Abstract
Bone morphogenetic protein (BMP) signaling pathways regulate multiple aspects of endochondral bone formation. The importance of extracellular antagonists as regulators of BMP signaling has been defined. In vitro studies reveal that the intracellular regulators, inhibitory Smads 6 and 7, can regulate BMP-mediated effects on chondrocytes. Although in vivo studies in which inhibitory Smads were overexpressed in cartilage have shown that inhibitory Smads have the potential to limit BMP signaling in vivo, the physiological relevance of inhibitory Smad activity in skeletal tissues is unknown. In this study, we have determined the role of Smad6 in endochondral bone formation. Loss of Smad6 in mice leads to defects in both axial and appendicular skeletal development. Specifically, Smad6−/− mice exhibit a posterior transformation of the seventh cervical vertebra, bilateral ossification centers in lumbar vertebrae, and bifid sternebrae due to incomplete sternal band fusion. Histological analysis of appendicular bones revealed delayed onset of hypertrophic differentiation and mineralization at midgestation in Smad6−/− mice. By late gestation, however, an expanded hypertrophic zone, associated with an increased pool of proliferating cells undergoing hypertrophy, was evident in Smad6 mutant growth plates. The mutant phenotype is attributed, at least in part, to increased BMP responsiveness in Smad6-deficient chondrocytes. Overall, our results show that Smad6 is required to limit BMP signaling during endochondral bone formation.
Keywords: BMP/Smad, growth plate, chondrocyte, rodent, growth and development
Introduction
Endochondral bone formation is the multistep process that is responsible for the development of the majority of the skeleton (reviewed in (1)). First, condensed mesenchymal cells in the developing limb bud differentiate into chondrocytes, which then contribute to the formation of the cartilage growth plate that is gradually replaced by bone. Growth plate chondrocytes undergo a complex program of proliferation, hypertrophic differentiation and apoptosis. The zone of hypertrophic chondrocytes is invaded by blood vessels, osteoblasts, and osteoclasts to initiate ossification of the cartilage matrix.
Bone morphogenetic protein (BMP) signaling pathways play important roles at multiple stages of endochondral bone formation. In particular, BMP signaling promotes chondrocyte proliferation, in part through upregulation of Sox9 and type II collagen expression (2,3). BMP signaling has also been shown to induce chondrocyte hypertrophy (4), but to delay terminal differentiation of chondrocytes (5). Thus, BMP signaling leads to increased chondrocyte proliferation and differentiation. The BMP signaling pathway is required for endochondral bone formation, as evidenced by severe chondrodysplasia in mice lacking BMP signaling components (6–8).
BMP signaling is initiated by the binding of ligands to heterotetrameric complexes of types I and II serine/threonine kinase receptors, leading to activation of downstream signaling mediators via two distinct mechanisms: the canonical Smad pathway and multiple noncanonical (non-Smad) pathways (9,10). In the canonical Smad pathway, BMP signals are transduced via phosphorylation of receptor-regulated Smads (R-Smads) 1, 5, and 8 by activated type I receptors. Once phosphorylated, R-Smads associate with the common partner Smad4, translocate into the nucleus, and interact with other transcription factors to induce the expression of downstream genes. In the noncanonical pathways, BMPs activate transforming growth factor β activated kinase 1 (TAK1), leading to the activation of the mitogen-activated protein kinases (MAPKs), including p38, JNK, and extracellular signal-regulated kinases (ERKs) (11).
The intensity and duration of BMP signaling can be regulated both extracellularly and intracellularly. Extracellular regulation occurs via binding of antagonists, such as noggin and chordin, to BMPs, thereby preventing them from binding to BMP receptor complexes. Intracellular regulation occurs, in part, through the actions of inhibitory Smads (I-Smads) 6 and 7. Smad7 can inhibit multiple pathways, including TGFβ/activin and BMP signaling, while Smad6 specifically inhibits BMP signaling (12,13). I-Smads block the phosphorylation of R-Smads by forming stable associations with activated type I receptors (14–16). In addition, I-Smads can recruit E3 ubiquitin ligases to type I receptors, leading to ubiquitination and subsequent degradation of these receptors (17,18). I-Smads can also bind to Smad1, thereby interfering with Smad1–Smad4 complex formation (19). The expression of I-Smads is directly induced by BMP signaling, thus forming a negative feedback loop (20,21).
The importance of extracellular BMP antagonists in regulating endochondral bone formation has been established (22,23). In fact, extracellular regulation by the BMP antagonist noggin is required, as noggin-deficient mice exhibit enlarged growth plates and joint fusions (22). Although gain- and loss-of-function studies reveal a role for I-Smads in chondrocytes in vitro (4,20,24), in vivo studies are limited to gain-of-function studies (25,26). Thus, whether I-Smads are required for normal endochondral bone formation is unknown. Here, we show that loss of Smad6 in mice results in both axial and appendicular skeletal defects that can be attributed to increased BMP responsiveness in chondrocytes. Thus, Smad6 limits BMP signaling for proper skeletal development.
Materials and Methods
Generation of Smad6−/− mice
Generation of Smad6−/− mice was previously described (27). Briefly, targeted disruption of the Madh6 (which encodes the Smad6 protein) was generated by insertion of a LacZ/neomycin resistance cassette at codon 342 of the exon encoding the MH2 domain. Embryos and mice were genotyped by PCR using the sense primer 5’-CCTTGCCATATCCTATGCTTGCG-3’ and anti-sense primer 5’-GCGCCGCACCGACTCACTGC -3’ to detect the wild-type allele, and the sense primer 5’-GCTTCCTCGTGCTTTACGGTATC-3’ and anti-sense primer as above to detect the mutant allele. Embryos and mice were on a mixed C57BL/6J/BALB/c background.
Skeletal preparation and histology
Skeletal preparations were performed as in (6). Briefly, embryos were eviscerated and fixed in 95 % EtOH overnight at 4 °C, followed by Alcian Blue staining (0.01 % Alcian Blue 8GX (Sigma-Aldrich, A5268) (w/v) in 95 % EtOH) overnight at room temperature. Samples were then stained for Alizarin Red (0.006 % Alizarin Red S (Sigma-Aldrich, A5533) (w/v) in 1 % KOH) for 3–4 hours and cleared in a series of graded KOH in glycerol.
For histology, embryos were fixed with 10 % buffered formalin (Fisher Scientific, SF100) overnight at 4 °C, decalcified with Immunocal (Decal Chemical Corp., Tallman, NY, USA) overnight at 4 °C, embedded in paraffin, and cut at a thickness of 5–7 µm. Sections were stained with Alcian Blue (Sigma-Aldrich, A5268) and Nuclear Fast Red (Sigma-Aldrich, N8002) as in (6). Von Kossa staining was performed by incubating sections with 1 % silver nitrate under UV light for 20 minutes and then counterstaining with Nuclear Fast Red.
Immunohistochemistry and immunoflourescence staining
For immunohistochemistry (IHC) and immunoflourescence staining, paraffin-embedded tissue sections were boiled in citrate buffer pH 6.0 for 15 min at 95 °C for antigen unmasking. Sections stained for extracellular matrix proteins (Type II Collagen, Type X Collagen, MMP-13 and osteopontin) were digested with 1 mg/ml hyaluronidase (Sigma-Aldrich, H3506) in Dulbecco’s PBS (Mediatech, Inc., Manassas, VA, USA) for 45 min at 37 °C prior to antigen unmasking.
For detection using the following primary antibodies: Ihh (Santa Cruz Biotechnologies, sc1196), MMP-13 (Abcam, ab84594), osteopontin (Thermo Scientific, RB-9097-P0), phospho-Smad1/5/8 (Cell Signaling, #9511), and Ptc1(Novus Biologicals, NB100-91923), sections were quenched in 3 % H2O2 in methanol, blocked with 0.5 % blocking reagent (TSA™ Biotin System, Perkin Elmer, NEL700A) in TBS (100 mM Tris pH 7.5, 150 mM NaCl), and incubated with primary antibody overnight at 4 °C. Detection of binding was performed using the TSA™ Biotin System according to the manufacturer’s protocol. Flourescence detection was conducted by using either the Streptavidin-AlexaFluor -555 (Invitrogen) secondary antibodies; sections were counterstained with DAPI (Invitrogen, D1306).
For detection of Type II Collagen (Abcam, ab21291) and Type X Collagen (Abcam, ab58632), sections were blocked and incubated with primary antibody, as described above, incubated with either the AlexaFluor-488 or -555 (Invitrogen) secondary antibodies for 30 min at room temperature, and then counterstained with DAPI. For detection of Smad6 (Santa Cruz Biotechnologies, sc-6034), sections were blocked, incubated with primary antibody, and quenched in 3 % H2O2 in methanol, as described above. Sections were then incubated with biotin anti-goat (Santa Cruz Biotechnologies) and Streptavidin-HRP (Perkin Elmer) secondary antibodies. Chromogenic detection was performed with the DAB Peroxidase Substrate Kit (Vector Laboratories, SK-4100) following the manufacturer’s instructions; sections were counterstained with Hematoxylin QS (Vector Laboratories, H-3404).
