly using physical force field-based scoring functions such as entropic contribution, pi-stacking, or environment-dependent polarization.
To improve the outcome of structure based drug discovery, there is a great need for an unbiased comprehensive test set to compare different scoring functions and identify their respective strengths and limitations, which may lead to novel ways to further improve the accuracy of binding affinity prediction. The recently established Community Structural-Activity Resources (CSAR)-National Research Council of Canada (NRC) high quality benchmark set9 (abbreviated as CSAR-NRC in the following sections) provides excellent opportunities to develop and benchmark different scoring functions. This benchmark set contains two diverse subsets (Set1 and Set2) of protein-ligands complexes whose experimental binding affinity as well as high-resolution x-ray structures are available.
In this study, we employ the CSAR-NRC benchmark set to test two scoring functions of very different nature. One is MedusaScore6, which is a force field based scoring function derived from Medusa force field10 and originally designed for protein folding simulations. To ensure the best transferability of MedusaScore, its parameters are based on physicochemical properties and no protein-ligand complex data are used for training. Second is based on quantitative structure binding affinity relationship (QSBAR) modeling,11 an approach that correlates special descriptors of protein-ligand interface to ligand binding affinity using statistical modeling approaches. In the previous study, QSBAR models were constructed from 264 x-ray protein-ligand complexes with known binding affinity using protein-ligand interfacial descriptors derived from Pauling electronegativity. Herein, we develop novel descriptors by incorporating conceptual DFT atomic properties12 into the generation of protein-ligand interfacial descriptors and use high-quality CSAR-NRC sets to construct and validate QSBAR models that are used to predict binding affinity of ligands in external datasets. These empirical QSBAR models may be able to capture implicitly some subtle interactions that are difficult to calculate and that may be ignored by physical force fields.
We find that both scoring functions, i.e., MedusaScore and QSBAR models afford reasonably good performance in binding affinity prediction for CSAR-NRC ligands. Moreover, when combining the two scoring functions together, we find that the consensus scoring function improves the prediction accuracy compared to each individual scoring function. We attribute this observation to the complementarity of the two types of scoring functions that employ completely different principles to capturing and representing protein-ligand interactions as well as to higher accuracy of consensus prediction versus individual components. More specifically, we find that sets of prediction outliers from each scoring function do not completely overlap. Also, by analyzing the prediction outliers for each scoring function based on their protein family membership and their chemical features, we identify several distinct chemical features and specific non-covalent interactions, which are associated with wrong predictions. Some of these traits are specific to outliers when using MedusaScore while others are characteristic of QSBAR model. Such analysis not only provides insights into complementarity between these two types of scoring functions but also gives possible clues for future improvement of their accuracy.
Methods
Dataset
The CSAR-NRC High Quality (CSAR-NRC HiQ) sets are downloaded from the CSAR web site.9 The two sets, Set1 and Set2, included in the package contain 176 and 167 complexes, respectively. The descriptive analysis of the two data sets, based on the binding affinity of complexes and the protein family, is shown in Table 1. For each of the downloaded complexes, the original Sybyl MOL2 format is converted to the PDB format using Openbabel 2.2.0.13 Due to current limitations of the MedusaScore program, we also remove all the capping residues from the protein structures using a Perl script.
Table 1.