Cell proliferation and TUNEL labeling
For detection of cell proliferation in vivo, immunofluorescence staining was performed by using either the anti-PCNA primary antibody (Zymed, #13-3900) or phospho-histone H3 (Ser10) antibody (Cell Signaling, #9701), and biotin-XX anti-mouse (Invitrogen) and Streptavidin-AlexaFluor-555 (Invitrogen) secondary antibodies. For TUNEL labeling, the fluorescein In Situ Cell Death Detection Kit (Roche Applied Sciences) was used according to the manufacturer’s instructions.
Cell culture and luciferase reporter assays
Primary chondrocytes were isolated from costal cartilage as previously described (28). For quantitative real-time PCR and semi-quantitative RT-PCR, cells were seeded at 1 × 106 in 6-well plates and maintained for 7 or 10 days in chondrogenic media (DMEM supplemented with 10 % FBS, 1 % antibiotic-antimycotic (Invitrogen) and 50 µg/ml ascorbic acid). Cells were serum starved overnight with DMEM supplemented with 1 % FBS and 1 % antibiotic-antimycotic (Invitrogen), then stimulated the next day with 100 ng/ml BMP2 (R & D Systems, 355-BM) for 2 hours or with an equal volume of DMSO for untreated cells. Each experiment was repeated twice (i.e. from two different cell isolations).
For western blot analysis, cells were seeded at 100 × 103 cells/well in 12-well plates and maintained for 10 days in chondrogenic media. Cells were serum starved overnight with DMEM supplemented with 1 % antibiotic-antimycotic (Invitrogen), then stimulated the next day with 50 ng/ml BMP2 (R & D Systems, 355-BM) for 2, 4, 12, or 24 hours or with an equal volume of DMSO for untreated cells (0 hour). Each experiment was repeated in triplicate (i.e. from three different cell isolations).
For luciferase reporter assays, cells were seeded at 100 × 103 cells/well in 12-well plates and then transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions with 0.5 µg of the 560 bp-Msx2-TKluc (29) and 0.05 µg of the pRL-TK (Promega) reporter plasmids. Cells were serum starved for 24 hours post-transfection with DMEM supplemented with 1 % antibiotic-antimycotic (Invitrogen), and then stimulated with the next day with 50 ng/ml BMP2 for 2 or 8 hours or with an equal volume of DMSO for untreated cells (0 hour). Firefly and Renilla luciferase activities were measured with the Dual-Luciferase Promoter Assay System (Promega). Firefly luciferase activities were obtained in triplicate for each experiment and were normalized to Renilla luciferase activities. Statistical significance was assessed via Within-Subjects Multifactorial ANOVA using the ezANOVA statistical software (http://www.cabiatl.com/mricro/ezanova/index.html).
Quantitative real-time PCR, Semi-quantitative RT-PCR, and Western blot analysis
RNA was extracted using the RNeasy Kit (Qiagen). Synthesis of cDNA was performed with Superscript III (Invitrogen). Quantitative real-time PCR reactions were performed with a SYBR Green Real-time PCR Master Mix (Fermentas) by using an Mx3005P QPCR System (Stratagene). The primer sequences were as follows: β-actin: forward 5’-CTGAACCCTAAGGCCAACCG-3’, reverse 5’- GTCACGCACGATTTCCCTCTC-3’; Ihh (from (30)): forward 5’-GACTCATTGCCTCCCAGAACTG-3’, reverse 5’-CCAGGTAGTAGGGTCACATTGC-3’; PPR: forward 5'- CTGGCCATTGGGGGCACCAG -3', reverse 5'-CGGCGCGCAGCATAAACGAC-3'; Ptc1 (from (31)): forward 5’-CAAGTGTCGTCCGGTTTGC-3’, reverse 5’-CTGTACTCCGAGTCGGAGGAA-3’; PTHrP (from (30)): forward 5'-GAACATCAGCTACTGCATGACAAG-3', reverse 5'-TCTGATTTCGGCTGTGTGGATC-3'; Smad6: forward 5’-ATTCTCGGCTGTCTCCTCCT-3’, reverse 5’-CCCTGAGGTAGGTCGTAGAA-3’; type II Collagen (from (32)): forward 5’-ACTGGTAAGTGGGGCAAGAC -3’, reverse 5’- CCACACCAAATTCCTGTTCA -3’. Semi-quantitative RT-PCR reactions were comprised of 25–42 cycles at 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. The primer sequences were as follows: GAPDH: forward 5’-CCAGAACATCATCCCTGCATC-3’, reverse 5’-GGTAGGAACACGGAAGGCC-3’; type II collagen: forward 5’-AATGGGCAGAGGTATAAAGATAAGGA-3’, reverse 5’-CATTCCCAGTGTCACACACACA-3’; type X collagen: forward 5’-CAAACGGCCTCTACTCCTCTGA-3’, reverse 5’- CGATGGAATTGGGTGGAAAG-3’.
For western blot analysis, cells were lysed in RIPA buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1 % NP-40, 1 % Na-deoxycholate, 0.1 % SDS) supplemented with protease inhibitors (complete Mini tablets, Roche Applied Science) and phosphatase inhibitors (Sigma-Aldrich, P5726). Whole cell lysates (5 µg) were run on 10 % SDS-polyacrylamide gels and transferred semi-dry onto PVDF membranes. The membranes were blocked with 5 % milk in TBS-tween (30 mM Tris pH 7.4, 300 mM NaCl, 0.2 % tween-20), incubated with primary antibody (from Cell Signaling: phospho-p38 (#9215), p38 (#9212), phospho-Smad1/5/8 (#9511) or Smad5 (#9517); Abcam: BMPR1A (#ab59947); Sigma-Aldrich: β-actin (A5316) or tubulin (T6793)) diluted in blocking buffer overnight at 4 °C, and then incubated with appropriate secondary antibody diluted in blocking buffer for 1 hour at room temperature. Binding was detected via enhanced chemiluminesence using the Amersham™ ECL Plus kit (GE Healthcare).
Results
Smad6 localization in cartilage elements during development
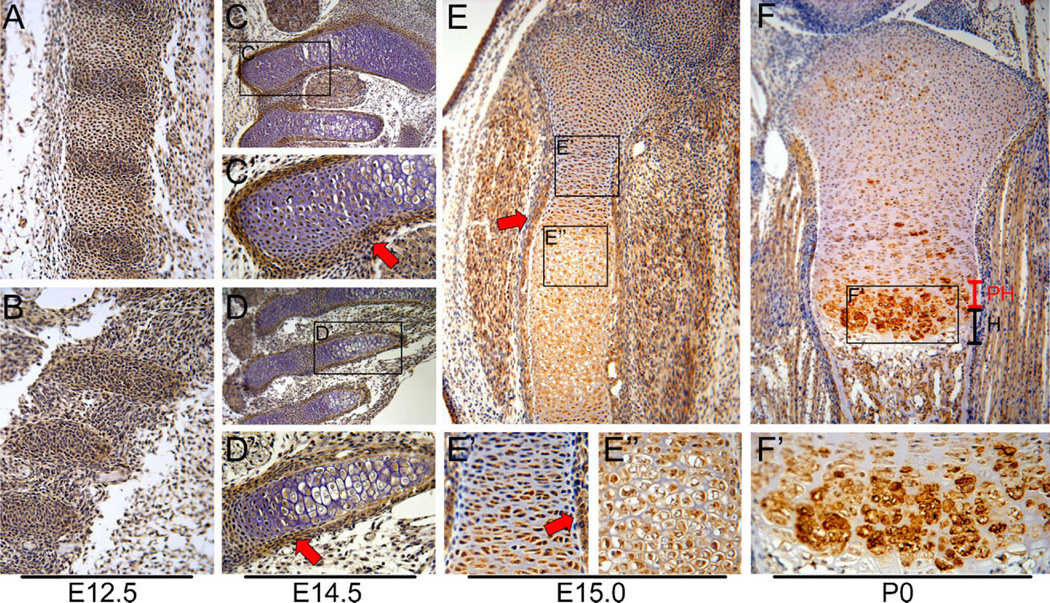
The expression of Smad6 in the developing axial skeleton was examined by IHC on E12.5 wild-type embryos. Smad6 was expressed in the primordia of vertebrae and intervertebral discs (Fig. 1A), ribs (Fig. 1B) and sternebrae (not shown). By E14.5, Smad6 was detected in the growth plates and perichondria of vertebrae (Fig. 1C, C’), ribs (Fig. 1D, D’) and sternebrae (not shown).
Figure 1.
Smad6 localization in cartilage during development. Immunohistochemistry of sagittal sections of wild-type embryos at E12.5 for Smad6 (brown color) demonstrates expression in (A) developing vertebral bodies and intervertebral discs of lumbar vertebrae and (B) developing anterior ribs. Staining of sagittal sections at E14.5 shows Smad6 expression in (C) cervical vertebrae and (D) anterior ribs. Higher magnification of the (C’) lateral region of C1 and (D’) 2nd rib shows Smad6 expression in the growth plate and perichondrium (red arrows). (E) Smad6 expression in tibiae at E15.0. (E’) Smad6 is localized in the cytoplasm and nucleus of proliferative cells at E15.0. Smad6 is also expressed in the perichondrium (red arrow) (E”) Smad6 is localized in the cytoplasm of hypertrophic cells at E15.0. (F) By postnatal day 0 (P0), Smad6 levels are highest in the prehypertrophic and upper hypertrophic zones of the tibial growth plate. (F’) Smad6 is localized in both the cytoplasm and the peripheral cell membrane of hypertrophic cells. For all stains, no detectable staining was observed in controls, from which primary antibodies were omitted. PH, prehypertrophic; H, hypertrophic zone.
In appendicular bones, Smad6 was expressed throughout the growth plate at E15.0 (Fig. 1E). Examination of subcellular localization revealed that Smad6 was localized in both the cytoplasm and nucleus of proliferating cells (Fig. 1E’), while it was localized predominantly in the cytoplasm of hypertrophic cells (Fig. 1E”). At P0, Smad6 expression persisted in the reserve and proliferative zones of growth plates, while higher levels were observed in the prehypertrophic and upper hypertrophic zones; no expression was seen in terminal hypertrophic chondrocytes (Fig. 1F). At this stage of development, Smad6 expression was localized in both the cytoplasm and the peripheral cell membrane of hypertrophic cells (Fig. 1F’). These results suggest that Smad6 may play a role in regulating chondrocyte proliferation and hypertrophic differentiation throughout chondrogenesis in vivo.
Skeletal defects in Smad6−/− mice
Smad6 knockout (Smad6−/−) mice were recovered at Mendelian ratios up to E18.5, while the majority of Smad6−/− mice died within 24 hours after birth (i.e. Smad6−/− mice represented 5 % of heterozygous intercross progeny (n = 112)). The mutant mice that did survive were slightly smaller compared to their wild-type littermates at P0 (Fig. 2A.) The early lethality of Smad6−/− mice in our hands is in contrast to the results of Galvin and colleagues, who reported that some Smad6−/− mice survive to early adulthood (i.e. 3–13 % of heterozygous intercross progeny); but that those mice that survive to this stage die of cardiovascular defects, including cartilaginous metaplasia of the aorta (27). The cause of the earlier lethality observed in our study is unknown, but may reflect differences in the background strain.
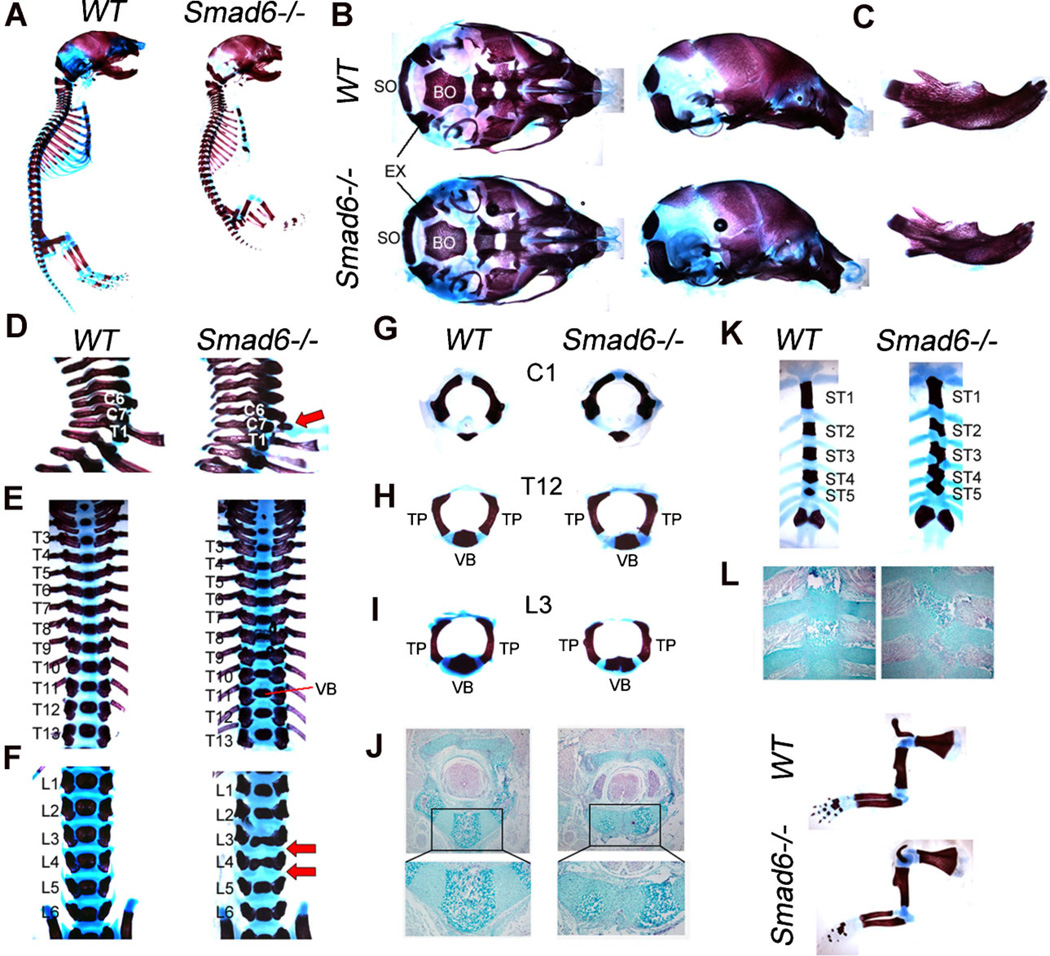
Figure 2.
Skeleton defects in Smad6−/− mice at P0. (A) Whole mount skeletal preparations of wildtype (WT) and Smad6−/− P0 embryos. (B) Basal (left) and lateral (right) view of skull. Mandibles and hyoid bones have been removed. (C) Lateral view of mandibles. (D) Lateral view of cervical vertebrae. Red arrow highlights rib indicative of posterior transformation of the seventh cervical vertebra in mutants. (E) Dorsal view of thoracic vertebrae. (F) Dorsal view of lumbar vertebrae. Red arrows highlight rounded intervertebral discs in mutants. Split vertebral bodies are evident in L3 and L4 of mutants. (G) Anterior view of C1. (H) Anterior view of T12. (I) Anterior view of L3 shows a split vertebral body in mutants. (J) Alcian blue staining of lumbar vertebra shows bilateral ossification centers in mutants, presumably as a result of incomplete fusion of somite pairs. (K) Dorsal view of sternum shows bifid sternebrae and fused ST3, ST4 and ST5 in mutants. (L) Alcian blue staining of sternum shows defects in sternal band fusion in Smad6−/− mice. (M) Forelimbs of WT and Smad6−/− embryos at P0. EX, exoccipital; BO, basioccipital bone; SO, supraoccipital; TP, transverse process; VB, vertebral body.
Analysis of whole-mount skeletal preparations at P0 revealed that Smad6−/− mice exhibit a domed skull and shortened snout (Fig. 2A,B). Analysis of the cranial base revealed that while the length of the cranial base in mutants was similar to that in wild-type littermates, the widths of both the supraoccipital bone and the posterior end of the basioccipital bone were narrow (Fig. 2B). These defects may contribute to the formation of the domed skull in mutants. Additionally, the mandibles exhibited more curvature and were shorter in mutants (Fig. 2C). No defects were seen in cranial sutures (data not shown).
Loss of Smad6 in mice led to defects in the axial skeleton. In particular, Smad6−/−mice exhibit a posterior transformation of the seventh cervical vertebra (C7), as evidenced by the presence of a small rib anlagen (Fig. 2D). Additionally, thoracic vertebral bodies are flatter in Smad6−/− mice compared to wild-type littermates (Fig. 2E), and bilateral ossification centers were seen in lumbar vertebrae (Fig. 2F, I). Intervertebral discs in these regions were rounded in mutants (Fig. 2F). Analyses of representative disarticulated vertebrae show normal development of the first cervical vertebra (C1) in mutants (Fig. 2G). Formation of pedicles and transverse processes of thoracic and lumbar vertebrae was also normal in mutants (Fig. 2H, I). Alcian blue staining of lumbar vertebrae shows that the bilateral ossification centers found in mutants are due to incomplete fusion of sclerotomal cells (Fig. 2J).
The sternum develops by the ventral migration of two lateral mesenchymal bands that fuse at the midline (33). Individual sternebrae form by endochondral ossification in intercostal regions, leading to segmentation of the sternum (33). The sterna of wild-type mice at P0 displayed six separate ossified sternebrae, with ribs attached to the cartilaginous intersternabrae. In contrast, Smad6−/− mice develop bifid sternebrae due to incomplete medial fusion (Fig. 2K, L). In addition, the third, fourth and fifth sternebrae (ST3, ST4 and ST5) of mutant mice were fused (Fig. 2K).
Skeletal preparations at P0 showed that appendicular bones were slightly shorter in mutant mice (Fig. 2M), proportionate with the overall smaller size of Smad6−/− mice. Overall, these results indicate that Smad6 regulates skeletal development.