The descriptive analysis of data sets based on protein-ligand binding pKd values and the protein families
| data set | Set1 | Set2 | |
|---|---|---|---|
| parameter | |||
| pKd values | Count | 176 | 167 |
| Mean | 6.23 | 6.07 | |
| Median | 6.25 | 6.19 | |
| Standard deviation | 2.31 | 2.18 | |
| Range/Lowest/Highest | 13.15/−0.15/13 | 10.7/1.4/12.1 | |
|
| |||
| sequence | # of families/# of singletons (90% sequence similarity) | 121/80 | 106/68 |
MedusaScore
MedusaScore6 is a physical force field-based scoring function that describes the major physical interactions between proteins and ligands, including van der Waals interaction, hydrogen bonding and solvation. It is calculated as a linear combination of various energy terms as:
| (1) |
where Evdw_attr, Evdw_rep are the attractive and repulsive part of the van der Waals (VDW) interaction; Esolv is the solvation energy; Ebb_hbond, Esc_hbond and Ebb_sc_hbond are the hydrogen bond energies formed between backbone atoms, between side chains, and between backbone and side chains, respectively. The design of the force field is similar to that of the Rosetta force field,14 which has also been widely used in protein folding and design. The VDW interaction model and parameters are adapted from CHARMM19.15 The solvation model is the EEF1 implicit solvent model proposed by Lazaridis and Karplus.16 We use the hydrogen bonding model proposed by Kortemme and Baker.17 When evaluating the non-bonded interactions, we use a cutoff distance of 9.0 Å. The van der Waals repulsion (VDWR) potentials are implemented with linear extrapolation to dampen the fast increase of the potential as:
Here, rij is the distance between two atoms i and j. The energy parameters ε and σ are taken from the CHARMM19 force field of united atoms.15 Since the energy terms originate from different sources, a set of weighting parameters is assigned in order to balance their respective contributions.
MedusaScore is an extension of the Medusa force field10, which was developed originally to describe physical interactions within proteins. The original weighing factors of the Medusa force field were trained on 34 high-resolution protein crystal structures with diverse sequences. Notably there were no protein-ligand data used in the development of MedusaScore, but it still exhibits remarkable accuracy in both docking pose discrimination and binding affinity prediction.6 Thus, by default MedusaScore is expected to be transferable and applicable to virtual screening of a variety of chemical compounds. During the pose rescoring by MedusaScore, we turn off the VDWR term because it was shown to be sensitive to small deviation in ligand poses.6 It is safe to remove the term in this case because all steric clashes have already been considered during the generation of docking poses.
Quantitative Structure-Binding Affinity Relationships (QSBAR) models
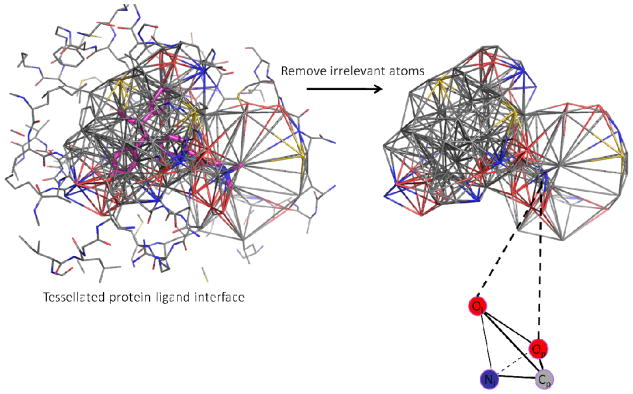
The QSBAR models derived from either Set1 or Set2 using novel descriptors of the protein-ligand interface are applied to predict either Set2 or Set1, respectively. The protein-ligand interfacial descriptors used in the QSBAR modeling are the combination of newly developed PL/MCT-Tess descriptors and the published EnTess descriptors.11 The PL/MCT-Tess descriptors are methodologically similar to the EnTess descriptors but are theoretically distinctive. The EnTess descriptors are obtained by using Pauling electronegativity (En) as atomic property and Delaunay Tessellation (Tess) to characterize the protein ligand interface as follows (Figure 1). When applied to protein-ligand complexes represented at the atomic resolution level, Delaunay tessellation partitions the protein ligand interface into an aggregate of space-filling, irregular tetrahedra where both protein and ligand atoms are vertices. Each Delaunay quadruplet is characterized by its unique four-atom composition, which defines the descriptor type (certainly, the same four-body compositions may occur in different or even, the same protein-ligand interfaces). Furthermore, for each quadruplet we calculate the sum of En values of the composing atom-vertices, which produces the descriptor value.
Figure 1.