Delayed onset of hypertrophy and increased matrix production in appendicular bones of Smad6−/− mice
Alcian blue staining of hindlimbs was performed to examine appendicular bone formation in more detail. While Alcian blue staining revealed no defects at E13.5 in Smad6−/− mice (Fig. 3A,B), a shorter hypertrophic zone was evident by E15.0 in long bones of mutant cartilage (Fig. 3C–F). The shortened hypertrophic zone could result from a delay in the onset of hypertrophy, and/or retention/recruitment of chondrocytes in the proliferative pool, or defective hypertrophic differentiation. To evaluate whether the smaller hypertrophic zones at E15.0 in Smad6−/− cartilage resulted from defective hypertrophic differentiation, we examined the expression level of type X collagen by immunofluorescence staining. Normal levels of type X collagen were evident in both wild-type and mutant littermates, but the domain of type X collagen expression was reduced in mutant cartilage (Fig. 3G,H). These results suggest that while the onset of hypertrophic differentiation appears to be delayed, there is no defect in the ability of mutant chondrocytes to undergo hypertrophy.
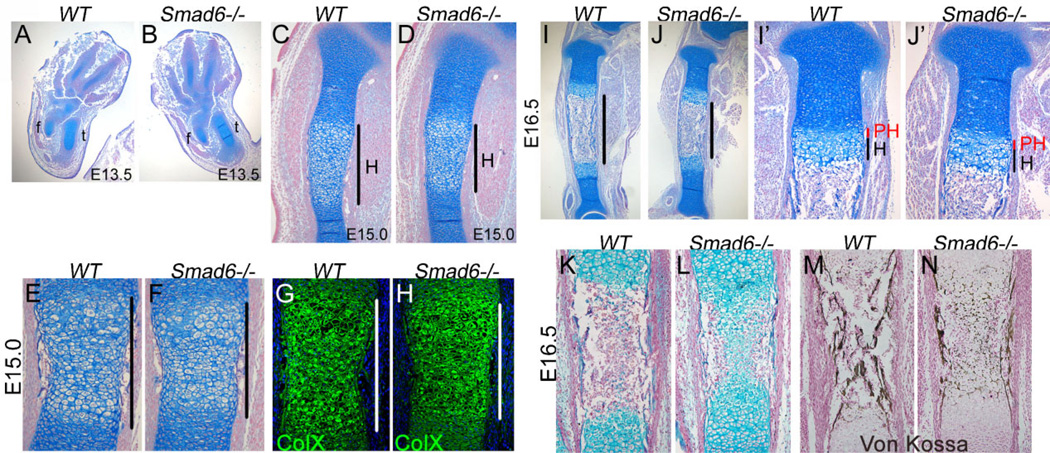
Figure 3.
Delayed onset of hypertrophic differentiation and mineralization at midgestation in Smad6−/− mice. (A,B) Formation of cartilage condensations in Smad6−/− limbs is similar to that in wild-type (WT) limbs at E13.5. (C,D) A shorter hypertrophic zone is evident in the Smad6−/− tibiae at E15.0. (E,F) Alcian blue staining of E15.0 femoral diaphyses. The hypertrophic zone in Smad6−/− mice is demarcated by black bars. (G,H) Immunofluorescence staining for type X collagen of E15.0 femurs. The hypertrophic zone is demarcated by white bars. No detectable staining was observed in negative controls, from which primary antibodies were omitted. (I,J) Shorter diaphyses are evident in the E16.5 Smad6−/− tibiae, as demarcated by black bars. (I’,J’) Higher magnification of proximal tibias at E16.5. (K,L) Alcian blue staining of E16.5 tibial diaphyses. (M,N) Von Kossa staining of E16.5 tibias shows defects in mineralization in Smad6−/− mice. f, fibula; t, tibia; PH, prehypertrophic zone; H, hypertrophic zone.
By E16.5, distinct growth plates separated by a marrow cavity were evident in cartilage of wild-type and Smad6−/− littermates (Fig. 3I,J). Histologically, the growth plate appeared indistinguishable between wild-type and mutant littermates at E16.5 (Fig. 3I’,J’). Consistent with delayed onset of hypertrophy seen at E15.0, the marrow cavity was smaller in mutants. Moreover, von Kossa staining at this stage revealed defects in mineralization in mutant cartilage (Fig. 3M, N). Taken together, these results indicate that loss of Smad6 leads to delayed onset of chondrocyte hypertrophy and ossification at midgestation, but once initiated, these processes can proceed normally in Smad6−/− mice.
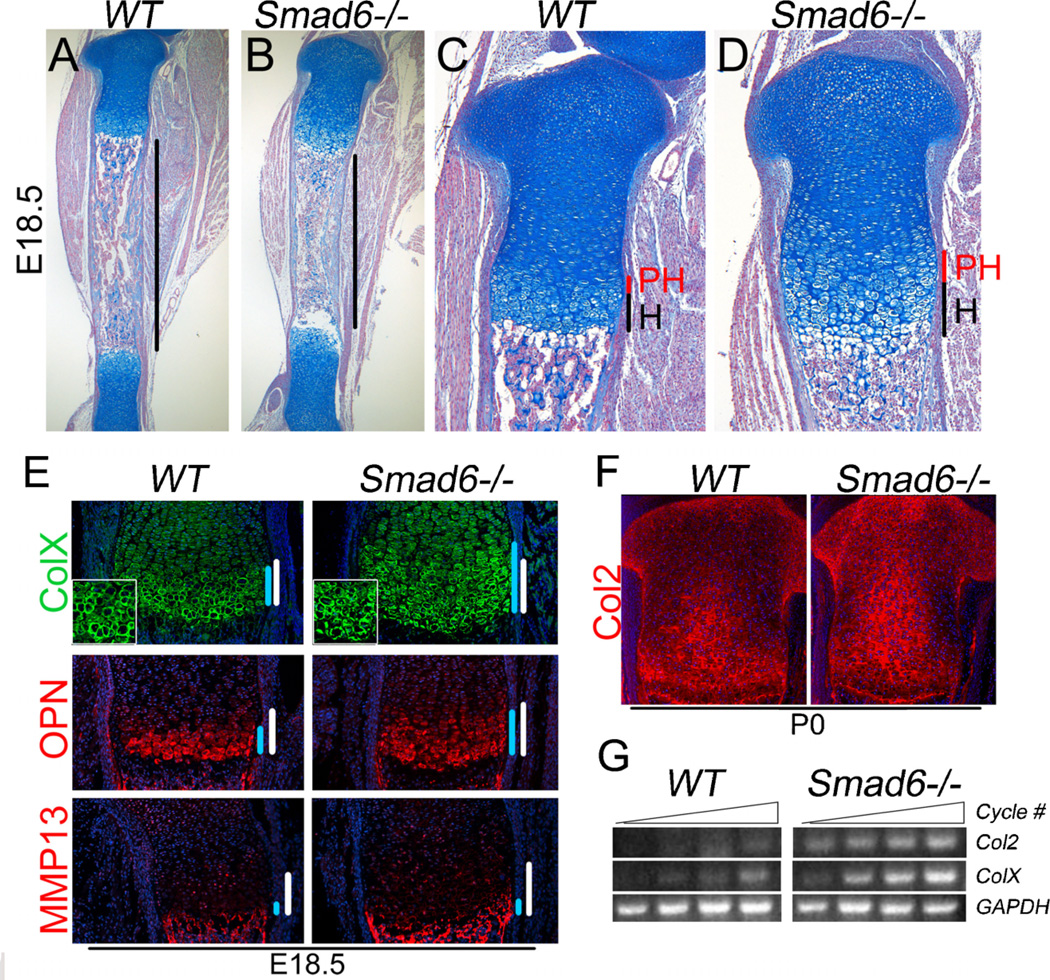
By E18.5, enlarged prehypertrophic and hypertrophic zones were clearly apparent in Smad6−/− growth plates (Fig. 4A–D). Immunofluorescence staining at this stage showed that the enlarged hypertrophic zones were accompanied by increased expression domains of type X collagen (Fig. 4E). Moreover, the intensity of staining for type X collagen was higher in mutants than in WT littermates, revealing increased biosynthetic activity of mutant hypertrophic chondrocytes (Fig. 4E, inset). Expression of osteopontin and MMP-13, markers of terminal differentiation, were examined to determine whether the enlarged expression domain of type X collagen reflected a delay in terminal maturation, in which case reduced expression of terminal differentiation markers would be expected. However, the expression domain of osteopontin, normally restricted to the lower hypertrophic zone, was expanded in mutants (Fig. 4E). Moreover, there was no difference in the expression domain of MMP-13, which is normally restricted to the most terminally differentiating chondrocytes (Fig. 4E). These results suggest that the enlarged hypertrophic zone is most likely attributed to an increase in the number of cells undergoing hypertrophic differentiation. Additionally, our results demonstrate that the pace of chondrocyte hypertrophy is not altered in mutants once differentiation is initiated.
Figure 4.
Enhanced hypertrophic differentiation at late gestation in Smad6−/− mice. (A,B) Shorter diaphyses are evident in E18.5 Smad6−/− tibias, as demarcated by black bars. (C,D) Higher magnification of proximal tibias at E18.5. (E) Immunofluorescence staining of adjacent sections of E18.5 distal femurs for the hypertrophic differentiation marker, type X collagen, and terminal differentiation markers, osteopontin (OPN) and MMP-13. The hypertrophic zone is demarcated by white bars. The region of expression is demarcated by blue bars. No detectable staining was observed in negative controls, from which primary antibodies were omitted. (F) Immunofluorescence staining of tibial growth plates for type II collagen shows enhanced collagen production at P0. No detectable staining was observed in negative controls. (G) Semiquantitative RT-PCR analysis of RNA isolated from WT and Smad6−/− primary chondrocytes cultured in chondrogenic media for 7 days.
As discussed above, analysis of type X collagen expression indicated increased matrix production in mutant hypertrophic chondrocytes. Evidence for increased activity of chondrocytes within the proliferative zone was provided by the observation that the domain of expression of type II collagen was expanded within the proliferative zone of mutant growth plates at P0 compared to WT littermates (Fig. 4F). Consistent with the model where loss of Smad6 leads to increased activity of proliferative and hypertrophic chondrocytes, the levels of type II and × collagen mRNA were elevated in Smad6-deficient chondrocytes (Fig. 4G). These results show that loss of Smad6 leads to enhanced activity of both proliferative and hypertrophic chondrocytes, associated with increased collagen production.
Increased rate of chondrocyte proliferation and apoptosis in Smad6−/− cartilage
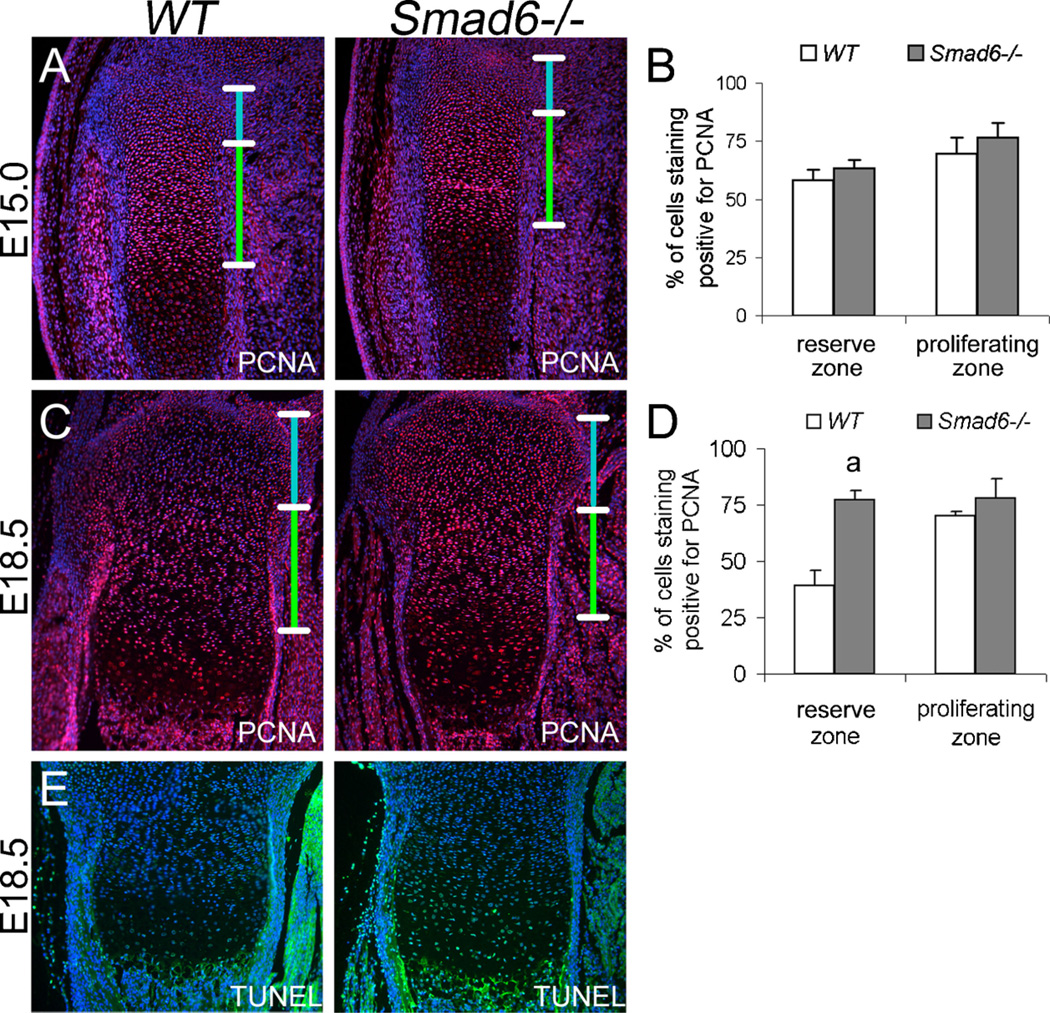
Immunofluorescence staining for PCNA, a marker for the G1 and S phases of the cell cycle, was performed to examine whether the delay in the onset of chondrocyte differentiation at midgestation and the enlarged hypertrophic zone at late gestation result from increased chondrocyte proliferation. A trend toward a moderate increase in proliferating cells in both the reserve and proliferative zones was observed in mutant growth plates at E15.0 (Fig. 5A,B), but the difference was not statistically significant. Similar results were found by immunofluorescence staining for phospho-histone H3 (Ser 10), a marker for M-phase (Supplemental Fig. 1). By E18.5, a significant increase in proliferating cells was evident in the reserve zone of mutant cartilage (Fig. 5C,D). Apoptosis, which is normally detectable in the most terminally differentiating chondrocytes, was elevated in Smad6−/− growth plates (Fig. 5E) Therefore, the enlargement of the hypertrophic zone in mutants is not due to prolonged retention of terminal hypertrophic chondrocytes. Rather, the data suggest that the increased pool of proliferating chondrocytes leads to the enlarged hypertrophic zone. We speculate that the increased level of apoptosis in Smad6−/− hypertrophic chondrocytes may be due to the elevated biosynthetic activity of these cells, which could lead to increased cellular stress.
Figure 5.
Increased proliferation and apoptosis at late gestation in Smad6−/− mice. (A) Immunofluorescence staining of E15.0 proximal tibiae for the proliferation marker, PCNA. The reserve and proliferative zones are demarcated by blue and green bars, respectively. No detectable staining was observed in negative controls, from which primary antibodies were omitted. (B) Quantification of the rates of proliferation in wild-type and mutant cells at E15.0. Values are expressed as percent labeled cells. (C) Immunofluorescence staining of E18.5 proximal tibiae for PCNA. The reserve and proliferative zones are demarcated by blue and green bars, respectively. (D) Quantification of the rates of proliferation in wild-type and mutant cells at E18.5. Values are expressed as percent labeled cells (Student’s t-test; ap < 0.05). (E) TUNEL staining of E18.5 proximal tibiae. No detectable staining was observed in negative controls.
Increased BMP activity and responsiveness in Smad6-deficient chondrocytes
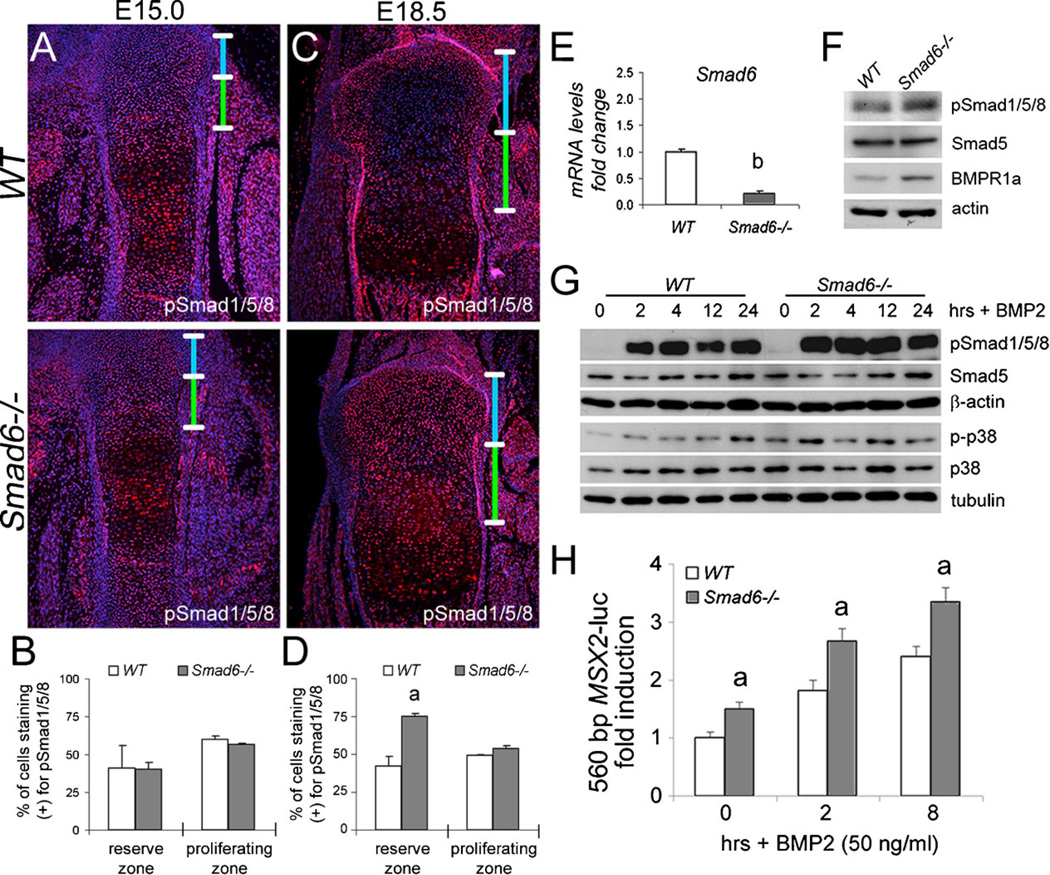
Given the known role for Smad6 as an intracellular inhibitor of BMP signaling (14,34), we examined the effect of loss of Smad6 on Smad1/5/8 activation in cartilage in vivo by immunofluorescence staining of growth plates. Phospho-Smad1/5/8 levels were similar in wild-type and mutant cartilage at E15.0 (Fig. 6A, B). By E18.5, phospho-Smad1/5/8 levels were significantly elevated in the resting zone of mutant cartilage (Fig. 6C,D). These results demonstrate that loss of Smad6 leads to elevated canonical BMP signaling in cartilage in vivo.