Illustration of the method to derive PL/MCT-Tess descriptors using the tesselated protein-ligand complex (3ERT, the ER/antagonists benchmarking dataset). The atom types for protein and ligand are treated differently. For instance, for the tetrahedron at the left corner, Cp and Op are carbon and oxygen atoms from the protein while Ol and Nl are oxygen and nitrogen atoms from the ligand.
In the implementation of PL/MCT-Tess descriptors, the new descriptors employ pairwise atomic potentials for the protein-ligand complexes (PL) based on maximal charge transfer (MCT)12 in place of Pauling electronegativities; thus, we call them PL/MCT-Tess. The values of PL/MCT-Tess descriptors are calculated from the following equation:
| (2) |
where PL/MCT-Tessm is the potential of the m-th tetrahedron type defined by its four-atom composition (i.e., individual descriptor type); n is the number of occurrences of this tetrahedron type in a given protein-ligand complex; p is the index of protein vertex-atoms, l is the index of ligand vertex-atoms, and dpl is the distance between a pair of protein and ligand atoms found in the same Delaunay tetrahedron.
Since the Pauling En and MCT values used in two distinct sets of descriptors represent chemical properties based on distinctive but related theories, it is sensible to test the modeling performance using the combined descriptor set. We have found that employing models built by the combined descriptor set (PL/MCT-Tess + ENTess descriptors), the prediction accuracy is much better than using models built by any single descriptor set (data not shown). The combined descriptor set is constructed by concatenating the ENTess and PL/MCT-Tess descriptor sets. We remove descriptors in the combined descriptor set that have low variance (all, or all but one value is constant) and high correlation (if pair-wise square correlation coefficient is greater than 0.99, one of the pair, chosen randomly, is removed). The remaining descriptors are range scaled (0 to 1).
This combined descriptor set is applied to Set1 or Set2 to construct QSBAR models, where absolute binding affinity is represented as a function of protein-ligand interfacial descriptors. We use kNN algorithm with our standard model development and validation workflow reviewed recently.18 In brief, an n-fold external validation protocol is employed when the entire dataset is randomly divided into n nearly equal parts and then n-1 parts are systematically used for model development and the remaining fraction of compounds is used for model evaluation. In this study, ten-fold protocol was used for Set1 and ninefold protocol was used for Set2 due to its smaller size. The Sphere Exclusion protocol implemented in our laboratory19, 20 is used to rationally divide the remaining subset of compounds (the modeling set) into multiple training and test sets that are used for model development and validation, respectively. The model acceptability thresholds are characterized by the lowest acceptable value of the leave-one-out cross validated R2 (q2) for the training set and by conventional R2 for the test set; our default values are 0.5 for q2 and 0.6 for R2. All validated models are finally assesses in an ensemble using the external evaluation set. The resulting models based on Set1 (Set2) are then used to predict the binding affinity of Set2 (Set1) complexes.
Consensus protocol
The multiple linear regression method is applied to combine predictions from QSBAR models and MedusaScore. The equation is as follows:
| (3) |
where Yc is the consensus predicted affinity of a ligand, YMedusaScore is the raw prediction of MedusaScore which in theory is supposed to be in linear relationship with the experimental binding affinity, and YQSBAR is the affinity of the same ligand predicted by QSBAR model. The coefficients (b1, b2 and b3) in the equation are optimized by training based on the predictions from Set1 (Set2) of protein-ligand complexes. The equation with optimized coefficients is then applied to predict binding affinity of Set2 (Set1) complexes, respectively.
Comparison metrics
We report the squared correlation coefficient (R2) and two rank correlation coefficients, Spearman rho and Kendall tau, to measure the performance of a scoring function in terms of the correlation between the predicted score and the experimental binding affinity. In addition, because the QSBAR models report the absolute predicted binding affinity, we could also calculate the coefficient of determination when the regression line is forced to go through origin (i.e., the R02 value) as well as corresponding root mean square error (RMSE0) and root median square error (RMDSE0) values, where the median of residuals is used instead.
Outlier analysis
We define the prediction outlier of a scoring function as the protein-ligand complex whose predicted score is one standard deviation (σ) of residuals larger or smaller than its fitted value from the regression line. The remaining complexes are categorized as normal. Furthermore, we subdivide prediction outliers of each scoring function into two groups, over-predicted and under-predicted. For each group, we analyze its distribution among protein families based on 90% sequence similarity threshold. We also identify chemical features specific for the ligands in the outlier complexes. To this end, we generate structural fragments and analyze their distribution between outlier and normal groups. The fragments (sequences of atoms and bonds from 2 to 6 atoms in length, ~1000 unique substructures in total) are generated by the ISIDA Fragmentor21 program, which we chose for its efficiency and availability (free of charge to academic investigators); but the same analysis is possible with other fragment-generating software. Same as for PL/MCT-tess and EnTess descriptors, we remove highly-intercorrelated and low-occurrence fragments. The statistical analysis of fragment distribution is done by permutation test in Matlab 7.7.0. Only the fragments that show significantly higher frequency of occurrence (z-score > 2) in outliers are kept for further analysis.
Results
The complete performance statistics of each scoring function against either Set1 or Set2 is reported in Table 2. The correlation plot of each scoring function and the distribution of predictions are shown in Figures 2 and 3. The IDs of complexes with their predicted scores (or absolute pKd values) are reported in Table S1 and the IDs of complexes whose binding affinities are under-predicted or over-predicted are reported in Table S2 and Table S3 for each scoring function. We will explain the performance of each scoring function in the following section and discuss the chemical moieties and protein families which tend to point to the complexes that are being under-predicted or over-predicted (Table 4).
Table 2.
The statistics (R2, R02, MAE, RMSE0, and RMDSE0 as well as number of complexes predicted) for predicting Set1 and Set2 with respective QSBAR models, MedusaScore, and the consensus approach. The description of metrics can be found in Methods.
| Set1 predictions
| |||||||
|---|---|---|---|---|---|---|---|
| Parameter | R2 | R02 | Spearman | Kendal | RMSE0 | RMDSE0 | # of complexes |
| Method | |||||||
| QSBAR | 0.44 | 0.44 | 0.50 | 0.68 | 1.75 | 1.09 | 176 |
| MedusaScore | 0.34 | NA | −0.42 | −0.59 | NA | NA | 175 |
| Consensus | 0.45 | 0.45 | 0.51 | 0.69 | 1.72 | 1.07 | 175 |
| Set2 predictions | |||||||
|---|---|---|---|---|---|---|---|
| QSBAR | 0.53 | 0.53 | 0.55 | 0.75 | 1.50 | 1.02 | 167 |
| MedusaScore | 0.47 | NA | −0.48 | −0.67 | NA | NA | 164 |
| Consensus | 0.58 | 0.57 | 0.57 | 0.77 | 1.43 | 0.97 | 164 |
RMSE0: Root mean square deviation
RMDSE0: Root median square deviation
Figure 2.
The distribution of predicted values for Set1 (or Set2) by QSBAR models, MedusaScore, or the combined scoring function. The x-axis is the predicted binding affinity (QSBAR models and the combined scoring function) or the MedusaScore. The y-axis is the experimental binding affinity. The black line is the linear regression line and the yellow line is the regression line forced through the origin. The red lines are parallel to the black regression line and stand one standard deviation of the residuals away from it. The points beyond or below the red lines are considered as outliers.
Figure 3.
The residual distribution plot. The x-axis is the x-error relative to the fitting line (i.e., residual) and the y-axis is the # of complexes. The red dotted lines represent the values which are ± one standard deviation of the residuals. The region between two red lines shows the density of complexes which have “normal” prediction errors.
Table 4.