Figure 6.
Increased BMP signaling in Smad6−/− chondrocytes. Immunofluorescence staining of (A) E15.0 distal femurs and (C) E18.5 proximal tibiae for phosphorylated forms of Smad1/5/8 (pSmad1/5/8). The reserve and proliferative zones are demarcated by blue and green bars, respectively. No detectable staining was observed in negative controls, from which primary antibodies were omitted. Quantification of pSmad1/5/8 staining in wild-type and mutant cells at (B) E15.0 and (D) E18.5. Values are expressed as percent labeled cells (Student’s t-test; ap < 0.05). (E) Real-time PCR analysis of RNA isolated from WT and Smad6−/− primary chondrocytes cultured in chondrogenic media. Expression levels for Smad6 were normalized to β-actin and are shown as fold change relative to WT mRNA levels. The data represent averages from triplicate reactions with the indicated s.d. and significant differences (Student’s t-test; bp < 0.001). (F) Western blot analysis of pSmad1/5/8 and BMPR1a levels in lysates isolated from WT and Smad6−/− primary chondrocytes cultured in chondrogenic media for 10 days. (G) Western blot analysis shows elevated levels of pSmad1/5/8 and phospho-p38 (p-p38) in lysates isolated from WT and Smad6−/− primary chondrocytes treated with BMP2 (50 ng/ml) for 0, 2, 4, 12, or 24 hours. (H) BMP2 induction of the 560 bp MSX2-luc promoter is enhanced in Smad6−/− primary chondrocytes. Cells were treated with BMP2 (50 ng/ml) for 0, 2, or 8 hours. The data represent an average from three wells with the indicated s.d. and significant differences (multifactorial ANOVA (Within-subjects); ap < 0.05 versus wild-type control).
Because the Smad6−/− mice are global knockouts, we investigated whether Smad6 deficiency has direct effects on BMP signaling by examining Smad6 function in primary chondrocytes. Loss of Smad6 in mutant chondrocytes was confirmed by quantitative real-time PCR analysis (Fig. 6E). Levels of phospho-Smad1/5/8 were elevated in Smad6-deficient cells cultured in chondrogenic media for 10 days as determined by western blot analysis (Fig. 6F). Increased levels of the BMP receptor BMPR1A (also known as ALK3) were observed in Smad6-deficient cells (Fig. 6F), consistent with the reported function of Smad6 in promoting type I BMP receptor degradation (17). Taken together, these results show Smad6 is required to limit the canonical Smad pathway in chondrocytes, in part, by regulating type I BMP receptor levels.
The effects of loss of Smad6 on the intensity and duration of canonical and noncanonical BMP signaling in chondrocytes were evaluated by treating isolated primary chondrocytes with BMP2. Elevated levels of phospho-Smad1/5/8 were observed up to 12 hours following BMP2 stimulation in Smad6-deficient chondrocytes (Fig. 6G). Similarly, phospho-p38 levels were elevated under basal conditions and for up to 12 hours post-stimulation with BMP2 in mutant chondrocytes (Fig. 6G). We then tested whether Smad6-deficient chondrocytes are more responsive to BMP2 using a Smad1/5-responsive 560 bp Msx2 promoter fragment (29) linked to luciferase (560 bp MSX2-luc) as a reporter. Levels of both basal and BMP2-mediated induction of the Msx2 promoter were greater in mutant chondrocytes than that in wild-type cells (Fig. 6H). Overall, these results show that loss of Smad6 enhances BMP2-induced activation of both canonical and noncanonical pathways, leading to increased BMP responsiveness in Smad6-deficient chondrocytes.
Increased Ihh expression and activity in Smad6-deficient chondrocytes
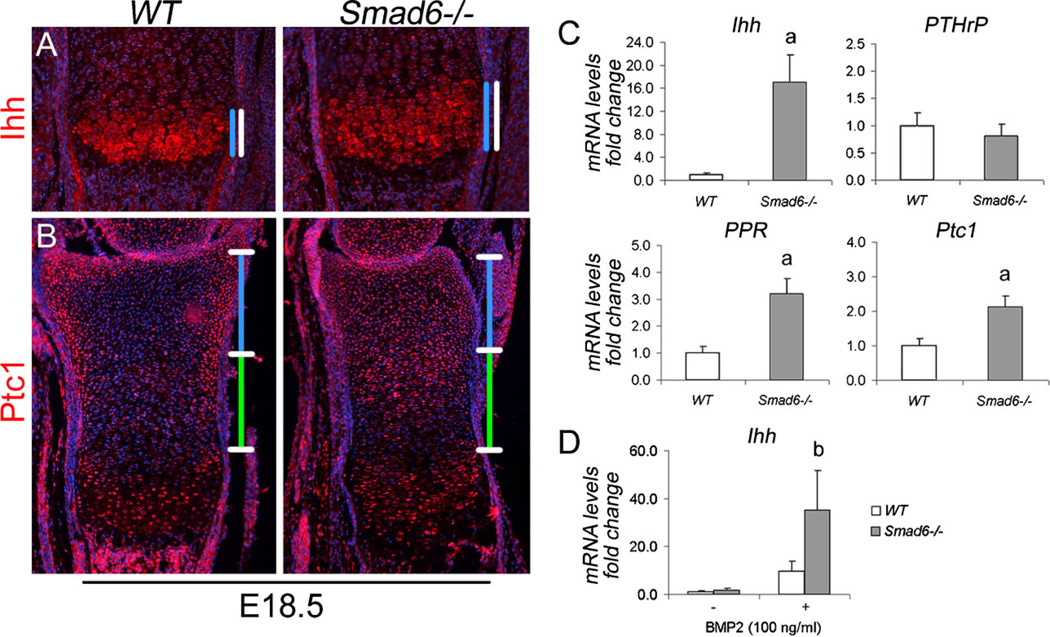
The Ihh/PTHrP pathway forms a negative-feedback loop to regulate the onset of chondrocyte hypertrophy (reviewed in (1)). We have shown previously that BMP signaling through the canonical Smad pathway positively regulates Ihh signaling in chondrocytes (6,7). To examine the consequences of Smad6 deficiency on Ihh signaling in cartilage in vivo, we evaluated the expression of Ihh via immunofluorescence staining of growth plates. The domain of Ihh protein expression was expanded in mutant cartilage (Fig. 7A), demonstrating that Smad6 impacts Ihh signaling in cartilage. We then evaluated Ihh activity in vivo via immunofluorescence staining of growth plates for its downstream target, Patched 1 (Ptc1). Ptc1 was expressed at low levels in the proliferative zone and at higher levels in the prehypertrophic and periarticular regions of wild-type growth plates (Fig .7B); in mutant growth plates, Ptc1 expression was elevated throughout the reserve and upper proliferative zones (Fig. 7B).
Figure 7.
Loss of Smad6 leads to Increased Ihh expression and activity in chondrocytes. (A) Immunofluorescence staining of E18.5 distal tibiae for Ihh. The prehypertrophic and hypertrophic zones are demarcated by white bars. The region of expression is demarcated by blue bars. (B) Immunofluorescence staining of E18.5 distal tibiae for patched1 (Ptc1). The reserve and proliferative zones are demarcated by blue and green bars, respectively. No detectable staining was observed in negative controls, from which primary antibodies were omitted. (C) Real-time PCR analysis of RNA isolated from WT and Smad6−/− primary chondrocytes cultured for 10 days in chondrogenic media. Expression levels for each gene of interest were normalized to β-actin and are shown as fold change relative to WT mRNA levels. The data represent averages from triplicate reactions with the indicated s.d. and significant differences (Student’s t-test; ap < 0.001). (D) Realtime PCR analysis of RNA isolated from WT and Smad6−/− primary chondrocytes cultured in chondrogenic media for 10 days, serum-starved overnight, and then treated with BMP2 (100 ng/ml) for 2 hours. Ihh mRNA levels were normalized to β-actin and are shown as fold change relative to WT mRNA levels. The data represent averages from triplicate reactions with the indicated s.d. and significant differences (Student’s t-test; bp < 0.05).