Some of the chemical features associated with the under-predicted or over-predicted complexes
| Features associated with under-predicted complexes | |||
|---|---|---|---|
| MedusaScore | |||
| ISIDA Fragments21 | Representation | Ratio** | Example*** |
| S-C=N | Thiazole | 7/7 |

|
| S-C-C=C-S | Sulfonamide connected to sulfur- containing heterocycle (e.g., thiophene) | 6/7 |

|
| QSBAR models | |||
| C=C-C-O-C=C | flavan-derivative | 3/4 |

|
| Features associated with over-predicted complexes | |||
| MedusaScore | |||
| P-O-C-O-C-C | Phosphate group + sugar (often in a nucleoside) | 3/4 |

|
| QSBAR models | |||
| C=C-C=C-C=C | Naphthalene ring | 4/8 |

|
# of complexes in the under-predicted (or over-predicted) group with the feature / # of total complexes with the feature
Example shows a fragment (solid lines) mapped onto the actual molecular scaffold (dashed lines).
MedusaScore
We calculate MedusaScore for both Set1 and Set2. We use the VDWR-excluded protocol with no additional parameter adjustment. There are four complexes that contain ligand atom types that are not yet parameterized by MedusaScore (trimethylsulfonium groups in complex #183 in set1, and #249 and #74 in set2, as well as the phosphoramide group in complex #18 in set2). The R2 values are 0.34 and 0.47, for Set1 and Set2, respectively. We also test the effect of adding the VDWR term in MedusaScore. The R2 are slightly decreased to 0.30 and 0.44 for Set1 and Set2, respectively. The slight decrease of accuracy is consistent with the previous observation6 that the VDWR term is more sensitive to small deviations in the complex structure, causing uncertainty for binding energy estimation. The observation that accuracy only slightly decreases after including the VDWR term for prediction also verifies that the CSAR data sets are of high quality and only minimal steric clashes exist in the structure of the complexes. The largest VDWR interaction energy is found to be 29.7 kcal/mol for complex #154 in Set1. There are other three complexes in Set2 (complex #225, #222, and #92) that also have VDWR term larger than 20 kcal/mol. These complexes are found to be under-predicted by MedusaScore.
In total, there are 31.3% of Set1 complexes and 34.7% of Set2 complexes considered as outliers based on the definition described in Methods. A majority of complexes belonging to the glutamate-related family (glutamate receptor 1, 2, 3, 4, 6) are under-predicted by MedusaScore. Closely inspecting the protein-ligand interactions in those complexes, we find salt-bridge interactions, which are ignored in the current version of MedusaScore, are dominant. Moreover, both of the two complexes in the family of ADAM17 are under-predicted. This might be due to ignoring of metal-mediated interactions (the catalytic Zinc) in the binding pocket, where metals directly contribute to the ligand binding.
We have also analyzed the structural fragments based on their tendency of occurring in outliers in comparison with the normal group. We find that the combination of thiolane/thiophene moiety and the sulfonamide (or amide) group tends to contribute to the under-prediction of certain complexes (Table 4). For example, the four protease complexes (1:158, 1:159, 1:160, 1:161) and three coagulation factor X complexes (1:52, 1:141, 1:196) are under-predicted by MedusaScore. The most interesting chemical scaffold is the thiazole group, which seems to be strongly associated with the under-prediction of binding affinity. The thiazole group can be found in the four protease complexes (vide supra) and two carbonic anhydrase-related complexes (1:206 and 1:222). Moreover, MedusaScore tends to over-predict complexes which contain phosphate groups connected to a sugar moiety (usually in a nucleoside ligand).
QSBAR models
After removing descriptors with high inter-correlation and low variance, there are 422 and 377 descriptors (out of 1108 descriptors) used in modeling building and validation of Set1 and Set2, respectively. The results of external n-fold cross validation from CSAR data set modeling are reported in Table 3. The average external n-fold cross-validation R2 is 0.45 for Set1 and 0.53 for Set2. Since each fold has rather small size (around 17 complexes), R2 values could have large fluctuation due to the random distribution of prediction outliers among folds. Therefore, we also take MAE and RMSE values into account in the evaluation of prediction accuracy. We analyze the outliers in the fold(-s) with the worst MAE and RMSE values (i.e., fold#2 in Set1 and fold#1 in Set2). We find that some of the outliers have special moieties (and thus, could be viewed as structural outliers), for example, the N5-[(R)-amino(sulfoamino)phosphoryl] group (2:18) and the hydroxy(oxo)phosphoniumolate group (1:25), or the whole family (Lipocalin) of complexes (1:207 and 1:208) is not present in the modeling set. On the other hand, in spite of having close neighbors in the descriptor space, some complexes are still predicted poorly (e.g., 2:126), suggesting that further improvement of protein-ligand interfacial descriptors is needed.
Table 3.
The statistics (R2, MAE, and RMSE) for external n-fold validation sets using QSBAR models built from Set1 and Set2.
| “Set1” data set modeling
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 |
| Parameter | ||||||||||
| R2 | 0.2 | 0.21 | 0.68 | 0.54 | 0.57 | 0.4 | 0.56 | 0.65 | 0.63 | 0.42 |
| MAE | 1.25 | 1.56 | 1.16 | 1.57 | 1.21 | 1.34 | 1.19 | 1.09 | 1.21 | 1.36 |
| RMSE | 1.58 | 1.85 | 1.48 | 1.84 | 1.53 | 2.01 | 1.5 | 1.36 | 1.49 | 1.71 |
| “Set2” data set modeling
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | NA |
| Parameter | ||||||||||
| R2 | 0.27 | 0.55 | 0.73 | 0.64 | 0.53 | 0.64 | 0.72 | 0.52 | 0.36 | NA |
| MAE | 1.63 | 0.89 | 1.18 | 1.11 | 1.11 | 1.04 | 0.9 | 1.51 | 1.4 | NA |
| RMSE | 2.18 | 1.15 | 1.46 | 1.21 | 1.34 | 1.23 | 1.2 | 2.04 | 1.73 | NA |
The validated Set1 (Set2) models are applied to predict Set2 (Set1). The results are reported in Table 2. The prediction accuracy of Set2 using Set1 models is higher than the prediction accuracy of Set1 using Set2 models (i.e., R2 value is 0.44 vs. 0.53, respectively). It is an expected outcome, because QSAR-based models have difficulty extrapolating data points under-represented in the training set, and, indeed, Set1 has more data points at the extremes of the binding affinity distribution.
We analyze the prediction outliers as described in Methods. About 29.5% of Set1 complexes and 23.3% of Set2 complexes are considered ill-behaved (i.e., outliers) by QSBAR models. Around 1000 ISIDA fragments are generated for Set1 and Set2. After removing fragments with low variance or high correlation, around 600 fragments in either Set1 or Set2 remain for the permutation test. Upon the analysis, we find that the ligands, which contain the flavan moiety or the combination of thiolane/thiophene moiety and the amide group, tend to be under-predicted by QSBAR models (Table 4). The flavan moiety occurs in the ligand complexes of particular protein families. For example, the complexes belong to the estrogen receptor-β (1:42, 1:43) and estrogen receptor (1:33) family. Coincidentally, the features of thiolane/thiophene and sulfonamide group are also found to contribute to the under-prediction by MedusaScore (vide supra). On the other hand, the ligands with naphthalene moiety tend to be over-predicted only by QSBAR models (e.g., 2:19, 2:23, 2:44, and 2:77). Moreover, the carboxyalkyl phosphate scaffold (with or without metal coordination) is found to be associated with over- or under-prediction. We find that complexes whose ligands contain large hydrophobic moieties (e.g., flavan and naphthalene) in a hydrophobic environment tend to be mis-predicted; this points to the underlying assumption of PL/MCT-tess descriptors that protein-ligand binding is driven mostly by charge transfer interactions. Moreover, the hybridization of carbon is not taken into account in the current implementation of PL/MCT-tess and EnTess descriptors. These factors may contribute to the low accuracy of prediction for compounds containing large hydrophobic moieties.