The direct effect of loss of Smad6 on Ihh signaling in chondrocytes was examined via analysis of Ihh, Pthrp, and PTHrP receptor (PPR or PTH1R) expression in wild-type and Smad6-deficient primary chondrocytes. Both Ihh and PPR mRNA levels were significantly increased in Smad6-deficient cells, while no difference in PTHrP mRNA levels was evident (Fig. 7C). Consistent with increased Ihh expression, Ptc1 mRNA levels were elevated in mutant chondrocytes (Fig. 7C). Moreover, BMP-induced Ihh expression was markedly increased in mutant chondrocytes (Fig. 7D). Taken together, these in vivo and in vitro results demonstrate that loss of Smad6 upregulates Ihh expression and activity, in part, due to enhanced BMP responsiveness in mutant chondrocytes.
Discussion
Mice deficient in Smad6 exhibit cardiovascular defects, including aortic ossification resulting from cartilaginous metaplasia (27). The skeletal phenotype of these mice, however, has not been investigated previously. Although it has been shown that overexpression of Smad6 in chondrocytes results in abnormal skeletal development (25), the physiological role of Smad6 during endochondral bone formation was unknown. In this work, we found that mice deficient in Smad6 exhibit craniofacial, axial and appendicular skeletal defects. Elevated BMP signaling in Smad6-deficient chondrocytes most likely causes the mutant phenotype. Thus, the results of our study provide the first in vivo evidence that Smad6 is required to limit BMP signaling for proper skeletal development.
Smad6 is required for axial skeleton development
Loss of Smad6 led to posterior transformation of cervical vertebrae, demonstrating a role for Smad6 in regulating anterior-posterior (A/P) patterning of the axial skeleton. BMP signaling has been shown previously to regulate A/P patterning, as mice carrying a hypomorphic type II BMP receptor exhibit anterior transformation of thoracic vertebrae (35). Our results here suggest that the increased BMP signaling resulting from loss of Smad6 causes posterior transformation of axial elements.
Smad6 can also interact directly with members of the Hox family of transcription factors (36), which regulate axial patterning. For example, loss of either Hoxa4 (37) or Hoxc8 (38) in mice results in the formation of a rib anlagen at C7, similar to that found in our Smad6−/− mice. In vitro studies showed that Smad6 interacts with Hoxc8 to inhibit Smad1-induced transcriptional activity (39), while Smad1 antagonizes the repressor function of Hoxc8 to promote osteopontin gene expression (40). Thus, Smad6 may play a role in mediating cross-talk between BMP signaling and Hox transcription factors to specify positional identity of vertebrae along the A/P axis.
The development of the sternum also involves both Hox transcription factors and BMP signaling (22,41,42). Defects in the fusion of sternal bands, as in Smad6 mutants, have been found in mice with either limited (41) or elevated BMP signaling (22). The results of these studies suggest that sternum development requires strict control of BMP signaling, in part through intracellular regulation by Smad6.
Smad6 is required to limit BMP signaling during endochondral bone formation
The stage-dependent expression pattern of Smad6 parallels that of pSmad1/5 in the growth plate (6,7), and is consistent with previous studies showing that Smad6 is a direct target of canonical BMP pathways (43). The subcellular localization of Smad6 agrees with the reported functions of Smad6 in the cytoplasm (i.e. binding to pSmad1 (19)) and at the cell membrane (i.e. binding to type I receptors (19,34)), and suggests that the inhibitory functions of Smad6 depend on whether the chondrocytes are undergoing proliferation or hypertrophic differentiation, as well as the stage of development. Overall, these results suggest a role for Smad6 in regulating proliferation and hypertrophic differentiation of chondrocytes in vivo.
We found that loss of Smad6 results in stage-specific defects in endochondral bone formation. Mesenchymal condensation occurs normally in Smad6−/− limbs, indicating that Smad6 does not play a major role in early stages of chondrogenesis. By midgestation, delayed onset of hypertrophic differentiation is evident in Smad6−/− growth plates. The limb phenotype of Smad6−/− mice differs from that in mice in which BMP signaling is elevated due to noggin-deficiency (22), as well as in developing limbs in which BMPs were overexpressed using transgenic approaches (44,45). In particular, increased BMP activity in noggin-null mice and in limbs overexpressing BMP-2 or BMP-4 led to massive cartilage overgrowth without any increase in chondrocyte proliferation; the overgrowth was attributed to increased recruitment of progenitor cells to the chondrocyte lineage. It is thus possible that the delay in onset of hypertrophy that we observe in mutants reflects an indirect effect of loss of Smad6 in a different cell type, such as the perichondrium. Elevated BMP signaling can also enhance chondrocyte proliferation, as demonstrated in cartilage explants treated with BMP-2 (5,46). In Smad6−/− growth plates, elevated BMP signaling correlated with cell cycle entry of resting chondrocytes. No difference in proliferation rates was found between proliferating chondrocytes of mutant and wild-type littermates. Our inability to detect increased proliferation in the proliferative zone in mutant growth plates may be because the rate of proliferation is already maximal, or because additional increases are beyond detection with PCNA or phospho-histone H3 staining. However, increased collagen production by proliferating chondrocytes was observed, indicating that Smad6 has direct effects on these cells. Taken together, it appears that noggin acts during the stage of recruitment of progenitor cells into cartilage elements, while Smad6 acts at the stage of cell cycle entry of reserve chondrocytes in the growth plate. Thus, regulation of BMP signaling both intracellularly and extracellularly is required for normal endochondral bone formation.
Interestingly, delayed onset of hypertrophy, as in the Smad6 mutants, was also seen in mice overexpressing Smad6 (25) and in chick limbs treated with chordin (23). Loss of BMP signaling due to Smad6 overexpression or chordin treatment also blocks the progression of hypertrophic differentiation of chondrocytes. Correspondingly, transgenic mice expressing constitutively active BMPR1A exhibited shortened proliferative zones. The authors concluded that the growth plate phenotype resulted from accelerated hypertrophic differentiation; however, no evidence for accelerated hypertrophic differentiation was provided (47). In the above model systems, BMP signaling is impacted in all of the cells in the growth plate. In contrast, our model reflects BMP-mediated effects on chondrocyte differentiation only in cells in which Smad6 is a physiological regulator of BMP activity. In particular, increased collagen production by both proliferating and hypertrophic chondrocytes was evident in mutant growth plates at late gestational stages. Direct actions of Smad6 on collagen production were confirmed in Smad6-deficient chondrocytes. Overall, our results suggest that elevated BMP signaling in the Smad6 mutants affects the onset of hypertrophic differentiation, as well as collagen production by proliferating and hypertrophic chondrocytes.
Enhanced chondrocyte proliferation, apparent in the reserve zone of E18.5 mutant growth plates, may result from the activation of both canonical and noncanonical BMP pathways, as the main mediators of these pathways (Smad1/5/8 and TAK1) are required for chondrocyte proliferation in vivo (6,48,49). A major consequence of canonical and noncanonical BMP pathway activation is the upregulation of Ihh expression and activity (6,50,51). Our results demonstrate that loss of Smad6 leads to upregulation of Ihh expression and activity, in part via enhanced BMP-responsiveness in Smad6-deficient chondrocytes. Given that the loss of Ihh leads to premature hypertrophy associated with reduced chondrocyte proliferation (52), elevated Ihh activity evident in Smad6−/− mice and in Smad6-deficient chondrocytes is consistent with delayed onset of hypertrophic differentiation.
PPR mediates chondrocyte proliferation to regulate the onset of hypertrophic differentiation, as evidenced by accelerated differentiation of chondrocytes in PPR−/− mice (53). We found that PPR expression is elevated in Smad6-deficient chondrocytes. This finding is also consistent with our conclusion that loss of Smad6 leads to delayed onset of hypertrophic differentiation. The transcription factors that directly regulate PPR expression in chondrocytes are unknown. It is possible that the canonical BMP pathway mediates PPR expression in chondrocytes, given that PPR expression is reduced in mice lacking Smads1 and 5 (6).
Smad6 inhibits BMP signaling intracellularly by associating with type I BMP receptors (19,34), thereby facilitating Smurf1-mediated receptor degradation (17). Our in vivo and in vitro studies demonstrate that loss of Smad6 results in elevated canonical BMP signaling, in part, by regulating type I receptor levels in chondrocytes. The enhanced activation of both the canonical and noncanonical pathways by BMP2 in Smad6-deficient chondrocytes is consistent with the increased levels of BMPR1A. Smad6 can also limit BMP signaling by associating with receptor-activated Smad1, thereby inhibiting its association with Smad4 (19). Thus, increased BMP signaling in Smad6-deficient chondrocytes may also be due to increased availability of Smad1.
In summary, we have identified a role for Smad6 as a regulator of multiple stages of chondrogenesis, including specification of A/P fate, entry and exit of resting chondrocytes into the proliferative pool, and ECM synthesis. We also show that Smad6 executes these functions, in part, through BMP-mediated regulation of Ihh expression and activity. Additional studies will be required to test whether Smad6 and the remaining inhibitory Smad, Smad7, have overlapping functions in the growth plate.
Supplementary Material
Acknowledgements
We thank Christina Hong and Xin Liu for technical assistance and are grateful to members of the laboratory for helpful discussions and critical comments on the manuscript. This work was supported by the National Institutes of Health Grants RO1-AR044528 (to K.M.L) and F31-AR060147 (to K.D.E) via NIAMS.
Footnotes
Conflicts of Interest The authors have no conflict of interest.
Additional Supporting Information may be found in the online version of this article.