Comparing prediction outliers from QSBAR models and from MedusaScore scoring function, we find that these groups do not completely overlap (Figure S1) and the corresponding structural features associated with QSBAR outliers are distinct from the ones for MedusaScore. This outcome is not unexpected since these two types of scoring function employ completely different principles toward representing protein-ligand interactions. This also implies the possibility of improving overall prediction accuracy by combining the two scoring functions.
Distribution of chemical fragments of ligands in CSAR data set
We also analyze the distribution of ligand chemical features (represented by ISIDA fragments) in the entire CSAR data set. Figure 4 shows the occurrence of each chemical fragment (in % to that of the CSAR data set) in Set1 and Set2 ligands. The fragments are sorted by predominance of occurring in Set1. Overall, Set1 is chemically more diverse than Set2. Approximately 70% of chemical fragments are more prominent in Set1 than in Set2, and around 4% are unique for Set1. On the contrary, all of the fragments predominant in Set2, though under-represented, can still be found in Set1. The fragments marked by circles or squares (Figure 4) are associated with previously identified prediction outliers (e.g. flavan, thiolane/thiophene and sulfonamide ligand features). As expected, these chemical fragments are not represented equally in Set1 and Set2. This analysis suggests that the predictive power of Set2 models can be improved by extending the Set2 data set.
Figure 4.
Distribution of chemical features (ISIDA fragments, see Methods) in Set1 (blue) and Set2 (red); circles and squares denote structural features predominantly found in the ligands of mispredicted complexes (i.e. prediction outliers).
Interpretation of descriptors selected by QSBAR models
We analyze the descriptors selected by either Set1 or Set2 models (q2 ≥ 0.5 and R2 ≥ 0.6) based on their frequency of occurrence in the respective models. For each descriptor we calculate Z-score based on the frequency distribution of all selected descriptors in Set1 (Set2) models. Figure 5 shows descriptors sorted by the difference of their Z-scores in Set1 and Set2. We find that the descriptors whose tetrahedral type includes a metal are frequent in Set1 models (i.e. high Z-score) but not in Set2 models. This can explain some mispredictions of Set1 by Set2 models, because metal interactions are under-represented in Set2 data set. Moreover, those descriptors, whose tetrahedral type is related to the under-predicted outliers of Set1 (see scaffolds in Figure 5), are selected less frequently by Set2 models. Therefore, we expect that by expanding Set2 the prediction accuracy of the corresponding QSBAR models should improve.
Figure 5.
Relative frequencies (z-score) of PL/MCT-Tess descriptors selected by either Set1 or Set2 models.
Consensus scoring function
We optimize the b1, b2, and b3 parameters of the combined scoring equation (see Methods: Eq. 3) using Set1 predictions by QSBAR models and by MedusaScore. The R2 value between the fitted combined score and the experimental binding affinity is 0.45 and the respective parameters (b1, b2, and b3) are 0.58, −0.03, and 0.82. Applying the trained scoring function to Set2 gives R2 of 0.58, which is higher than the R2 value by using QSBAR models and significantly higher than MedusaScore alone (p < 0.05 by permutation test, N=10,000) ). This suggests the complementarity of these two types of scoring functions. Consequently, we apply the same procedure to optimize the combined scoring equation using Set2 predicted scores. The resulting R2 value is 0.58 and b1, b2, and b3 parameters are 0.002, −0.03, and 0.87, respectively. Applying the trained scoring function to Set1 gives R2 of 0.45, which is slightly higher than R2 by QSBAR and significantly higher than MedusaScore alone (p < 0.05). The relatively limited improvement over the individual QSBAR model might be due to the poorer performance on Set1 by each of the individual scoring functions.
We also analyze the prediction outliers of the combined scoring function. There are about 27.2% of Set1 complexes and 33.5% of Set2 complexes considered as prediction outliers. The percentage of outliers in Set2 for the combined scoring function is not as low as in the case of QSBAR models despite the fact that the overall performance of the combined scoring function for Set2 is better. By analyzing chemical features of outliers, we find characteristic moieties that correspond to those obtained for QSBAR models. For example, the thiolane/thiophene moiety with sulfonamide group and flavan-related scaffolds are found in the ligands of under-predicted complexes and the naphthalene moiety is in over-predicted ones.