Contributor Information
Kristine D. Estrada, Email: kestrada@ucla.edu.
Kelsey N. Retting, Email: Kelsey.Retting@pfizer.com.
Alana M. Chin, Email: alanachin07@ucla.edu.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Zehentner BK, Dony C, Burtscher H. The transcription factor Sox9 is involved in BMP-2 signaling. J Bone Miner Res. 1999;14(10):1734–1741. doi: 10.1359/jbmr.1999.14.10.1734. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q, Yu Y, Chen Q, Li C, Wu H, Wan Y, Ma J, Sun F. Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol. 2008;217(1):228–241. doi: 10.1002/jcp.21496. [DOI] [PubMed] [Google Scholar]
- 4.Valcourt U, Gouttenoire J, Moustakas A, Herbage D, Mallein-Gerin F. Functions of transforming growth factor-beta family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J Biol Chem. 2002;277(37):33545–33558. doi: 10.1074/jbc.M202086200. [DOI] [PubMed] [Google Scholar]
- 5.Minina E, Wenzel HM, Kreschel C, Karp S, Gaffield W, McMahon AP, Vortkamp A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128(22):4523–4534. doi: 10.1242/dev.128.22.4523. [DOI] [PubMed] [Google Scholar]
- 6.Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136(7):1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133(23):4667–4678. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 8.Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102(14):5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 10.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 11.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, Nishida E, Hagiwara M. A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J Biol Chem. 1996;271(23):13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 12.Ishisaki A, Yamato K, Hashimoto S, Nakao A, Tamaki K, Nonaka K, ten Dijke P, Sugino H, Nishihara T. Differential inhibition of Smad6 and Smad7 on bone morphogenetic protein- and activin-mediated growth arrest and apoptosis in B cells. J Biol Chem. 1999;274(19):13637–13642. doi: 10.1074/jbc.274.19.13637. [DOI] [PubMed] [Google Scholar]
- 13.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol. 2001;155(6):1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389(6651):622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 15.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 17.Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14(7):2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99(11):2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12(2):186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Ionescu AM, Schwarz EM, Zhang X, Drissi H, Puzas JE, Rosier RN, Zuscik MJ, O'Keefe RJ. Smad6 is induced by BMP-2 and modulates chondrocyte differentiation. J Orthop Res. 2003;21(5):908–913. doi: 10.1016/S0736-0266(03)00008-1. [DOI] [PubMed] [Google Scholar]
- 21.Benchabane H, Wrana JL. GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol Cell Biol. 2003;23(18):6646–6661. doi: 10.1128/MCB.23.18.6646-6661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280(5368):1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D, Ferguson CM, O'Keefe RJ, Puzas JE, Rosier RN, Reynolds PR. A role for the BMP antagonist chordin in endochondral ossification. J Bone Miner Res. 2002;17(2):293–300. doi: 10.1359/jbmr.2002.17.2.293. [DOI] [PubMed] [Google Scholar]
- 24.Scharstuhl A, Diepens R, Lensen J, Vitters E, van Beuningen H, van der Kraan P, van den Berg W. Adenoviral overexpression of Smad-7 and Smad-6 differentially regulates TGF-beta-mediated chondrocyte proliferation and proteoglycan synthesis. Osteoarthritis Cartilage. 2003;11(11):773–782. doi: 10.1016/s1063-4584(03)00165-1. [DOI] [PubMed] [Google Scholar]
- 25.Horiki M, Imamura T, Okamoto M, Hayashi M, Murai J, Myoui A, Ochi T, Miyazono K, Yoshikawa H, Tsumaki N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J Cell Biol. 2004;165(3):433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwai T, Murai J, Yoshikawa H, Tsumaki N. Smad7 Inhibits chondrocyte differentiation at multiple steps during endochondral bone formation and down-regulates p38 MAPK pathways. J Biol Chem. 2008;283(40):27154–27164. doi: 10.1074/jbc.M801175200. [DOI] [PubMed] [Google Scholar]
- 27.Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Jr, Falb D, Huszar D. A role for smad6 in development and homeostasis of the cardiovascular system. Nat Genet. 2000;24(2):171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994;14(4):329–335. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 29.Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, Cho JY, Dobias SL, Yi SE, Lyons K, Bell JR, Arora K, Warrior R, Maxson R. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development. 2004;131(20):5153–5165. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- 30.Amano K, Hata K, Sugita A, Takigawa Y, Ono K, Wakabayashi M, Kogo M, Nishimura R, Yoneda T. Sox9 family members negatively regulate maturation and calcification of chondrocytes through up-regulation of parathyroid hormone-related protein. Mol Biol Cell. 2009;20(21):4541–4551. doi: 10.1091/mbc.E09-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14(5):700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Clark CA, Li TF, Kim KO, Drissi H, Zuscik MJ, Zhang X, O'Keefe RJ. Prostaglandin E2 inhibits BMP signaling and delays chondrocyte maturation. J Orthop Res. 2009;27(6):785–792. doi: 10.1002/jor.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JM. Studies on the morphogenesis of the mouse sternum. I. Normal embryonic development. J Anat. 1952;86(4):373–386. [PMC free article] [PubMed] [Google Scholar]
- 34.Goto K, Kamiya Y, Imamura T, Miyazono K, Miyazawa K. Selective inhibitory effects of Smad6 on bone morphogenetic protein type I receptors. J Biol Chem. 2007;282(28):20603–20611. doi: 10.1074/jbc.M702100200. [DOI] [PubMed] [Google Scholar]
- 35.Delot EC, Bahamonde ME, Zhao M, Lyons KM. BMP signaling is required for septation of the outflow tract of the mammalian heart. Development. 2003;130(1):209–220. doi: 10.1242/dev.00181. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Nie S, Chang C, Qiu T, Cao X. Smads oppose Hox transcriptional activities. Exp Cell Res. 2006;312(6):854–864. doi: 10.1016/j.yexcr.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Horan GS, Wu K, Wolgemuth DJ, Behringer RR. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc Natl Acad Sci U S A. 1994;91(26):12644–12648. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juan AH, Lei H, Bhargava P, Lebrun M, Ruddle FH. Multiple roles of hoxc8 in skeletal development. Ann N Y Acad Sci. 2006;1068:87–94. doi: 10.1196/annals.1346.046. [DOI] [PubMed] [Google Scholar]
- 39.Bai S, Shi X, Yang X, Cao X. Smad6 as a transcriptional corepressor. J Biol Chem. 2000;275(12):8267–8270. doi: 10.1074/jbc.275.12.8267. [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Yang X, Chen D, Chang Z, Cao X. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J Biol Chem. 1999;274(19):13711–13717. doi: 10.1074/jbc.274.19.13711. [DOI] [PubMed] [Google Scholar]
- 41.Solloway MJ, Dudley AT, Bikoff EK, Lyons KM, Hogan BL, Robertson EJ. Mice lacking Bmp6 function. Dev Genet. 1998;22(4):321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73(2):279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 43.Ishida W, Hamamoto T, Kusanagi K, Yagi K, Kawabata M, Takehara K, Sampath TK, Kato M, Miyazono K. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J Biol Chem. 2000;275(9):6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 44.Duprez D, Bell EJ, Richardson MK, Archer CW, Wolpert L, Brickell PM, Francis-West PH. Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech Dev. 1996;57(2):145–157. doi: 10.1016/0925-4773(96)00540-0. [DOI] [PubMed] [Google Scholar]
- 45.Tsumaki N, Nakase T, Miyaji T, Kakiuchi M, Kimura T, Ochi T, Yoshikawa H. Bone morphogenetic protein signals are required for cartilage formation and differently regulate joint development during skeletogenesis. J Bone Miner Res. 2002;17(5):898–906. doi: 10.1359/jbmr.2002.17.5.898. [DOI] [PubMed] [Google Scholar]
- 46.De Luca F, Barnes KM, Uyeda JA, De-Levi S, Abad V, Palese T, Mericq V, Baron J. Regulation of growth plate chondrogenesis by bone morphogenetic protein-2. Endocrinology. 2001;142(1):430–436. doi: 10.1210/endo.142.1.7901. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi T, Lyons KM, McMahon AP, Kronenberg HM. BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc Natl Acad Sci U S A. 2005;102(50):18023–18027. doi: 10.1073/pnas.0503617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gunnell LM, Jonason JH, Loiselle AE, Kohn A, Schwarz EM, Hilton MJ, O'Keefe RJ. TAK1 regulates cartilage and joint development via the MAPK and BMP signaling pathways. J Bone Miner Res. 2010;25(8):1784–1797. doi: 10.1002/jbmr.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, Gygi S, Glimcher LH. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28(14):2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai LP, DaSilva KA, Mitchell J. Regulation of Indian hedgehog mRNA levels in chondrocytic cells by ERK1/2 and p38 mitogen-activated protein kinases. J Cell Physiol. 2005;203(1):177–185. doi: 10.1002/jcp.20208. [DOI] [PubMed] [Google Scholar]
- 51.Seki K, Hata A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. J Biol Chem. 2004;279(18):18544–18549. doi: 10.1074/jbc.M311592200. [DOI] [PubMed] [Google Scholar]
- 52.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LH, Ho C, Mulligan RC, Abou-Samra AB, Juppner H, Segre GV, Kronenberg HM. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273(5275):663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.