Conclusions
We found that applying QSBAR models or MedusaScore individually can only afford predictions with relatively modest accuracy for the CSAR-NRC set. Interestingly, after combining the results from QSBAR models and MedusaScore, we found that the accuracy of binding affinity prediction improves (especially, for Set2), suggesting the complementary nature of the two types of scoring functions. By analyzing prediction outliers for each scoring function, we have identified distinct chemical features associated with mispredictions. Some of these features lead only to MedusaScore errors, while several others were indicative of mispredictions solely by QSBAR models. This analysis not only highlights the complementarity between these two types of scoring functions but also suggests further directions for improvement, such as the parameterization of metals and salt-bridge interactions for MedusaScore and the application of extended data sets for training QSBAR models.
Supplementary Material
Acknowledgments
These studies are supported by the National Institute of Health Grant R01GM080742 and the ARRA supplement 3R01GM080742-03S1 (to N.V.D.), GM066940 and its ARRA supplement GM066940-06S1 (to A.T.). The authors thank Dr. Denis Fourches for his assistance with using the ISIDA Fragmentor software and data analysis.
Footnotes
Supporting Information Available: predicted values by each of scoring functions and ID/family of under/over-predicted complexes from each of scoring function. This information is available free of charge via the Internet at http://pubs.acs.org/.
Reference List
- 1.Good A. Structure-based virtual screening protocols. Curr Opin Drug Discov Devel. 2001;4:301–307. [PubMed] [Google Scholar]
- 2.Lyne PD. Structure-based virtual screening: an overview. Drug Discov Today. 2002;7:1047–1055. doi: 10.1016/s1359-6446(02)02483-2. [DOI] [PubMed] [Google Scholar]
- 3.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 4.Cheng TJ, Li X, Li Y, Liu ZH, Wang RX. Comparative Assessment of Scoring Functions on a Diverse Test Set. J Chem Inf Model. 2009;49:1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 5.Meng EC, Shoichet BK, Kuntz ID. Automated Docking with Grid-Based Energy Evaluation. J Comput Chem. 1992;13:505–524. [Google Scholar]
- 6.Yin S, Biedermannova L, Vondrasek J, Dokholyan NV. MedusaScore: An accurate force field-based scoring function for virtual drug screening. J Chem Inf Model. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions .1. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput-Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 8.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J Mol Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 9. [assessed Sept 24, 2010];Community Structural-Activity Resources (CSAR) http://www.csardock.org/
- 10.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:725–733. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang SX, Golbraikh A, Tropsha A. Development of quantitative structure-binding affinity relationship models based on novel geometrical chemical descriptors of the protein-ligand interfaces. J Med Chem. 2006;49:2713–2724. doi: 10.1021/jm050260x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parr RG, Von Szentpaly L, Liu SB. Electrophilicity index. J Am Chem Soc. 1999;121:1922–1924. [Google Scholar]
- 13. [assessed July 4, 2008];The Open Babel Package, version 2.2.0. http://openbabel.sourceforge.net/
- 14.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 15.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 16.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins-Struct Funct Genet. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 17.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol Inf. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 19.Golbraikh A, Tropsha A. Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput-Aided Mol Des. 2002;5:231–243. doi: 10.1023/a:1020869118689. [DOI] [PubMed] [Google Scholar]
- 20.Golbraikh A, Shen M, Xiao Z, Xiao YD, Lee KH, Tropsha A. Rational selection of training and test sets for the development of validated QSAR models. J Comput-Aided Mol Des. 2003;17:241–253. doi: 10.1023/a:1025386326946. [DOI] [PubMed] [Google Scholar]
- 21.Varnek A, Fourches D, Hoonakker F, Solov’ev VP. Substructural fragments: an universal language to encode reactions, molecular and supramolecular structures. J Comput-Aided Mol Des. 2005;19:693–703. doi: 10.1007/s10822-005-9008-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.